Abstract
Notch and Wnt signaling function together to regulate colonic progenitor cell division and differentiation. Studies in mice have also shown that Notch signaling is required for adenoma formation in response to elevated Wnt-pathway signaling that occurs in the APCMin mouse model of human adenomatous polyposis coli. We therefore used in situ hybridization to analyze expression of Notch ligands, receptors and fringe genes, as well as the Notch target gene, HES1, in human colorectal cancer (CRC). In a small cohort of tumors, JAGGED ligands, NOTCH1, LFNG and HES1 were expressed at levels similar to, or higher than, levels observed in the crypt. To explore the possibility that Notch signaling may play a quantitative role in human CRC we next analyzed HES1 mRNA expression in 130 tumors, each associated with outcome data. The vast majority of these tumors expressed HES1, although at varying levels. Absolute expression levels did not correlate with patient survival. These results establish that JAG ligands and NOTCH1, as well as Notch receptor activation are consistent features of human CRC and support the notion that many of these tumors, like the APCMin mouse, may respond to anti-Notch therapeutic regimes.
Keywords: Notch, Hes1, colon cancer, signaling, prognosis
Introduction
Colorectal cancer (CRC) is a common malignancy, accounting for example, for 9% of all cancer deaths in the United States (1). Mutations (both hereditary and somatic) that contribute to adenoma formation or progression have been identified in a number of genes (2). Most significantly, mutations in genes involved in Wnt signaling, such as the adenomatous polyposis coli (APC) gene cause colon cancer through constitutive formation of a nuclear ß-catenin/TCF transcription complex in colonic epithelial cells (3). A number of genetic and developmental studies, primarily in mice, have helped to identify gene products and pathways that are essential for colon tumor formation induced by aberrant Wnt signaling. Indeed, modifier loci have been identified that determine the rate of intestinal tumor formation/progression in APCMin mutant mice on different strain backgrounds (4). Also, studies to identify Wnt/APC/ß-catenin/TCF-4 pathway transcription targets in the normal colon led to the identification of a number of genes that control tumor formation downstream of this pathway (5,6). Finally, studies on the basic mechanisms of cell fate specification and differentiation of stem and progenitor cells in the GI tract led to the discovery that Notch activation is essential for adenoma formation in APCMin mice (7).
The Notch genes code for transmembrane receptors that regulate cell division, differentiation and survival of stem and/or progenitor cells in a wide range of tissues (8–10). In vertebrates there are four Notch receptors: Notch1, 2, 3 and 4, whose activity are controlled by Delta-like (Dll1, 3 and 4) and Jagged (Jagged1 and 2) family ligands. The specificity of ligand/receptor interaction is determined in many contexts through restricted expression of a sugar transferase from the Fringe gene family: Lfng, Mfng and Rfng (11–13). Activation of Notch by ligands results in sequential cleavage of the receptor by TACE or Kuzbanian proteases followed by γ-secretase. This complex cleavage reaction releases an intracellular domain fragment of Notch, termed NotchIC, which translocates into the nucleus. Nuclear NotchIC binds to the conserved DNA-binding protein, RBPJκ/CBF-1, and to transcriptional regulatory proteins to form a complex which activates transcription of genes suppressed in the absence of a Notch signal (9,14).
During lineage specification in a number of developmental contexts, Notch and Wnt signaling pathways function together to coordinate bHLH transcription factor-dependent cell fate specification (15,16). For example, Math1 is a bHLH transcriptional activator that is required for differentiation of intestinal and colonic goblet, enteroendocrine and paneth cells (17). Deletion of the Notch pathway transcription factor RBPJκ/CBF-1 or inhibition of γ-secretase-mediated Notch activation causes elevated Math1 expression and premature conversion of proliferative crypt cells into post mitotic goblet cells. These data indicate that Notch signaling is required for proliferation of intestinal progenitor cells and to suppress their Math1-dependent differentiation (7). Indeed, Notch1 and Notch2 function redundantly in this context to induce Hes1 (18). This bHLH repressor functions to directly suppresses transcription of CDK inhibitors (p27Kip1 and p57Kip) thereby facilitating proliferation of progenitors (18), and to antagonize Math1 in order to suppress goblet cell fate specification (7,18–22).
In the APCMin mutant mouse, Notch receptor signaling cooperates with the APC mutation to induce adenoma formation (7). Interestingly, human CRCs are typically devoid of Goblet secretary cells and express very low levels of HATH1 (the human orthologue of Math1) (23,24), suggesting that Notch receptor signaling might be activated in most human CRC. Here we report that JAGGED ligands, NOTCH1, LFNG and HES1 are expressed at levels similar to or higher than those observed in the crypt base in a small CRC cohort. Furthermore, in a survey of 130 human CRC, HES1 was expressed in most tumors indicating that Notch receptors are activated. Taken together with the low level of HATH1 expression in CRC noted by others, and the observation that Notch activation is a requirement for adenoma maintenance in the APCMin mutant mouse model, these data implicate Notch signaling in adenocarcinoma of the human colon.
Materials and methods
Slide preparation and in situ hybridization
Slide preparation and in situ hybridization was performed essentially as described previously (25). Briefly, 6 µm microtome sections of paraffin-embedded colon tissue were cut and placed onto glass slides. Slides were warmed to 42°C for 30 min, and then incubated with desiccant at RT overnight (ON). Tumor tissue sections were de-waxed in xylene twice for 10 min. Xylenes were removed through two 5-min incubations in 100% ethanol. Tissues were rehydrated by serial incubation in 95%, 85%, 70%, 50% and 30% ethanol in saline. Tissues were then re-fixed in 4% PFA/PBS for 20 min and washed twice in PBS. Next, tissues were treated with 20 µg/ml of proteinase K (Invitrogen) for 7.5 min, followed by a wash in PBS, another fixation in 4% PFA/PBS, and a final PBS wash. To prevent non-specific binding of probe, tissues were twice acetylated for 5 min in 0.1 M triethanolamine-HCl containing 500 µl of acetic anhydride and 448 µl of 10 N NaOH. After 5-min in PBS, followed by saline, tissues were dehydrated through an inverse rehydration process (see above) and air-dried. Radio-labeled probe was placed in hybridization mixture (50% deionized formamide, 0.3 M NaCl, 20 mM Tris-HCl pH 8.0, 5 mM EDTA, 10 mM NaPO4 pH 8.0, 10% dextran sulfate, 1X Denhardt's solution, 0.5 mg/ml yeast tRNA and 10 mm DTT) at a final concentration of 1.5×105 cpm/µl and denatured at 80°C for 2 min. Prepared tissue sections were covered with 60 µl of probe/hybridization mixture under a cover slip and allowed to incubate in a sealed container ON at 55°C. Cover slips were removed with a brief incubation in 5X SSC/0.1% 2-mercaptoethanol (2-ME) at 55°C. Slides were then placed in 50% formamide/2X SSC/2-ME at 65°C for 30 min. Next, slides were washed 3 times in 0.5 M NaCl/10 mM Tris-HCl/5 mM EDTA at 37°C followed by incubation in 20 µg/ml RNase A (Roche) for 30 min in the same buffer. After a final wash in NaCl/Tris-HCl/EDTA buffer, a repeat high-stringency incubation in formamide/2X SSC/2-ME was performed for 30 min. Slides were then washed in 2X SSC/2-ME followed by 0.1X SSC/2-ME for 30 min, each at 65°C. Finally, tissue sections were dehydrated as described above. Slides were treated with Kodak NTB-2 nuclear emulsion and stored at 4°C for approximately three weeks prior to development. Slides were developed in Kodak D-19 solution, fixed in Kodafix and counterstained with 0.1% toluidine blue.
Colon Cancer Family Registry and tissue microarrays
Participants were recruited to a multicenter Colon Cancer Family Registry as previously described (26,27). All samples were obtained following informed consent, and this study was approved by the Colon CFR Institutional Review Board. The specific tissue microarrays blocks used in this study were comprised of up to 100 cores from individual blocks, each obtained from colorectal cancer normal and tumor tissue from participants of the Ontario Familial Colon Cancer Registry (OFCCR). A pathologist reviewed H&E slides to choose 3–5 cores most representative of normal and tumor tissue in each block that was most suitable for coring. Trained histology technologists used a Tissue Arrayer (Beecher Instruments) to remove 0.6 nm cores from the donor blocks and reimbed these into new microarray blocks. Four micron slides were then cut for gene expression analysis. Tumor staging, grading, family history and microsatellite instability data were obtained for these samples according to standard protocols (26,27). Patient and tumor characteristics for the specific samples studied herein were obtained (data not shown).
Generation of probes for in situ hybridization
33P-UTP-labeled antisense Notch receptor and ligand gene probes were prepared as described previously (25). The human LFNG, MFNG, RFNG and HES1 probes correspond to nucleotides 655-315 (LFNG - coding sequence nt 89–841), nt 905-457 (MFNG - coding sequence nt 171–1136), nt 955-612 (RFNG - coding sequence nt 195–812) and nt 1–658 (HES1 - coding sequence nt 1–462). 33P-UTP-labeled antisense Fringe and HES1 probes were prepared in the same way as all other Notch pathway probes (25).
Statistical analysis
Overall survival was measured from diagnosis to last follow-up or death. Kaplan-Meier curves were calculated for the high and low expression of HES1 mRNA. ‘High’ expression represented the top quartile of Allred scores. This was determined separately for each TMA since the efficiency of hybridization was not necessarily the same in each experiment. Survival between groups was compared using the log-rank test. Cox proportional hazard regression was used to look for a dose-response relationship between level of HES1 expression and survival. P≤0.05 was considered statistically significant.
Results and Discussion
Expression of Notch pathway genes in human CRC
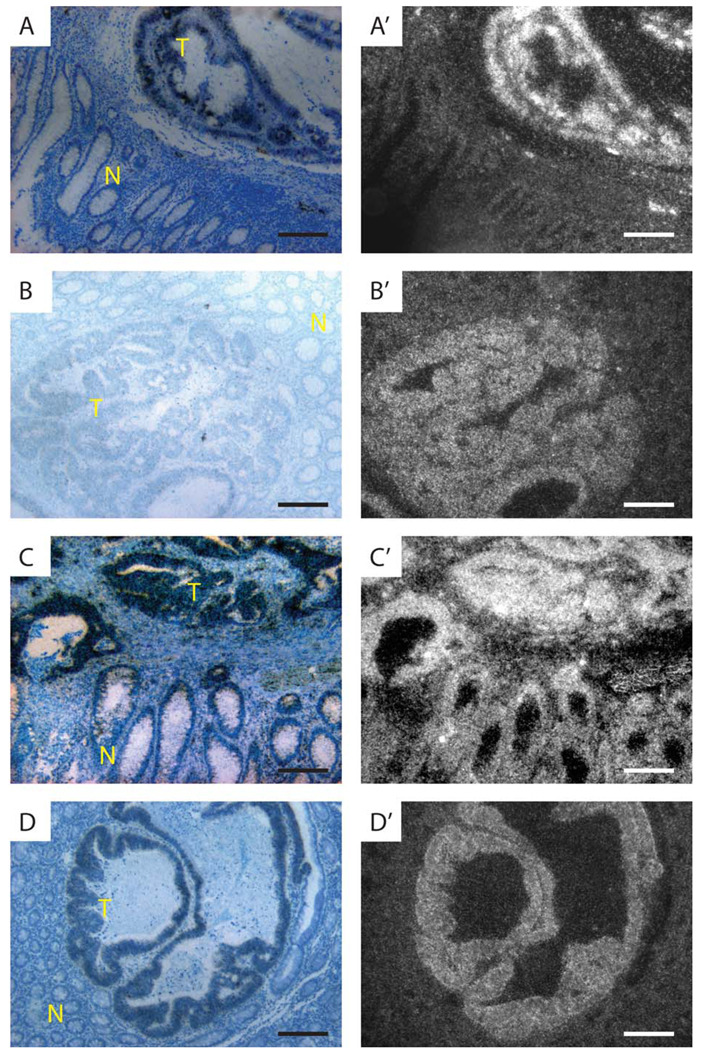
To analyze Notch pathway gene expression in human CRC, we performed in situ hybridization with probes previously used to study expression of this system in breast cancer (25). As a first step, we analyzed expression of JAGGED and DLL ligands in 10–20 random CRC cases, some with surrounding normal tissue. JAG1 was expressed at variable levels. Most tumors expressed this gene at a level comparable to expression seen in the crypt base and normal epithelium. In a few tumors, JAG1 was only marginally expressed or its expression was elevated (Table I and Fig. 1A). In contrast, JAG2 was only marginally expressed in the normal crypt base and differentiated epithelium, but expressed at an elevated level in all 20 CRC cases examined (Table I and Fig. 1B). DLL1 and DLL3 showed low expression whereas DLL4 expression was marginal. This situation was comparable to expression levels observed in normal mucosa (Table I).
Table I.
Expression of Notch activation system genes in human CRC and normal adjacent tissue.
| Adenocarcinoma | Epithelium | Proliferating crypt |
Vasculature a | Lamina propria |
Lymphoid follicle |
|
|---|---|---|---|---|---|---|
| JAG1 | 4/20 +/−; 11/20 +; 5/20 ++ | + | + | ++ | − | − |
| JAG2 | 18/20 +; 2/20 ++ | +/− | +/− | + | − | +/− |
| DLL1 | 10/10 + | + | + | +/− | + | + |
| DLL3 | 10/10 + | + | + | − | − | − |
| DLL4 | 10/10 +/− | +/− | +/− | ++ | − | − |
| NOTCH1 | 17/20 ++; 3/20 +++ | + | ++ | + | + | ++ |
| NOTCH2 | 10/10 + | +/−b | +/−b | − | +/− | +/− |
| NOTCH3 | 10/10− | − | − | +++ | − | − |
| NOTCH4 | 10/10 − | − | − | + | − | − |
| LFNG | 14/20 ++; 6/20 +++ | + | ++ | − | +/− | +/− |
| MFNG | 10/10 − | − | − | − | − | +/− |
| RFNG | 10/10 + | + | + | − | − | − |
| HES1 | 8/12 ++; 4/12 +++ | + | ++ | − | − | − |
(−), no expression; (+/−), marginal expression; (+), low; (++), intermediate; (+++), high.
Expression was seen in the endothelium with the exception of NOTCH3, which was seen in the VSMC.
5–10 individual cells with +++ expression were seen per gland (with the majority in the lower 1/3 of the gland).
Figure 1.
Expression of Notch ligands, receptors and fringes in human CRC. In situ hybridization using antisense probes for JAG1 (A,A'), JAG2 (B,B'), NOTCH1 (C,C'), and LFNG (D,D'). Bright field images are shown on the left (A–-D) and dark filed images on the right (A'–D'). High level Notch ligand, receptor and Fringe mRNA expression occurs in some invasive adenenocarcinomas of the colon. Scale bars in each panel are 200 microns in length.
Next we used probes to determine Notch receptor gene expression. NOTCH1, which was expressed at higher levels in the crypt than in differentiated colonic epithelium (28,29), was expressed at intermediate or high levels in all 20 cases (Table I). In most cases, expression levels were similar to that observed in the crypt base of adjacent non-tumor tissue (17/20). Three of the CRC cases expressed elevated levels of NOTCH1 in comparison to the crypt (Fig. 1C). NOTCH2 was expressed at low levels in 10 of 10 tumors examined and expressed at very high levels in a small subset of cells scattered throughout the crypt and villus (Table I) (28). In contrast, NOTCH3 and NOTCH4 were not detectably expressed in CRC, nor in the crypt or villus. These probes were effective at detecting expression in vasculature as expected (Table I).
Fringe genes control Notch activation in many developmental contexts and are expressed in the developing intestine (30). We therefore tested for expression of these genes in human CRC. LFNG was expressed in all 20 CRC cases examined, at levels comparable to (14/20) or higher than that observed in the crypt base (6/20) (Fig. 1D and Table I). As with NOTCH1, LFNG expression is higher in the crypt than in differentiated epithelial cells of the villus (Table I). In contrast, MFNG expression was not observed in CRC or in adjacent normal colonic tissue, with the exception of lymphoid follicles, where marginal expression was noted. RFNG was expressed at low and comparable levels in CRC, crypt and villus cells. Notch pathway genes were also expressed in a number of non-tumor cell types as noted above (Table I). For example, vasculature within the tumor and in adjacent non-tumor tissue expressed JAG1, JAG2, DLL1, DLL4, NOTCH1, NOTCH3, and NOTCH4 as expected (31). Lamina propria expressed DLL1, NOTCH1, NOTCH2 and LFNG (Table I). Finally, lymphoid follicles expressed JAG2, DLL1, NOTCH1, NOTCH2, LFNG and MFNG (Table I). Expression in these structures helped facilitate comparison between tumors and provided positive controls for genes that were either not expressed in CRC, or expressed at marginal levels. In sum, human CRC is similar to the proliferating crypt with respect to Notch pathway gene expression. For example, NOTCH1 and LFNG are expressed at a higher level in the crypt than in the villus, and these two genes are expressed at levels comparable to the crypt in most tumors with adjacent normal tissue. In addition, JAGGED ligands, NOTCH1 and LFNG are expressed at elevated levels in a number of CRC cases suggesting that these genes may play a role in tumor development and/or progression.
Activation of Notch signaling in most human CRC
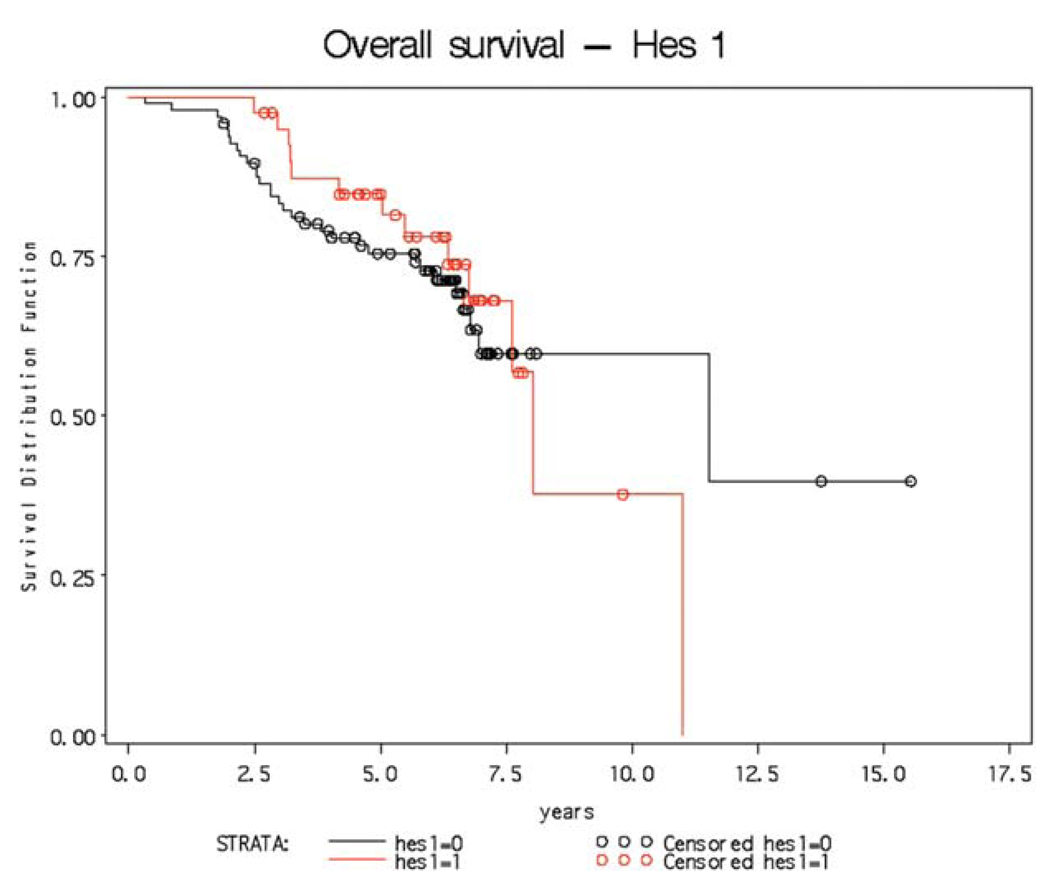
We next analyzed HES1 gene expression as a marker for Notch receptor signaling (7,19,32). In the same small group of tumors, HES1 was expressed in all 12 analyzed (Table I). In eight of these, expression was comparable to the level observed in crypts. In four of the twelve, CRC expression was elevated. Hes1 is expressed in tumors from the APCMin mutant mouse (7) and was detected by immunohistochemistry in a small cohort of human CRC (33). To test for any association between Notch activation and outcome, we therefore screened a cohort of 130 well-characterized colon tumors with associated comprehensive clinical database (Table II). This cohort was obtained from the US National Cancer Institute Colon Cancer Family Registry/Ontario Familial Colon Cancer Registry and included a mixture of familial and sporadic colon cancers (26). Blocks from these tumors were used to generate three tissue micro-arrays, which we screened for HES1 expression and scored using an Allred scoring system modified to score for radioactive in situ hybridization (34). The Allred scores ranged from 2 to 8, with detectable expression noted in every tumor indicated that Notch signaling is a consistent feature of human CRC. For each tumor, HES1 expression was either designated as low or high, with tumors designated as high, expressing at levels within the top quartile of tumors on each array. We next tested for, and failed to find, a correlation between HES1 expression and survival (Table II and Fig. 2). Since each TMA section could potentially hybridize to probe with a different efficiency, we also tested for a correlation between HES1 expression and survival within each of the three TMA datasets separately. Using this approach we also failed to detect a correlation between HES1 expression and survival (data not shown).
Table II.
Patient characteristics and survival data.
| Patients | n (%) | Hes1 Low (n) |
Hes1 Hi (n) |
P-valuea |
|---|---|---|---|---|
| Stage | ||||
| 1 | 21 | 16 | 9 | |
| 2 | 46 | 41 | 15 | 0.69 (NS) |
| 3 or 4 | 33 | 28 | 12 | |
| Grade | ||||
| Well-differentiated | 9 | 7 | 3 | |
| Moderate-differentiated | 81 | 65 | 30 | 0.23 (NS) |
| Poorly-differentiated | 10 | 11 | 1 | |
| Family history | ||||
| Sporadic | 6 | 5 | 4 | |
| Some | 68 | 70 | 24 | 0.72 (NS) |
| Strong | 26 | 23 | 13 | |
| Microsatellite instability | ||||
| Stable | 65 | 50 | 25 | |
| Low | 11 | 11 | 2 | 0.57 (NS) |
| High | 23 | 19 | 8 | |
| n=139 | ||||
| Stratum | Hes1 | Total | Deceased | Alive (%) |
5 year survival % (95% CI) |
|---|---|---|---|---|---|
| 1 | Low (n=0) | 97 | 31 | 66 (68.0) | 75 (65–83) |
| 2 | Hi (n=1) | 41 | 13 | 28 (68.3) | 85 (69–93) |
| 138 | 44 | 94 (68.1) |
Cochran-Armitage test for trend; NS, not significant.
There was one individual with a missing value. Test for difference between groups: p=0.81.
Figure 2.
Kaplan-Merier curve shows no relationship between high level HES1 gene expression and survival in patients with colorectal cancer.
In conclusion, a number of recent studies have shown that Notch signaling controls stem or progenitor cell proliferation, as well as cell fate specification and differentiation in the colon (7,18,19,21,22,35). In addition, Notch signaling is necessary for adenoma formation in the APCMin mouse (7). Here we provide evidence that Notch signaling is activated in human CRC. In a small cohort of colon tumors, we observed JAG ligand, NOTCH1, LFNG and HES1 gene expression at levels comparable to, or greater than, normally observed in cells of the crypt base. Indeed, with respect to expression of NOTCH1, LFNG and HES1, CRC resembles the crypt. Interestingly, JAG2 expression was elevated in every tumor examined. Finally, we analyzed Notch activation, using HES1 expression as a surrogate, in 130 tumors with associated patient follow-up data. HES1 expression was variable, although it was observed in every tumor. This is consistent with results from a recent report by Veenendaal et al on activation of Wnt, Ras, TGFß and Notch signaling in a cohort of 60 colorectal tumors (33). While survival data were not provided in this report, HES1 expression was compared between primary tumors and metastatic lesions, were it was frequently reduced. Here we report that the absolute level of HES1 mRNA did not correlate with survival indicating that activation of Notch signaling occurs in human colorectal cancer, but that the level of signaling is not prognostic. In conclusion, our data reveal that human CRC patients may indeed benefit from γ-secretase-inhibitors or other Notch targeted therapeutics as suggested from studies in the mouse (36) and with human CRC cell lines (37).
Acknowledgements
This work was supported by the National Cancer Institute, National Institutes of Health under RFA#CA-96-011 and through cooperative agreements with members of the Colon Cancer Family Registry and P.I.s. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating institutions or investigators in the Colon CFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the Colon CFR. The authors would like to thank members of the Egan lab for valuable advice and support. This study was supported by funds to S.E.E. from the Canadian Cancer Society and National Cancer Institute of Canada. M.R. is supported by the Society of University Surgeons. Finally, we would like to thank the patients for their generous donation of the tumor samples used in this study.
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 4.Dove WF, Cormier RT, Gould KA, Halberg RB, Merritt AJ, Newton MA, Shoemaker AR. The intestinal epithelium and its neoplasms: genetic, cellular and tissue interactions. Philos Trans R Soc Lond B Biol Sci. 1998;353:915–923. doi: 10.1098/rstb.1998.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 6.Batlle E, Bacani J, Begthel H, Jonkheer S, Gregorieff A, van de Born M, Malats N, Sancho E, Boon E, Pawson T, Gallinger S, Pals S, Clevers H. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435:1126–1130. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]
- 7.Van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 9.Callahan R, Egan SE. Notch signaling in mammary development and oncogenesis. J Mammary Gland Biol Neoplasia. 2004;9:145–163. doi: 10.1023/B:JOMG.0000037159.63644.81. [DOI] [PubMed] [Google Scholar]
- 10.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 11.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 12.Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, Whiting E, Ryan D, Zinyk D, Boulianne G, Hui CC, Gallie B, Phillips RA, Lipshitz HD, Egan SE. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- 13.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 14.Kopan R. Notch: a membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- 15.Hayward P, Kalmar T, Martinez Arias A. Wnt/Notch signalling and information processing during development. Development. 2008;135:411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 16.Gridley T. The long and short of it: somite formation in mice. Dev Dyn. 2006;235:2330–2336. doi: 10.1002/dvdy.20850. [DOI] [PubMed] [Google Scholar]
- 17.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 18.Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27(Kip1) and p57(Kip2) EMBO Rep. doi: 10.1038/embor.2008.7. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 20.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 21.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci USA. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132:1093–1104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- 23.Leow CC, Romero MS, Ross S, Polakis P, Gao WQ. Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res. 2004;64:6050–6057. doi: 10.1158/0008-5472.CAN-04-0290. [DOI] [PubMed] [Google Scholar]
- 24.Leow CC, Polakis P, Gao WQ. A role for Hath1, a bHLH transcription factor, in colon adenocarcinoma. Ann NY Acad Sci. 2005;1059:174–183. doi: 10.1196/annals.1339.048. [DOI] [PubMed] [Google Scholar]
- 25.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level co-expression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 26.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 27.Poynter JN, Figueiredo JC, Conti DV, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 28.Sander GR, Powell BC. Expression of notch receptors and ligands in the adult gut. J Histochem Cytochem. 2004;52:509–516. doi: 10.1177/002215540405200409. [DOI] [PubMed] [Google Scholar]
- 29.Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, Leung SY, Chen X. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroder N, Gossler A. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr Patterns. 2002;2:247–250. doi: 10.1016/s1567-133x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 31.Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–2718. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- 32.Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, van den Born M, Stappenbeck T, Wu Y, Clevers H, Kopan R. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veenendaal LM, Kranenburg O, Smakman N, Klomp A, Borel Rinkes IH, van Diest PJ. Differential Notch and TGFbeta signaling in primary colorectal tumors and their corresponding metastases. Cell Oncol. 2008;30:1–11. doi: 10.1155/2008/839076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickson BC, Mulligan AM, Zhang H, Lockwood G, O'Malley FP, Egan SE, Reedijk M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 35.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 2005;19:1686–1691. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Es JH, Clevers H. Notch and Wnt inhibitors as potential new drugs for intestinal neoplastic disease. Trends Mol Med. 2005;11:496–502. doi: 10.1016/j.molmed.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Akiyoshi T, Nakamura M, Yanai K, Nagai S, Wada J, Koga K, Nakashima H, Sato N, Tanaka M, Katano M. Gamma-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology. 2008;134:131–144. doi: 10.1053/j.gastro.2007.10.008. [DOI] [PubMed] [Google Scholar]