Abstract
Glucose and glutamine are abundant nutrients required for cell growth, yet how cells sense and adapt to changes in their levels is not well understood. The MondoA transcription factor forms a heterocomplex with its obligate partner Mlx to regulate ≈75% of glucose-dependent transcription. By mediating glucose-induced activation of thioredoxin-interacting protein (TXNIP), MondoA:Mlx complexes directly repress glucose uptake. We show here that glutamine inhibits transcriptional activation of TXNIP by triggering the recruitment of a histone deacetylase-dependent corepressor to the amino terminus of MondoA. Therefore, in the presence of both glucose and glutamine, TXNIP expression is low, which favors glucose uptake and aerobic glycolysis; the Warburg effect. Consistent with MondoA functioning upstream of TXNIP, MondoA knockdown reduces TXNIP expression, elevates glucose uptake and stimulates cell proliferation. Although glutamine has many intracellular fates, a cell permeable analog of a tricarboxylic acid cycle (TCA) intermediate, α-ketoglutarate, also blocks the transcriptional activity of MondoA at the TXNIP promoter and stimulates glucose uptake. Together our data suggest that glutamine-dependent mitochondrial anapleurosis dictates glucose uptake and aerobic glycolysis by blocking MondoA:Mlx-dependent transcriptional activation of TXNIP. We propose that this previously unappreciated coordination between glutamine and glucose utilization defines a metabolic checkpoint that restricts cell growth when subthreshold levels of these essential nutrients are available.
Keywords: metabolism, mitochondria, transcription, bHLHZIP
Cell growth is controlled by a coordinated response to both growth factors and nutrients. For example, growth factor stimulation triggers amino acid and glucose uptake. Further, in many circumstances, nutrients and growth factor signaling coordinate cell growth and survival by controlling the activity of TORC1 complex (1, 2). Alterations in both nutrient sensing and growth factor signaling pathways in cancer are extensive and well documented. In contrast, less is known about how cells respond transcriptionally to changes in nutrient availability and the outcome of this adaptive response in coordinating cell growth.
Glucose and glutamine are abundant nutrients absolutely required for cell division that feed into multiple pathways required to support cell growth (3). In actively growing cells, most of the glucose is secreted as lactate; the synthesis of lactate generates ATP required to maintain intracellular bioenergetics. The remaining glucose enters TCA cycle where it is metabolized to citrate. Citrate leaves the mitochondria to support the synthesis of fatty acids. High citrate efflux from the mitochondria “empties” the TCA cycle. Glutamine “refills” the TCA cycle by a process termed anapleurosis (4, 5). A functional TCA cycle supports growth by providing precursors for the biosynthesis of nucleotides and certain amino acids. Finally, a significant fraction of glutaminolytic flux results in the production of lactate via malic enzyme. This results in the generation of NADPH, which is required for reductive biosynthetic reactions (5).
We are interested in how cells sense and respond transcriptionally to glucose via the MondoA:Mlx transcriptional activator (6, 7). MondoA and Mlx are members of the basic helix-loop-helix leucine zipper (bHLHZIP) family of transcription factors, which bear superficial similarity to the Myc/Max/Mad family of transcriptional regulators (8). This family plays critical roles in controlling cell growth, differentiation, and death (9), yet how and whether MondoA:Mlx complexes function in these critical processes has not yet been evaluated.
A unique feature of MondoA:Mlx complexes is that they reside at the outer mitochondrial membrane (OMM), yet they shuttle between the OMM and the nucleus, suggesting that they facilitate communication between these organelles (7). In response to elevations in glucose concentration, MondoA:Mlx complexes accumulate in the nucleus where they occupy promoters of target genes and activate their expression. Nuclear accumulation of MondoA:Mlx complexes is also triggered by 2-deoxyglucose and requires the enzymatic activity of hexokinases, suggesting that MondoA:Mlx respond to changes in intracellular levels of glucose-6-phosphate (7).
MondoA:Mlx complexes are broadly expressed and are required for >75% of glucose-induced transcription in an epithelial cancer model, suggesting a predominant role for the complex in how cells respond to glucose (7). One direct glucose-induced transcriptional target of MondoA:Mlx complexes is thioredoxin-interacting protein (TXNIP). TXNIP has pleiotropic roles in cells by controlling redox status via binding and inhibition of thioredoxin (10). TXNIP also plays a more direct role in repressing cell proliferation by enhancing the stability of the cell cycle inhibitor p27 by sequestering components of the COP9 signalosome (11). Finally, although not examined in the context of cell growth, TXNIP may also negatively regulate cell division by restricting glucose uptake (7, 12). Consistent with TXNIP being a MondoA:Mlx effector, overexpression and knockdown of MondoA:Mlx complexes indicate that they are potent negative regulators of glucose uptake, which suggests they may also regulate cell growth (7). Whether utilization of glucose and glutamine is coordinated is unknown. Given the predominant role of MondoA in controlling glucose-dependent transcription and glucose uptake, we examined whether glutamine regulates MondoA activity.
Results
Glucose and Glutamine Are Required for Cell Growth.
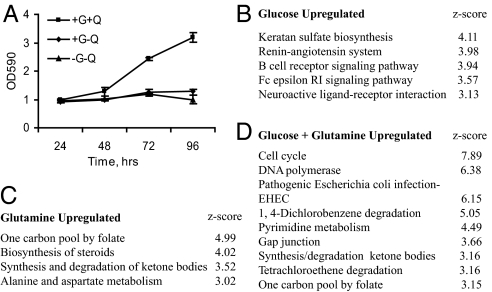
To better understand how glucose and glutamine coordinate cell growth and proliferation, we determined gene expression profiles in the pancreatic cancer cell line, BxPC-3, grown under different nutrient conditions. BxPC-3 only proliferated in the presence of glucose and glutamine, confirming that both nutrients are required for cell division (Fig. 1A). Expression profiles from glucose and glutamine starved cells (-G-Q) were compared with those from cells grown in glucose only (+G-Q), glutamine only (-G+Q), or glucose plus glutamine (+G+Q). This analysis identified 73, 662, and 1014 genes that were up-regulated 2-fold or greater in +G-Q, -G+Q, and +G+Q growth medium, respectively. Pathway analysis revealed several important differences between the three growth conditions (Fig. 1 B–D and Table S1). Consistent with their combined requirement for cell division, neither glucose nor glutamine alone induced pathways important for cell growth or proliferation. Second, there was no significant overlap in the biological pathways induced by glucose or glutamine alone, indicating that they also have independent roles. Third, the combination of glucose and glutamine together induced multiple pathways required for cell division (e.g., cell cycle, DNA polymerase, pyrimidine metabolism, nitrogen metabolism, and folate biosynthesis). Similar pathways were repressed in B lymphoma cells after glutamine deprivation (13). Together these findings suggest that coordinated changes in gene expression driven by the combined action of glucose and glutamine induce critical biosynthetic pathways and cell cycle regulators required for cell division.
Fig. 1.
Glucose and glutamine are required for cell division. (A) BxPC-3 cells were plated in complete media and grown for 24 h. Cells were then grown in medium lacking glucose “G” and/or glutamine “Q” as indicated. Relative cell numbers were determined over a 4-day time period. No growth was observed in media lacking glucose but containing glutamine. The data are the average ± SD of biological triplicates. (B–D) Expression and pathway analysis were used to identify specific biological pathways enriched under the indicated growth conditions. Only pathways with z-scores >3 are shown.
Glutamine Represses TXNIP Expression.
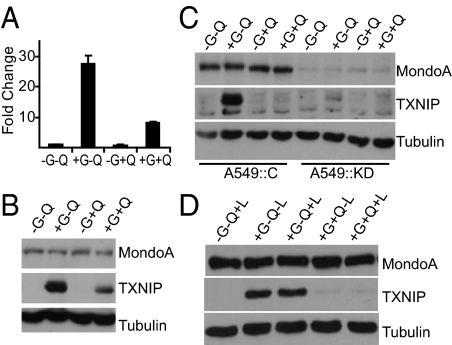
One gene highly induced by glucose alone, but repressed by the addition of glutamine in the array experiment was TXNIP. TXNIP is a potent negative regulator of glucose uptake (7, 12, 14), therefore its repression by glutamine suggests a mechanism that coordinates the response to glucose and glutamine. qRT-PCR analysis revealed that TXNIP was up-regulated by 27.6-fold in +G-Q growth medium. By contrast, it was only induced 7.9-fold in +G+Q growth medium, indicating that glutamine suppressed glucose-induced TXNIP levels (Fig. 2A). The activity of a TXNIP-promoter-luciferase reporter was also induced by glucose, and this induced level was repressed by glutamine (Fig. S1A), suggesting that glutamine represses TXNIP transcription rather than triggering degradation of TXNIP message. TXNIP protein was also dramatically induced by glucose, and this induction was repressed by glutamine (Fig. 2B). TXNIP expression was not detectable in starved cells nor was it induced by glutamine alone. Together, these data indicate that glutamine negatively regulates the glucose-mediated transcriptional induction of TXNIP.
Fig. 2.
Glucose and glutamine regulate TXNIP. (A) qRT-PCR was used to determine the relative expression of TXNIP, normalized to β-actin, in BxPC-3 cells cultured under the indicated growth conditions. Data are presented as fold change relative to expression in medium lacking glucose “G” and glutamine “Q.” Expression of MondoA, TXNIP, and Tubulin in BxPC-3 cells (B and D) and in A549::C (control) and A549::KD (MondoA knockdown) cells (C) cultured under the indicated growth conditions was determined by western blotting. (L) leucine.
MondoA:Mlx complexes are potent glucose-dependent activators of TXNIP expression (7). To investigate whether MondoA also contributes to glutamine dependent-repression of TXNIP, we knocked down MondoA in A549 human lung cancer cells and in HA1ER transformed embryonic human kidney epithelial cells. Similar to BxPC-3 cells, in control A549 and HA1ER cells, TXNIP expression was dramatically up-regulated by glucose in the absence of glutamine, but this induced level was strongly attenuated by glutamine (Fig. 2C and Fig. S1B and C). Glutamine addition to A549 cells reduced TXNIP expression nearly to uninduced levels, whereas the effect was less pronounced, but readily apparent, in HA1ER cells. As expected (7), in A549 and HA1ER MondoA knockdown cells, glucose-dependent induction of TXNIP expression was completely eliminated. This dramatic dependence of TXNIP expression on MondoA suggests that glutamine-dependent repression of TXNIP requires MondoA, but the contribution of other transcription factors cannot be ruled out. Finally, because glutamine inhibited TXNIP expression in multiple cell lines, we suggest that this regulatory mechanism is relatively widespread.
It is possible that elevated MondoA activation of TXNIP is a response to general amino acid depletion, rather than a specific response to glutamine removal. Leucine depletion is typically used to monitor effects of generalized amino acid starvation; leucine signals through the Rag GTPases to activate the growth-promoting TORC1 complex (15). We found no evidence that TXNIP expression was dependent on leucine levels either in the presence or absence of glutamine (Fig. 2D). As such, MondoA transcriptional activation activity does not appear to be generally responsive to amino acid starvation, but appears to be coupled specifically to changes in glutamine levels.
Glutamine Blocks MondoA Transcriptional Activity.
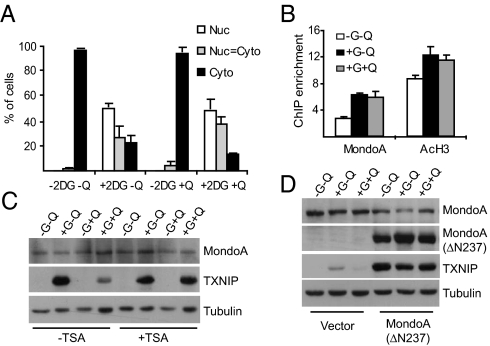
The levels of MondoA mRNA and protein were not significantly reduced by glutamine supplementation (Fig. 2 and data not shown). Therefore, we investigated other mechanisms to explain how glutamine blocks MondoA transcriptional activity. The occupancy of MondoA at the TXNIP promoter is controlled by its nuclear accumulation under high glucose growth conditions (7). However, glutamine inhibited neither the 2-deoxy-glucose-dependent nuclear accumulation of MondoA nor its occupancy of the TXNIP promoter at previously characterized carbohydrate response elements (Fig. 3 A and B). These findings suggest that glutamine blocks the transcriptional activity of promoter-bound MondoA:Mlx complexes, rather than controlling it's nuclear accumulation or DNA binding. Previous studies demonstrated a role for histone deacetylase-containing corepressor complexes at the TXNIP promoter (16). Consistent with these reports, the HDAC inhibitor trichostatin A (TSA) completely blocked glutamine-dependent repression of TXNIP expression (Fig. 3C). Growth of cells in medium containing only glucose led to an increased acetylation of histone H3 at the TXNIP promoter, suggesting that activation of TXNIP requires the recruitment of a histone acetyltransferase. Surprisingly, glutamine addition did not reduce histone H3 acetylation (Fig. 3B), suggesting the putative corepressor functions by modifying the amino-terminal tails of core histones other than H3 or perhaps by modifying other promoter-bound nonhistone substrates. Together, these experiments suggest that glutamine represses TXNIP expression by triggering recruitment of an HDAC-dependent corepressor to the TXNIP promoter.
Fig. 3.
Glutamine modulates the transcriptional activity of MondoA. (A) Subcellular localization of MondoA was determined in A549 cells treated with 2-DG for 3 h in the presence or absence of glutamine “Q.” (B) ChIP was used to determine MondoA occupancy and the acetylation status of Histone 3 (AcH3) at the TXNIP promoter in BxPC-3 cells cultured under the indicated growth conditions. The data shown are average ± SE of three biological replicates. (C) Expression of MondoA, TXNIP, and Tubulin in control and TSA-treated (100 ng/mL, 12 h) BxPC-3 cells cultured under the indicated growth conditions was determined by western blotting. (D) Expression of MondoA, TXNIP, and Tubulin in HA1E cells expressing vector alone or ΔN237NLSMondoA cultured under the indicated growth conditions was determined by western blotting.
To map the glutamine-dependent repression domain in MondoA, we expressed ΔN237NLSMondoA in HA1E cells and determined TXNIP expression under different growth conditions. ΔN237NLSMondoA lacks the N-terminal 237 residues of the ORF, localizes to the nucleus, and is constitutively active (6). ΔN237NLSMondoA drove high TXNIP expression in the presence of glutamine (Fig. 3D), suggesting that glutamine represses TXNIP by stimulating the recruitment of the HDAC-dependent corepressor, directly or indirectly, to MondoA, rather than functioning through MondoA-independent mechanisms. This constitutively active form of MondoA also drove TXNIP transcription in medium lacking glucose, which is consistent with our previous observation that endogenous MondoA accumulates in the nucleus and occupies the TXNIP promoter in response to glucose (7).
Glutamine Stimulates Glucose Uptake.
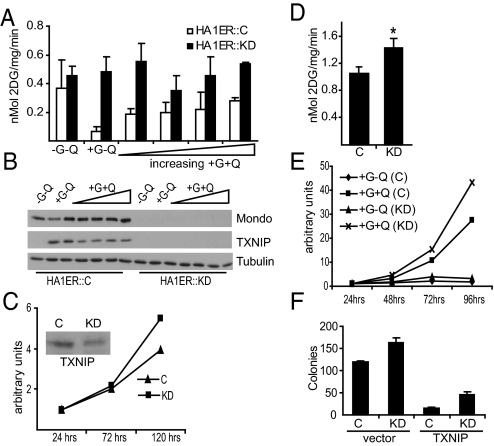
MondoA is a negative regulator of glucose uptake and this is likely via its direct transcriptional regulation of TXNIP (7). Because glutamine repressed TXNIP expression, we determined if glutamine stimulated glucose uptake. HA1ER cells grown in the absence of glucose and glutamine had undetectable TXNIP expression and, as expected, had elevated glucose uptake (Fig. 4 A and B). By contrast, cells grown in medium containing glucose but lacking glutamine showed high TXNIP expression and low glucose uptake. Addition of increasing concentrations of glutamine to the growth medium resulted in a dose-dependent decrease in TXNIP expression and a corresponding increase in glucose uptake. HA1ER cells with MondoA knockdown lacked TNXIP expression and had elevated glucose uptake at all glutamine concentrations tested. Similar results were observed in BxPC-3 cells (Fig. S2A and B). To test whether TXNIP is required downstream of MondoA to block glucose uptake, we reduced its levels in BxPC-3 cells using stable shRNA-mediated knockdown. As expected from previous reports, TXNIP loss increased growth rate and increased glucose uptake (Fig. 4 C and D and Fig. S2C).
Fig. 4.
MondoA-dependent regulation of TXNIP controls glucose uptake and cell proliferation. (A) Glucose uptake was determined in control HA1ER::C and MondoA HA1ER::KD cells cultured in different concentrations of glutamine. (B) Expression of MondoA, TXNIP, and Tubulin in HA1ER::C and HA1ER::KD cells cultured under the indicated growth conditions was determined by western blotting. In panels A and B, glutamine was used at 0.0, 0.1, 0.5, 1.0, 2.0, or 5.0 mM. Growth rate (C) and glucose uptake (D; *, P < 0.01) were measured in control “C” and TXNIP knockdown “KD” BxPC-3 cells. (E) Cell proliferation was performed as described in Fig. 1A in HA1ER::C and HA1ER::KD cells cultured under the indicated growth conditions. (F) Anchorage-independent growth of HA1ER::C and HA1ER::KD cells expressing either mCherry or mCherry-TXNIP. The data shown is a representative of two independent experiments done in triplicate (average ± SD).
MondoA Is a Negative Regulator of Cell Proliferation.
Our data predict that MondoA knockdown should provide cells with a proliferation advantage because of their enhanced glucose uptake. To test this hypothesis, we performed cell proliferation and colony formation assays in control and MondoA knockdown HA1ER cells in the presence or absence of glutamine. Neither cell type proliferated in the absence of glutamine, indicating that increased glucose uptake resulting from MondoA knockdown was not sufficient to overcome the proliferation defect caused by glutamine deprivation (Fig. 4E). By contrast, in medium containing both glucose and glutamine, MondoA knockdown cells had a growth advantage over control cells in both proliferation and in soft agar colony formation assays (Fig. 4 E and F).
Our data suggest that TXNIP is a critical downstream negative growth effector of MondoA in controlling glucose uptake and cell growth. Consistent with this hypothesis TXNIP overexpression reduced the proliferation rate of control and MondoA knockdown cells. TXNIP overexpression also reduced colony number of control and knockdown cells in soft agar assays (Fig. 4F and Fig. S2D and E).
Glutamine Repression of TXNIP Requires Mitochondrial Anapleurosis.
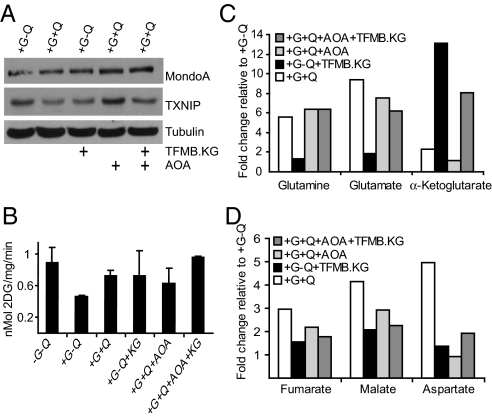
Because MondoA activity is not regulated by amino acid depletion (Fig. 2D), we next examined whether mitochondrial anapleurosis by glutamine might contribute to its repressive effect on TXNIP expression. Glutamine enters the TCA cycle by being converted to glutamate and subsequently to α-ketoglutarate (α-KG). α-KG is a TCA intermediate but cannot enter cells. To test whether glutamine-dependent anapleurosis is required for repression of TXNIP, we treated BxPC-3 cells with the cell permeable 3-trifluoromethylbenzyl (TFMB)-α-KG (17). Once TFMB-α-KG enters the cell, it is cleaved by cytosolic esterases yielding α-KG. We confirmed that glutamine starvation depleted glutamine, glutamate, α-KG, and the levels of several TCA intermediates; addition of glutamine restored levels of these intermediates (Fig. 5 C and D). Consistent with TFMB-α-KG feeding directly into the TCA cycle, its addition dramatically increased the level of α-KG and the level of other TCA intermediates, but did not change the levels of glutamine or glutamate. TFMB-α-KG reduced TXNIP expression to a level comparable to that observed with glutamine, suggesting that mitochondrial anapleurosis contributes to glutamine-dependent repression of TXNIP (Fig. 5A). Furthermore, TFMB-α-KG stimulated glucose uptake consistent with a model where mitochondrial anapleurosis is required for glucose uptake (Fig. 5B).
Fig. 5.
Mitochondrial anapleurosis blocks TXNIP expression. (A) Expression of MondoA, TXNIP, and Tubulin in BxPC-3 cells treated with 2 mM TFMB-αKG “KG” and/or 2 mM AOA for 16 h under the indicated growth conditions was determined by western blotting. (B) Glucose uptake was determined in BxPC-3 cells treated for 16 h as indicated. (C and D) BxPC-3 cells were treated as in A, and the concentrations of indicated metabolites were determined by GC-MS analysis (see SI Methods). The data are an average of quadruplicate samples. Each bar indicates the fold change relative to metabolite levels in cells grown in medium containing glucose but lacking glutamine.
To further substantiate the contribution of mitochondrial anapleurosis to TXNIP-repression, we treated cells with aminooxyacetate (AOA), which inhibits the transaminases that convert glutamate to α-KG (18, 19). Consistent with our working model, AOA reduced intracellular levels of α-KG, blocked glutamine-dependent repression of TXNIP and reduced glucose uptake (Fig. 5 A–C). Each of these AOA-dependent effects were suppressed by the addition of TFMB-α-KG, providing further evidence that TFMB-α-KG functions downstream of glutamate to repress TXNIP. Finally, addition of AOA did not reduce glucose uptake to the same extent as glutamine removal, suggesting that it influences glucose uptake by additional mechanisms.
Discussion
Our work defines a key role for MondoA:Mlx complexes in coupling glutamine-dependent anapleurosis of the TCA cycle with elevated glucose uptake and consequently cell growth. A key MondoA effector in this regulatory circuit is TXNIP. Our work and that of others establishes that repression of TXNIP stimulates glucose uptake and aerobic glycolysis and also blocks fatty acid oxidation (7, 14), which are features of rapidly dividing cells (3). Further, TXNIP reduction likely also drives cell division and survival more directly by triggering the degradation of the cyclin-dependent kinase inhibitor p27 (11) and inhibition of the PTEN tumor suppressor (14). Together our data supports a model where glutamine-dependent repression of TXNIP helps establish metabolic and nonmetabolic programs that support cell growth.
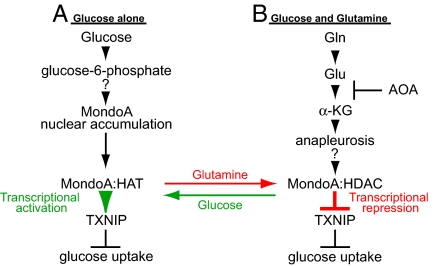
Within mitochondria of rapidly growing cells, most of the glucose-derived citrate is exported to support the synthesis of fatty acids required for cell growth. The resulting reduction in TCA intermediates, which are required for the synthesis of a number of other growth-sustaining biomolecules, is replenished by glutaminolysis (3, 5). Our experiments extend these findings by demonstrating a significant and previously unappreciated coordination between glucose and glutamine utilization pathways. Furthermore, we provide evidence that glutamine-dependent anapleurosis functions upstream of glucose uptake and utilization. Such a mechanism ensures that in the presence of low glutamine levels cell growth is held in check by limiting the availability of intracellular glucose (Fig. 6A). By contrast, elevated glutamine levels support cell growth by stimulating additional glucose uptake and aerobic glycolysis by repressing the glucose-induced and MondoA-dependent activation of TXNIP (Fig. 6B). We suggest that this coordination between glutamine and glucose represents a metabolic checkpoint that restricts cell division until threshold levels of both glutamine and glucose sufficient to support cell growth are available.
Fig. 6.
MondoA is a nutrient-dependent transcription factor. See text for details. As indicated, the mechanisms controlling how MondoA accumulates in the nucleus in response to glucose and how glutamine-dependent mitochondrial anapleurosis converts MondoA to a transcriptional repressor are not known. HAT, histone acetyl-transferase; HDAC, histone deacetylase; α-KG, α-ketoglutarate; AOA, aminooxyacetate; Gln, glutamine; Glu, glutamate.
We suggest that glutamine triggers MondoA-dependent repression of TXNIP. Most compelling, a cell permeable analog of α-KG drives repression of TXNIP in glutamine-free media. Further, glutamine-dependent repression of TXNIP is blocked by AOA, and this blockade is rescued by α-KG. Together these experiments strongly indicate that glutamine metabolism is required for repression of TXNIP. An alternative model is that glutamine deprivation drives TXNIP activation, perhaps through elevations in reactive oxygen species (ROS). We do not favor this model for several reasons. First, TXNIP expression is elevated by TSA in presence of glutamine, demonstrating the involvement of HDAC-dependent corepressors. Second, TXNIP is not induced by leucine deprivation as might be expected if TXNIP was regulated by general amino acid starvation. Finally, modulating ROS levels only has a minor impact on TXNIP expression (Fig. S3 A–C).
How might glutamine-dependent anapleurosis lead to repression of TXNIP? Given the myriad pathways that emanate from mitochondria, many possibilities exist. In glioblastoma cells, more that 50% of glutamine is converted to lactate and alanine, with the concomitant production of NADPH via the malic enzyme. Much of the remaining glutamine contributes to the synthesis of fatty acids and aspartate, which is required for the synthesis nucleotides and other amino acids (5). MondoA activity may be controlled, directly or indirectly, by one of these biosynthetic products. Alternatively, the activities of the prolyl hydroxylase enzymes, which modify the hypoxia-inducible factor family of transcriptional regulators, require α-KG as a cofactor (20). This finding raises the possibility that MondoA senses flux through the TCA cycle more directly.
Cells overexpressing a conditional allele of the Myc are hypersensitive to glutamine deprivation and die when glutamine is removed from the medium (19, 21). This glutamine “addiction” may result because Myc stimulates glutaminolysis by activating, directly or indirectly, the expression of genes involved in glutamine uptake and utilization (19, 22). We suggest that the elevation of glutamine-dependent anapleurosis in Myc overexpressing cells will also result in reduced TXNIP expression and the subsequent increase in glucose uptake and aerobic glycolysis. Consistent with this, it has been known for many years that Myc overexpression can stimulate lactate production (23). Although we have not yet tested this directly, we propose that glutamine removal triggers cell death both by reducing glutaminolysis and restricting glucose uptake by derepressing TXNIP expression. We further note that different Myc-overexpressing cell lines are variably sensitive to glutamine withdrawal (21), whereas other Myc-overexpressing cells are sensitive to glucose withdrawal (23). The mechanisms that account for the differential nutrient requirements in Myc-overexpressing cells are unknown; however, it almost certainly depends on varying rates of glycolysis and glutaminolysis between cell types. Related to this, we do not observe glutamine-dependent repression in all cell types tested. For example, in contrast to our observations above using transformed human cell lines, glutamine does not influence TXNIP levels in immortalized murine embryonic fibroblasts, and murine myoblasts (data not shown). This finding raises the possibility that glutamine may only repress TXNIP expression in highly glycolytic tumor cells, thereby serving to match intracellular glucose availability with glycolytic rate.
Our previous work showed that MondoA:Mlx complexes activate glucose-dependent transcription by accumulating in the nucleus, and occupying the promoters of targets like TXNIP, by sensing intracellular levels of glucose-6-phosphate (7) (Fig. 6A). The mechanisms that control glucose-dependent nuclear accumulation of MondoA are not known, but almost certainly involve modulation of the CRM1 nuclear export factor (24). Furthermore, how MondoA:Mlx activates glucose-dependent transcription remains to be determined; however, our data indicate a requirement for a histone acetyl-transferase (Fig. 3B). Glutamine, via mitochondrial anapleurosis, converts MondoA:Mlx from a transcriptional activator to a transcriptional repressor (Fig. 6B). Rather than controlling MondoA:Mlx function indirectly by modulating subcellular localization or promoter occupancy, glutamine triggers the recruitment of an HDAC-dependent corepressor to promoter-bound MondoA:Mlx complexes (Fig. 6B). Glutamine-dependent repression of MondoA activity is not restricted to TXNIP, as another glucose-activated and MondoA-dependent target, ARRDC4, is also repressed by glutamine. ARRDC4 is a paralog of TXNIP and can suppress growth similarly to TXNIP (Fig. S4A and B). As such, it seems likely that ARRDC4 and TXNIP function redundantly downstream of MondoA to negatively regulate cell growth by restricting glucose uptake. At this time, we cannot rule out the possibility that other MondoA transcriptional targets work in conjunction with TXNIP and ARRCD4 to restrict glucose uptake and cell growth.
Finally, our work provides insight into how one might eradicate highly glycolytic tumor cells by targeting central metabolic pathways. First, it has been argued that inhibiting glutaminolysis might be efficacious in restricting cancer cell growth by limiting the availability of one key nutrient required for cell growth (19, 21). Our work extends this concept by showing that blockade of glutamine metabolism also restricts glucose uptake. As such, the combined inhibition of glycolysis and glutaminolysis may be especially effective at driving apoptosis of highly glycolytic tumor cells. Second, given that flux through the TCA cycle is required to stimulate glucose uptake via MondoA-dependent repression of TXNIP expression, we suggest that small molecule inhibition of TCA enzymes is an attractive approach to sensitize cells to inhibition of glycolysis.
Methods
Cell Culture and Nutrient Depletion.
Cells were maintained at 37 °C in 5% CO2 in medium containing penicillin/streptomycin, glutamine, and 10% standard FBS (HyClone) unless otherwise indicated. BxPC-3 and A549 cells were grown in RPMI and F12K, respectively. HA1E and HA1ER cells (William Hahn, Dana-Farber Cancer Institute) were grown in α-MEM. HA1E are human embryonic kidney epithelial cells expressing hTERT and the SV40 early region. HA1ER cells are HA1E cells with additional transgenic expression of the activated H-RASG12V allele (7). Nutrient depletion studies were performed using glucose and glutamine-free DMEM that contained neither phenol red nor pyruvate or DMEM media that lacked glutamine and leucine. Reconstituted media for all experiments was supplemented with 10% FBS, 2 mM sodium pyruvate, and when needed, glucose or glutamine or leucine was added into the media to the final concentrations, 25, 2, and 1 mM, respectively, or as indicated in the figures.
Cell Proliferation Assays.
Equal number of BxPC-3 or HA1ER cells were plated in complete media and allowed to grow for 24 h. Cells were washed and starved in glucose and glutamine-depleted (-G-Q) media. Twenty-four hours later, fresh media with or without glucose and/or glutamine was provided, and cell proliferation was assessed by crystal violet stain as described previously (25). The data shown are the average ± SD of triplicate samples.
Reagents and Antibodies.
2-Deoxy-D-[3H] glucose was from New England Nuclear. 2-Deoxyglucose and AOA were from Sigma. Trichostatin A was from BioMol. Luciferase reporter assay system was from Promega. Primary antibodies were used at 1:500 for anti-Mondo (7), 1:1,000 for anti-V5 (Sigma), 1:1,000 for anti-VDUP1 (TXNIP) (Medical and Biological Laboratories), and 1:10,000 for anti-Tubulin (Sigma). Secondary antibodies were used at 1:5,000 (Amersham Biosciences and R and D Systems). Western Lightning Chemiluminescence Plus (Perkin-Elmer) was used for detection.
Preparation of 4,5-Dioxo-5-[3-(trifluoromethyl)benzyloxy]pentanoic Acid (TFMB-αKG).
Reagents for the synthesis of TFMB-αKG were from Sigma. A microwave vessel was charged with α-ketoglutaric acid (292 mg, 2.00 mmol), di-isopropylamine (230 μL, 1.64 mmol, 0.8 equiv), and DMF (2 mL). After dissolution, 3-trifluormethylbenzyl bromide (250 μL, 1.64 mmol, 0.8 equiv) was added and the tube sealed and subjected to microwave irradiation (max power, 200 W; temp, 50 °C; time, 15 min). The mixture was diluted with Et2O and washed three times (1:1 10% HCl:brine). The crude organics were dried (Na2SO4). Concentration gave a clear oil that was triturated three times 6:1 Hex:EtOAc to give the ester as a colorless oil (349 mg, 70%), which was used without further purification. 1HNMR (500 MHz, CDCl3): δ 7.65 (s, 1H); 7.60 (app t, J = 8.5 Hz, 2H); 7.51 (t, J = 7.5 Hz, 1H); 5.32 (s, 2H); 3.16 (t, J = 6.0 Hz, 2H); 2.71 (t, J = 6.0 Hz, 2H).LRMS (ESI+) m/z calculated C13H12F3O5 (M+H) 305.1, obsd. 305.1. Also found: m/z 327.0 (M+Na); 343.0 (M+K); 368.1 (M+ACN).
Plasmids and Viruses.
Plasmids expressing MondoA-V5, Mlx-Flag, and the retroviral vectors expressing MondoA shRNA and ΔN237NLSMondoA have been described (6, 7, 24). The TXNIP-luciferase reporter construct was generated by amplifying a 1518-bp promoter fragment, containing the previously described carbohydrate response element (26), from genomic DNA. This fragment was then cloned into pGL3 basic vector (Promega). mCherry was subcloned into the BamHI and NotI sites of pCDH-EF1-MCS1-puro (System Biosciences) to create Cherry-CDH. Mouse TXNIP was subcloned from TXNIP-V5-His (27) into the XbaI and EcoRI sites of Cherry-CDH to create TXNIP-Cherry-CDH. Lentiviral production was essentially as described previously (27, 28), with modification: Transgene lentiviral vectors were cotransfected into 293T cells with the packaging plasmids psPAX2 and pMD2.G (AddGene) in a 2:1:1 ratio. Supernatant containing pseudoviral particles were harvested at 48 h, 0.45-μm filtered, aliquoted, and stored at −80 °C.
Transient Transfections.
Luciferase reporter assays, immunofluorescence, and microscopy were conducted in HA1ER and A549 cells as previously described (7, 29).
ChIP and Expression Analysis.
BxPC-3 cells were cultured for 24 h in the presence or absence of glucose (25 mM) and/or glutamine (2 mM). ChIP experiments were conducted as described earlier. For expression analysis, total RNA was extracted from cells using RNeasy Mini kit (Qiagen), and cDNA was generated from 1 μg RNA using SuperScript III RT system (Invitrogen) (7).
Quantitative PCR (qPCR).
qPCR analysis was performed as described previously (7). Measurements represent the average of two biological replicates for the RNA analysis and an average of three biological replicates for ChIP experiments. Primer sequences are available on request.
Microarray Analysis.
BxPC-3 cells were plated in quadruplicate and starved overnight in glucose-free and glutamine-free media. For glucose and/or glutamine induction, cells were incubated for 6 h in media containing glucose (25 mM) or glutamine (2 mM) or glucose plus glutamine (25 and 2 mM). Total RNA was isolated, microarray hybridization and data analysis were described previously (7). Microarray (Agilent) hybridization was conducted on BxPC-3 samples harvested from -G-Q, +G-Q, -G+Q, and +G+Q growth conditions. Comparative gene expression profiling was done using Genesifter (Geospiza), and genes that were up-regulated by 2-fold or greater were identified. Specific biological enriched pathways were determined by querying the Kyoto Encyclopedia of Genes and Genome database.
Glucose Uptake Assays.
BxPC-3 and HA1ER cells were seeded in six-well dishes and grown overnight to ≈70% confluence. After a PBS wash, cells were starved for 6 h in glucose- and glutamine-deprived media and then supplemented with fresh media containing 25 mM glucose with the indicated amount of glutamine or 2 mM TFMB-α-KG. After 16 h, 2-deoxy-D-[3H] glucose uptake assays were performed as described previously (7).
Anchorage-Independent Growth.
Growth of cells in soft agar was performed as described previously (25). Briefly, HA1ER::C and HA1ER::KD cells were infected with mCherry, mCherry-TXNIP, or mCherry-ARRDC4 lentivirus, and 24 h later, the cells were plated in soft agar.
Supplementary Material
Acknowledgments.
We thank W. Chutkow and P. Patwari for providing the lentiviruses expressing mCherry-TXNIP and mCherry-ARRDC4; L. Owen for advice on soft agar colony assays; and E. Leibold, D. Tantin, and the Ayer lab for reviewing the manuscript. This work was supported by National Institutes of Health Grants GM55668 and GM60387 (to D.E.A.) and the Huntsman Cancer Foundation. DNA sequencing and oligonucleotide synthesis were supported by the Cancer Center Support Grant 2P30 CA42014.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901221106/DCSupplemental.
References
- 1.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Corless M, Kiely A, McClenaghan NH, Flatt PR, Newsholme P. Glutamine regulates expression of key transcription factor, signal transduction, metabolic gene, and protein expression in a clonal pancreatic beta-cell line. J Endocrinol. 2006;190:719–727. doi: 10.1677/joe.1.06892. [DOI] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: Mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoltzman CA, et al. Glucose sensing by MondoA:Mlx complexes: A role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci USA. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billin AN, Ayer DE. The Mlx network: Evidence for a parallel Max-like transcriptional network that regulates energy metabolism. In: Eisenman RN, editor. The Myc/Max/Mad Transcription Factor Network. Germany: Springer, Heidelberg; 2006. pp. 255–278. [DOI] [PubMed] [Google Scholar]
- 9.Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Chung JW, Jeon JH, Yoon SR, Choi I. Vitamin D3 upregulated protein 1 (VDUP1) is a regulator for redox signaling and stress-mediated diseases. J Dermatol. 2006;33:662–669. doi: 10.1111/j.1346-8138.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- 11.Jeon JH, et al. Tumor suppressor VDUP1 increases p27(kip1) stability by inhibiting JAB1. Cancer Res. 2005;65:4485–4489. doi: 10.1158/0008-5472.CAN-04-2271. [DOI] [PubMed] [Google Scholar]
- 12.Parikh H, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng T, Golub TR, Sabatini DM. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hui ST, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA. 2008;105:3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler LM, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci USA. 2002;99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKenzie ED, et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27:3282–3289. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. J Biol Chem. 1984;259:6215–6221. [PubMed] [Google Scholar]
- 19.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 21.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci USA. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eilers AL, Sundwall E, Lin M, Sullivan AA, Ayer DE. A novel heterodimerization domain, CRM1, and 14–3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol Cell Biol. 2002;22:8514–8526. doi: 10.1128/MCB.22.24.8514-8526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith R, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 27.Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. The interaction of thioredoxin with TXNIP. Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chutkow WA, Patwari P, Yoshioka J, Lee RT. Thioredoxin-interacting protein (TXNIP) is a critical regulator of hepatic glucose production. J Biol Chem. 2008;283:2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- 29.Pickett CL, Breen KT, Ayer DE. A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev Biol. 2007;310:226–239. doi: 10.1016/j.ydbio.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.