Abstract
The transcriptional coactivator host cell factor 1 (HCF-1) is critical for the expression of immediate-early (IE) genes of the alphaherpesviruses herpes simplex virus type 1 (HSV-1) and varicella-zoster virus. HCF-1 may also be involved in the reactivation of these viruses from latency as it is sequestered in the cytoplasm of sensory neurons but is rapidly relocalized to the nucleus upon stimulation that results in reactivation. Here, chromatin immunoprecipitation assays demonstrate that HCF-1 is recruited to IE promoters of viral genomes during the initiation of reactivation, correlating with RNA polymerase II occupancy and IE expression. The data support the model whereby HCF-1 plays a pivotal role in the reactivation of HSV-1 from latency.
Host cell factor 1 (HCF-1) is a cellular transcriptional coactivator that is essential for the induced expression of the immediate-early (IE) genes of the alphaherpesviruses herpes simplex virus type 1 (HSV-1) and varicella-zoster virus (VZV) during initiation of lytic infection (12, 19). The protein is recruited to the IE enhancer domains via interactions with the viral IE activators (VP16 for HSV-1 and ORF10 for VZV), where it forms stable multiprotein enhanceosome complexes on the IE enhancer core elements (12, 18, 31, 32). While these enhancer elements play a dominant role in the induction of IE gene expression, HCF-1 may also be recruited to IE promoters and enhancers through interactions with other viral (i.e., VZV IE62) as well as cellular (i.e., Sp1 and GA-binding protein [GABP]) factors that can mediate the expression of the IE genes (19, 23, 28, 29). HCF-1 is a component of chromatin modification complexes, including the MLL/Set histone methyltransferases (33, 34) and the histone demethylase lysine-specific demethylase 1 (LSD1) (Y. Liang, J. L. Vogel, A. Narayanan, H. Peng, and T. M. Kristie, submitted for publication), and functions to coordinate activities to modulate chromatin-mediated repression and promote the installation of activating chromatin marks (7, 20; Liang et al., submitted).
In addition to its role in the regulation of viral lytic replication (4, 7, 9, 10, 20, 24-26; Liang et al., submitted), chromatin plays a role in the latency-reactivation cycles of HSV-1, where repressive or activating histone modifications at the IE gene promoters correlate with latency or reactivation, respectively (1-3, 5, 10, 15, 21, 22, 30). That HCF-1-dependent complexes might play a role in the reactivation process, in a manner similar to their role in viral lytic infection, is suggested by (i) the unique cytoplasmic localization of HCF-1 in unstimulated sensory neurons and its nuclear translocation in response to signals that promote viral reactivation, both in vivo and ex vivo (11, 13), and (ii) the requirement of the histone demethylase LSD1, an HCF-1-associated component, for viral reactivation in ex vivo ganglion reactivation studies (Liang et al., submitted).
Recruitment of HCF-1 to IE promoters upon reactivation ex vivo.
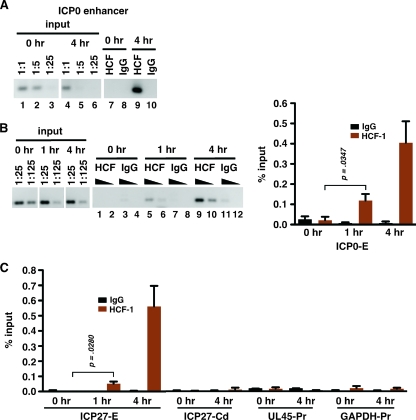
To further investigate the role of HCF-1 in HSV-1 reactivation, chromatin immunoprecipitations (ChIP) were used to assess HCF-1 occupancy at viral IE promoters during reactivation in neurons of trigeminal ganglia (TG). TG from latently infected mice were removed and immediately processed (0 h) or were explanted into tissue culture for 4 h to stimulate efficient viral reactivation prior to processing as described in the supplemental material. Aliquots of isolated chromatin corresponding to 10 latently infected ganglia were subjected to ChIP using control immunoglobulin G (IgG) or HCF-1 antibodies (Fig. 1A). Due to the limited amount of material and the low frequency of viral reactivation, a nested PCR method was developed to increase the sensitivity of the assay (see the supplemental material). As shown in Fig. 1A, no HCF-1 could be detected at the ICP0 enhancer domain at 0 h (lane 7) but the protein was readily detected after explant for 4 h (lane 9).
FIG. 1.
Recruitment of HCF-1 to viral IE promoters during reactivation. HCF-1 occupancy of the indicated promoter and control domains was analyzed by semiquantitative PCR using nested primer sets (see the supplemental material) along with dilutions of DNA isolated from chromatin prior to immunoprecipitation (input). (A) HCF-1 and control IgG ChIP assays were done using chromatin isolated from TG explanted for 0 or 4 h and assessed for HCF-1 occupancy at the ICP0 enhancer domain (ICP0-E). (B) Time course of HCF-1 occupancy at the ICP0 enhancer domain. (C) HCF-1 and control IgG ChIP assays analyzed for HCF-1 occupancy of the ICP27 enhancer domain (ICP27-E) and control regions (ICP27-Cd, ICP27 coding region; UL45-Pr, UL45 promoter; GAPDH-Pr, GAPDH promoter). The graphs represent data from four independent experiments. Occupancy of viral sequences was analyzed by nested PCR or by nested qPCR (conventional PCR followed by qPCR using a nested primer set). Occupancy of the cellular GAPDH promoter was analyzed via qPCR. Error bars are standard errors of the means. Statistical analyses used two-tailed t tests with statistical significance at a P value of <0.05.
In sensory neurons of unstimulated TG, HCF-1 is sequestered in the cytoplasm. Upon stimulation, the percentage of neurons that exhibit nuclear HCF-1 localization increases over the course of 6 h poststimulation (11, 13). To determine whether the occupancy of HCF-1 could be detected at earlier times postinduction of reactivation, the ChIP assays were done using TG explanted for 0, 1, or 4 h. As shown in Fig. 1B and C, HCF-1 occupancy was reproducibly detected as early as 1 h postexplantation and occupancy increased significantly by 4 h at both the ICP0 and ICP27 IE gene enhancer domains. In contrast, no significant occupancy was detected within control regions (ICP27 coding region, UL45 promoter, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH] promoter) (Fig. 1C). Thus, specific HCF-1 occupancy of the viral IE promoter correlates with the previously described time course of transport and nuclear localization of HCF-1.
HCF-1 recruitment correlates with RNAPII occupancy and induction of HSV-1 IE gene transcription.
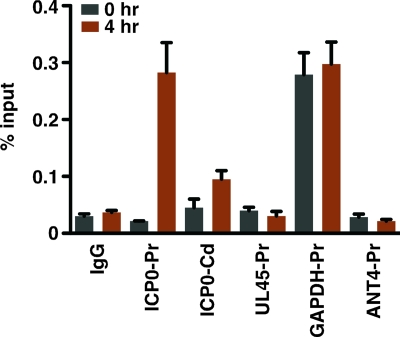
The recruitment of HCF-1 to the viral IE gene enhancer-promoter domain during the initial stages of reactivation is anticipated to correlate with the recruitment of RNA polymerase II (RNAPII) and the accumulation of IE mRNAs. As shown in Fig. 2, RNAPII occupancy of the ICP0 promoter was not significant relative to that of the IgG control or that of the promoters of the nontranscribed ANT4 gene or the viral late UL45 gene at 0 h poststimulus but was detected at the promoter of the transcribed GAPDH gene. At 4 h poststimulus, RNAPII occupancy of the viral IE promoter was clearly evident. Additionally, a low-level occupancy was reproducibly detected within the coding sequences (Fig. 2, ICP0-Cd). No change in the status of occupancy of the control UL45, ANT4, or GAPDH promoter was detected.
FIG. 2.
HCF-1 recruitment correlates with RNAPII occupancy at viral IE promoters. ChIP analyses of RNAPII occupancy of the indicated promoter and control domains at 0 and 4 h after TG explant. Occupancy was analyzed as described in the legend to Fig. 1. The GAPDH gene is a positive (transcribed gene) control while the ANT4 gene is a negative (repressed gene) control. The UL45 gene is an HSV-1 late gene. ICP0-Pr, ICP0 promoter domain; ICP0-Cd, ICP0 coding region; UL45-Pr, UL45 promoter; GAPDH-Pr, GAPDH promoter; ANT4-Pr, ANT4 promoter.
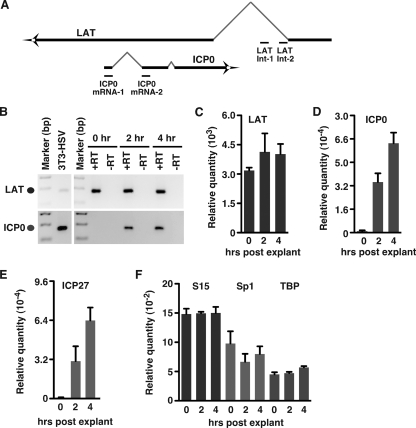
To confirm that HCF-1 and RNAPII occupancy correlates with transcriptional activation and accumulation of viral IE mRNAs, cDNAs were produced from total RNAs isolated from latently infected TG at 0, 2, and 4 h after reactivation explant. The levels of viral (latency-associated transcript [LAT], ICP0, and ICP27) and cellular control (S15, Sp1, and TATA-binding protein [TBP]) mRNAs were determined by nested PCR or quantitative PCR (qPCR). The LAT RNA and cellular control mRNAs were readily detected at zero time (Fig. 3B, C, and F). In contrast, ICP0 and ICP27 mRNAs were not detected at zero time but were detected at 2 h postexplant, and their relative abundance increased by 4 h postexplant (Fig. 3B, D, and E). The cellular controls S15, Sp1, and TBP showed no significant variance between TG harvested at 0, 2, and 4 h postexplant (Fig. 3F).
FIG. 3.
IE mRNA accumulation after reactivation stimulation. (A) Schematic representation of the ICP0/LAT transcripts illustrating the primer sets used to distinguish the ICP0 mRNA from LAT stable intron RNA. (B) PCR analysis of LAT stable intron RNA (top panel) and nested PCR analysis of ICP0 mRNA (bottom panel) from TG explanted for 0, 2, and 4 h. +RT, with reverse transcriptase; −RT, without reverse transcriptase. (C) PCR quantitation of the levels of LAT stable intron RNA. (D) Quantitation of nested PCR analyses of ICP0 mRNA. The graphs represent data from three independent experiments. Error bars are standard errors of the means. (E) Nested qPCR analysis of the levels of ICP27 mRNA in TG explanted for 0, 2, and 4 h. (F) qPCR analysis of the levels of control mRNAs (S15, Sp1, and TBP) at 0, 2, and 4 h postexplantation. All quantitation is relative to that of cDNAs produced from RNA of NIH 3T3 cells infected with 0.1 PFU/cell HSV-1 for 6 h.
HCF-1 occupancy localizes to IE distal promoter-enhancer domains.
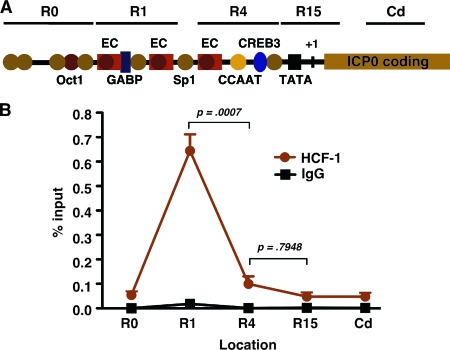
The promoter-enhancer domains of the HSV-1 IE genes are complex. These regions contain reiterated enhancer core elements that nucleate the assembly of the Oct-1/VP16/HCF-1 enhancer complex during initiation of lytic infection (Fig. 4A). In addition, these domains contain binding sites for other cellular transcription factors, such as GABP, Sp1, and CREB3, that interact directly with HCF-1 and are capable of recruiting the coactivator (6, 8, 12, 16, 17, 29).
FIG. 4.
Localization of HCF-1 occupancy within the ICP0 promoter-enhancer domain. (A) Schematic representation of the ICP0 promoter-enhancer domain. HCF-1 occupancy was assessed at the five indicated regions by ChIP followed by nested qPCR using region-specific primer sets. The location of the three enhancer core (EC) elements and the binding sites for several defined transcription factors that impact the level of IE gene expression are shown. (B) Results of HCF-1 and control IgG ChIP assays using chromatin from TG explanted for 4 h. The data were derived from four independent experiments. Error bars are standard errors of the means. R0, region upstream of the defined enhancer domain; R4, promoter-proximal region; R15, promoter initiation region, Cd, coding region.
To gain insight into the component(s) that might be involved in the recruitment of HCF-1 to viral IE genes during reactivation, the occupancy of HCF-1 was localized using primer sets across the ICP0 promoter-enhancer domain. In these experiments, isolated chromatin from 4-h-induced TG was sonicated to approximately 300 bp to increase the resolution of the ChIP. As shown in Fig. 4B, immunoprecipitation with anti-HCF-1 antibodies resulted in the recovery of IE promoter fragments nearly exclusively representing the distal promoter-enhancer region (Fig. 4A). In contrast, no significant levels of fragments representing any other region were detected. The results indicate that stable HCF-1 association is dependent upon factors recognizing elements within the distal enhancer region, a region that minimally contains binding sites for GABP and Sp1 in addition to two enhancer core elements.
HCF-1, a component of the regulatory mechanisms governing HSV-1 reactivation.
HCF-1 is an essential component of the HSV-1 IE regulatory paradigm. The protein is recruited to viral IE promoter-enhancer domains via complex interactions with multiple factors, including viral IE activators, that contribute to the induced expression of these genes during initiation of viral lytic infection. HCF-1 functions, at least in part, by coordination of chromatin modification complexes, such as the Set1/MLL1 histone methyltransferase and the histone demethylase LSD1, to prevent the accumulation of repressive chromatin and promote the installation of positive chromatin marks.
In addition to its role in lytic infection, chromatin plays a significant role in the regulation of HSV-1 latency and reactivation cycles. Activating marks are evident on the transcribed LAT gene while repressive marks are prevalent on the repressed IE gene promoter regions (1, 14, 15, 21, 30). Conversely, activating marks accumulate on the viral IE gene promoter domains during the initial stages of reactivation (1, 21). Strikingly, HCF-1 is sequestered in the cytoplasm of sensory neurons, where the virus establishes latency. The protein is rapidly relocalized to the nucleus upon in vivo and ex vivo stimulation, suggesting that HCF-1 may also play a critical role in reactivation of HSV-1 from latency. However, a direct involvement in viral reactivation has not yet been demonstrated.
In this study, a ChIP approach was used to demonstrate that HCF-1 is recruited to HSV-1 IE gene promoter-enhancer domains upon stimulation of reactivation of HSV-1 from latency in sensory neurons. Recruitment was detected as early as 1 h postinduction of reactivation, and occupancy increased by 4 h postinduction. This time course correlates well with the nuclear transport of HCF-1, where the number of neurons exhibiting nuclear localization increases over the course of 6 h poststimulation. Additionally, HCF-1 occupancy of viral IE promoter domains correlated with occupancy by RNAPII and accumulation of viral IE mRNAs. This study represents the first report of the direct recruitment of a critical component of the IE gene regulatory circuit during viral reactivation.
HCF-1 is not a direct DNA binding component, and its recruitment is dependent upon interactions with site-specific transcription factors. The protein interacts with members of numerous transcription factor families, including several activators (GABP, Sp1, and VP16) that contribute to the induced transcription of viral IE genes during lytic infection. Occupancy of the IE promoter-enhancer domain by HCF-1 during reactivation localizes to the distal promoter region of the enhancer domain. This region contains several defined factor binding sites, including two TAATGARAT enhancer core elements, a consensus GABP, and several Sp1 sites. In contrast, no significant association of HCF-1 with the promoter-proximal enhancer core element or the putative CREB3 site was detected, a factor that was proposed to be involved in HCF-1 recruitment during reactivation (16). While this suggests that the factor(s) responsible for HCF-1 recruitment is localized distal to the promoter domain, this may also be a reflection of multiple interactions that result in a highly stable association at this region. It is important to note that due to the limitations of the ChIP assay, the data do not exclude the possibility that HCF-1 is transiently or nonstably recruited via factors whose recognition sites are within other regions of the IE enhancer-promoter domains.
It is also possible that the factors that recruit HCF-1 could depend upon the specific reactivation stimuli. Of note, a recent study has indicated that the viral IE activator VP16, a protein whose function is dependent upon HCF-1, may be critical for viral IE transcription and reactivation (27). The recruitment of HCF-1 to the region containing two enhancer core elements is consistent with this model. However, it remains to be determined which factors are directly responsible for HCF-1 recruitment and whether those factors reflect distinct signaling pathways. Regardless, the demonstration of the direct recruitment of HCF-1 to viral IE genes during the initial stages of reactivation promotes the model that HCF-1 complexes play a critical role in the viral reactivation process.
Supplementary Material
Acknowledgments
We thank N. Fraser for HSV-1 strain 17, M. Jang, T. Pierson, A. McBride, L. Lagenaur, and members of the Laboratory of Viral Diseases and the Molecular Genetics Section, NIAID, for helpful advice and discussions, J. Skinner (Bioinformatics and Computational Biosciences Branch, NIAID) for expert statistical analysis of the experimental data, and T. Pierson, J. Vogel, and Y. Liang for critical readings of the manuscript.
This study was supported by the Laboratory of Viral Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 1 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amelio, A. L., N. V. Giordani, N. J. Kubat, J. E. O'Neil, and D. C. Bloom. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80:2063-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amelio, A. L., P. K. McAnany, and D. C. Bloom. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 80:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Q., L. Lin, S. Smith, J. Huang, S. L. Berger, and J. Zhou. 2007. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J. Virol. 81:5192-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cliffe, A. R., and D. M. Knipe. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordani, N. V., D. M. Neumann, D. L. Kwiatkowski, P. S. Bhattacharjee, P. K. McAnany, J. M. Hill, and D. C. Bloom. 2008. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J. Virol. 82:6056-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunther, M., M. Laithier, and O. Brison. 2000. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell. Biochem. 210:131-142. [DOI] [PubMed] [Google Scholar]
- 7.Huang, J., J. R. Kent, B. Placek, K. A. Whelan, C. M. Hollow, P.-Y. Zeng, N. W. Fraser, and S. L. Berger. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 80:5740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, K. A., and R. Tjian. 1985. Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature 317:179-182. [DOI] [PubMed] [Google Scholar]
- 9.Kent, J. R., P. -Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78:10178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knipe, D. M., and A. Cliffe. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6:211-221. [DOI] [PubMed] [Google Scholar]
- 11.Kolb, G., and T. M. Kristie. 2008. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J. Virol. 82:9555-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristie, T. M. 2007. Early events pre-initiation of alphaherpesvirus viral gene expression, p. 112-127. In A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, NY. [PubMed]
- 13.Kristie, T. M., J. L. Vogel, and A. E. Sears. 1999. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl. Acad. Sci. USA 96:1229-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubat, N. J., A. L. Amelio, N. V. Giordani, and D. C. Bloom. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, R., and V. Misra. 2000. Potential role for luman, the cellular homologue of herpes simplex virus VP16 (α gene trans-inducing factor), in herpesvirus latency. J. Virol. 74:934-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, R., P. Yang, P. O'Hare, and V. Misra. 1997. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 17:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1995. Proteins and cis-acting elements associated with transactivation of the varicella-zoster virus (VZV) immediate-early gene 62 promoter by VZV open reading frame 10 protein. J. Virol. 69:4693-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narayanan, A., M. L. Nogueira, W. T. Ruyechan, and T. M. Kristie. 2005. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J. Biol. Chem. 280:1369-1375. [DOI] [PubMed] [Google Scholar]
- 20.Narayanan, A., W. T. Ruyechan, and T. M. Kristie. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. USA 104:10835-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann, D. M., P. S. Bhattacharjee, N. V. Giordani, D. C. Bloom, and J. M. Hill. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 81:13248-13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann, D. M., P. S. Bhattacharjee, and J. M. Hill. 2007. Sodium butyrate: a chemical inducer of in vivo reactivation of herpes simplex virus type 1 in the ocular mouse model. J. Virol. 81:6106-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nogueira, M. L., V. E. Wang, D. Tantin, P. A. Sharp, and T. M. Kristie. 2004. Herpes simplex virus infections are arrested in Oct-1-deficient cells. Proc. Natl. Acad. Sci. USA 101:1473-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh, J., and N. W. Fraser. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82:3530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Placek, B. J., J. Huang, J. R. Kent, J. Dorsey, L. Rice, N. W. Fraser, and S. L. Berger. 2008. The histone variant H3.3 regulates gene expression during lytic infection by herpes simplex virus type 1. J. Virol. 83:1416-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva, L., A. Cliffe, L. Chang, and D. M. Knipe. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, R. L., C. M. Preston, and N. M. Sawtell. 2009. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog. 5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triezenberg, S. J., K. L. LaMarco, and S. L. McKnight. 1988. Evidence of DNA: protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 2:730-742. [DOI] [PubMed] [Google Scholar]
- 29.Vogel, J. L., and T. M. Kristie. 2000. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 19:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Q. Y., C. Zhou, K. E. Johnson, R. C. Colgrove, D. M. Coen, and D. M. Knipe. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 102:16055-16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, A. C., M. A. Cleary, J. S. Lai, K. LaMarco, M. G. Peterson, and W. Herr. 1993. Combinatorial control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp. Quant. Biol. 58:167-178. [DOI] [PubMed] [Google Scholar]
- 32.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294-304. [DOI] [PubMed] [Google Scholar]
- 33.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.