Abstract
Various studies have detailed the role of E2F proteins in both transcription activation and repression. Further work has shown that distinct promoter elements, but comprising the same E2F recognition motif, confer positive or negative E2F control and that this reflects binding of either activator or repressor E2F proteins respectively. We now show that the specificity of binding of an activator or repressor E2F protein is determined by adjacent sequences that bind a cooperating transcription factor. We propose that the functional E2F element is a module comprising not only the E2F binding site but also the adjacent site for the cooperating transcription factor.
Keywords: E2F, transcription, gene regulation
Introduction
The control of normal cell growth and cell fate requires a coordination of cell signaling events leading to regulation of transcription of key genes affecting the phenotypes. One of the signaling pathways that play a central role in the regulation of cellular proliferation, differentiation, development, as well as apoptosis is the retinoblastoma RB/E2F pathway (DeGregori and Johnson, 2006; Dyson, 1998; Nevins, 1998). Indeed, E2Fs are critical for the control of genes associated with DNA replication and mitosis and the control of these transcription factors is frequently disrupted in cancer cells (Dyson, 1998; Ishida et al., 2001b; Nevins, 1998; Polager et al., 2002; Ren et al., 2002; Sears and Nevins, 2002; Sherr, 1996; Zhu et al., 2004). The E2F family of transcription factors consist of eight members (E2F1−8) (DeGregori and Johnson, 2006).
Previous studies suggest that the individual members of the E2F family play independent roles, including evidence that the E2F genes exhibit distinct expression profiles during the cell cycle, individual members of the E2F family associate with different members of the Rb family, the E2F1, E2F2, and E2F3a proteins function primarily as transcriptional activators, whereas the E2F3b, E2F4, E2F5, E2F6, E2F7, and E2F8 proteins function primarily as transcriptional repressors, and the abilities of the individual members of the E2F family to mediate proliferation, to regulate differentiation and development, and to induce apoptosis differ (Christensen et al., 2005; de Bruin et al., 2003; DeGregori and Johnson, 2006; Di Stefano et al., 2003; Logan et al., 2005; Sears and Nevins, 2002; Trimarchi et al., 2001). For example, previous studies have shown that E2F1 and E2F3 are both required for cell cycle entry whereas E2F3 alone plays an important role in successive cell cycle entries in asynchronously growing cells (Kong et al., 2007). Consistent with these distinct roles, genes whose regulation is affected by knockdown of E2F1 versus E2F3 differ markedly (Kong et al., 2007). In addition, E2F1 possesses a unique ability to induce apoptosis (DeGregori et al., 1997; Hallstrom et al., 2007; Hallstrom and Nevins, 2003; Hallstrom and Nevins, 2006; Kowalik et al., 1998; Kowalik et al., 1995; Vigo et al., 1999; Ziebold et al., 2001). These distinct signaling events mediated by E2F1 are reflected in the activation of distinct sets of genes (Hallstrom et al., 2007). These observations point to the critical role of specificity of transcriptional control by the E2F proteins and raise the important question of the mechanism by which related but distinct family members differentially control transcription of target genes. Of course, the E2Fs are but an example of the broader question of transcriptional control specificity and in particular, the mechanisms by which transcription factors identify functional binding sites within the human genome.
One possible mechanism that could be utilized to achieve transcriptional specificity within the E2F family of transcription factors is the recognition of slightly different nucleotide sequences by E2F family members. While there is one report suggesting the E2F proteins might recognize distinct DNA sequences in promoters (Tao et al., 1997), a previous study of the crystal structure of an E2F4-DP2 complex bound to the adenovirus E2 promoter has revealed that the amino acids of the E2F and DP proteins that contact the E2F consensus DNA-binding site are conserved among all of the E2F and DP family members (Zheng et al., 1999). Moreover, in vitro site selection experiments and in vivo chromatin immunoprecipitation (ChIP) experiments have also shown that different E2F-DP complexes recognize the same E2F binding sequences (Takahashi et al., 2000; Trimarchi and Lees, 2002; Wells et al., 2000). It is therefore unlikely that the DNA sequence recognized by E2F is solely responsible for distinguishing E2F target genes.
An alternative basis for promoter specificity is the concept of combinatorial transcription control in which pairs of transcription factors function in combination with one another to synergistically activate the given promoters (Stanojevic et al., 1991; Yamamoto et al., 1998). In this scenario, the specificity is determined not just by a single DNA binding element but rather the combined sequence and context of the elements. Our previous work has suggested a mechanism for transcriptional specificity among the E2F family of transcription factors that involves distinct protein-protein interactions to regulate cellular target genes. This includes roles for the YY1 and TFE3 transcription factors in interactions with activator E2Fs to form specific promoter complexes important for expression of genes at G1/S (Giangrande et al., 2004; Giangrande et al., 2003; Schlisio et al., 2002). Combinatorial gene control may not only be an important mechanism through which specific members of the E2F family activate particular E2F target genes, but also may be an important mechanism through which specific members of the E2F family repress particular E2F target genes. As an example, previous work has shown, that the repressor E2F4 and the protein that binds to the CHR element play a role in combinatorial gene control of the Cdc2 gene (Zhu et al., 2004).
We have now focused on the mechanisms underlying the specificity of E2F transcription activation versus repression by directly demonstrating that the specificity of binding of an activator or repressor E2F protein is dictated by adjacent promoter sequences that bind cooperating transcription factors.
Results
Distinct E2F binding elements determine positive and negative E2F transcription control
Previous work has shown that specific members of the E2F family mediate either transcriptional activation or transcriptional repression (Trimarchi and Lees, 2002). For instance, the E2F1−3 proteins are identified as activators whereas the E2F4 and E2F5 proteins mediate transcription repression. Importantly, ChIP assays have shown that these distinct classes of E2Fs also bind to distinct promoter sequences (Araki et al., 2003; Schlisio et al., 2002; Zhu et al., 2004). As an example, only the activator E2Fs (E2F2 and E2F3) bind to the positive-acting −1 E2F site within the Cdc6 promoter (Schlisio et al., 2002) (Figure 1A). In addition, activator E2Fs (E2F1, E2F2, and E2F3) have been shown to bind to the positive-acting distal E2F site within the Cdc2 promoter, whereas the repressor E2F (E2F4) binds to the negative-acting proximal E2F site within the Cdc2 promoter (Zhu et al., 2004) (Figure 1A). Although it is clear from these studies that there are distinct interactions between specific members of the E2F family of transcription factors and particular E2F binding sites within E2F target gene promoters, the mechanism underlying such transcriptional specificity is not understood.
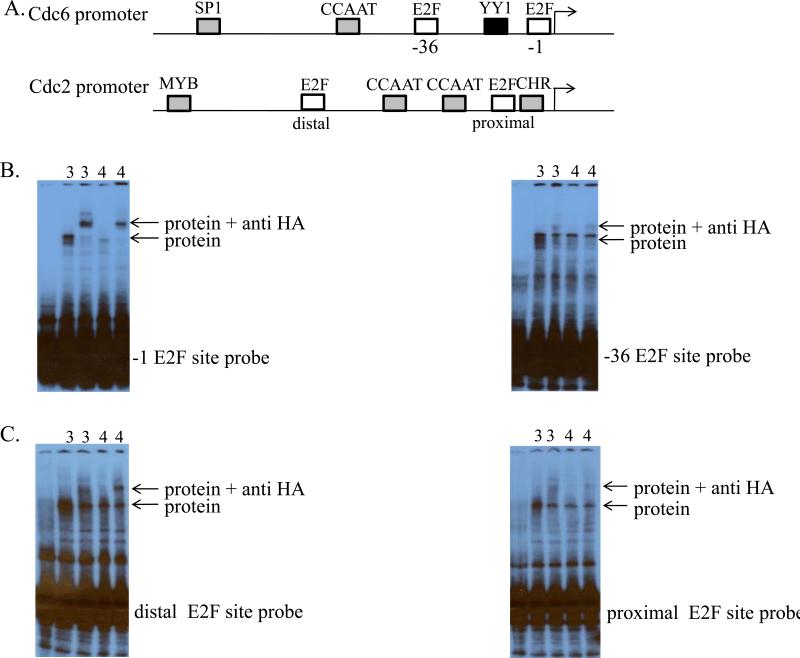
Figure 1. Binding of E2F proteins to E2F promoter elements.
A. Schematic of the human Cdc6 and Cdc2 promoters. Arrows depict the transcription start sites. Transcription factor binding sites relevant to this study are as indicated (Schlisio et al., 2002; Tommasi and Pfeifer, 1995; Yan et al., 1998; Zhu et al., 2004). B. C. Binding of E2F3 and E2F4 to E2F sites within the Cdc6 promoter and Cdc2 promoter, respectively. EMSAs used to detect binding of in vitro transcribed and translated HA-E2F3 and HA-E2F4 proteins to a Cdc6 promoter fragment containing the −1 E2F site, to a Cdc6 promoter fragment containing the −36 E2F site, to a Cdc2 promoter fragment containing a wild type distal E2F site and a mutated proximal E2F site, or to a Cdc2 promoter fragment containing a wild type proximal E2F site and a mutated distal E2F site, as indicated next to each gel. Lane 1 depicts the control reaction containing only the radiolabeled DNA probe. Lanes 2 and 4 depict binding reactions using 4 μg of in vitro transcribed and translated HA-E2F3 or HA-E2F4 protein, respectively. Lanes 3 and 5 depict antibody supershift assays using 0.2 μg of a polyclonal antibody against HA.
One possible mechanism by which specific E2Fs could distinguish between particular E2F binding sites could involve the recognition of distinctions among the nucleotide sequences of the E2F binding sites within the promoters. To address this directly, we employed electrophoretic mobility shift assays (EMSAs) to measure the binding of activator E2Fs (e.g. E2F3) versus repressor E2Fs (e.g. E2F4) to positive- and negative-acting E2F sites within the Cdc6 and Cdc2 promoters. As shown in Figure 1B, both E2F3 and E2F4 bind to the positive-acting (−1 E2F) site as well as the negative-acting (−36 E2F) site within the Cdc6 promoter. Similarly, as shown in Figure 1C, both E2F3 and E2F4 bind to the positive-acting (distal) E2F site as well as the negative-acting (proximal) E2F site within the Cdc2 promoter. These data show that members of the E2F family of transcription factors are capable of binding to each of the E2F binding sites and that the subtle distinctions in the nucleotide sequences of E2F binding sites do not appear to play a role in distinguishing E2F binding sites within promoters.
Specificity of promoter interaction by an E2F activator or repressor is determined by promoter context
An alternate mechanism by which specific E2Fs could distinguish between particular E2F binding sites could involve the interaction with transcription factors that are binding partners of specific E2Fs. Previous ChIP assays demonstrating a role for such interactions in allowing the formation of functional complexes in vivo suggest that binding partners of specific E2Fs could direct specific activator or repressor E2Fs to particular E2F binding sites. For instance, E2F3 was shown to interact specifically with the TFE3 transcription factor to allow the formation of a functional promoter complex with the DNA polymerase α p68 promoter (Giangrande et al., 2003). To directly address the contribution of cooperating transcription factors in directing specific E2Fs to particular E2F sites within the promoters of E2F target genes, we used ChIP assays with transfected plasmid-borne promoter constructs. Specifically, these assays were done to assess whether changing the partner element co-occurring with an E2F binding site in a given promoter could result in changing the specificity of the individual member of the E2F family that interacts with the particular E2F binding site in the given promoter.
Previous work has demonstrated that activator E2Fs (E2F2 and E2F3) bind to the positive-acting −1 E2F site in the Cdc6 promoter (Schlisio et al., 2002). As shown by the data in Figure 2A, we demonstrate that E2F4, but not E2F3, binds to the negative-acting −36 E2F site within the Cdc6 promoter. This promoter thus provides a model for studying interactions between specific members of the E2F family and particular E2F sites within a given promoter. We have utilized ChIP assays to examine the interactions of E2Fs on transfected plasmid-borne Cdc6 promoter constructs to determine if the transcription factors that participate in combinatorial gene regulation with E2Fs influence which specific members of the E2F family bind to particular E2F sites within the promoter. This type of ChIP assay has been used previously by our laboratory to evaluate the effect of promoter mutations on binding of transcription factors to promoters in vivo (Giangrande et al., 2003; Zhu et al., 2004). T98G cells were transfected with plasmid-borne Cdc6 promoter constructs, as described below, and the cells were harvested for ChIP at a time point representing the G1/S phase of the cell cycle.
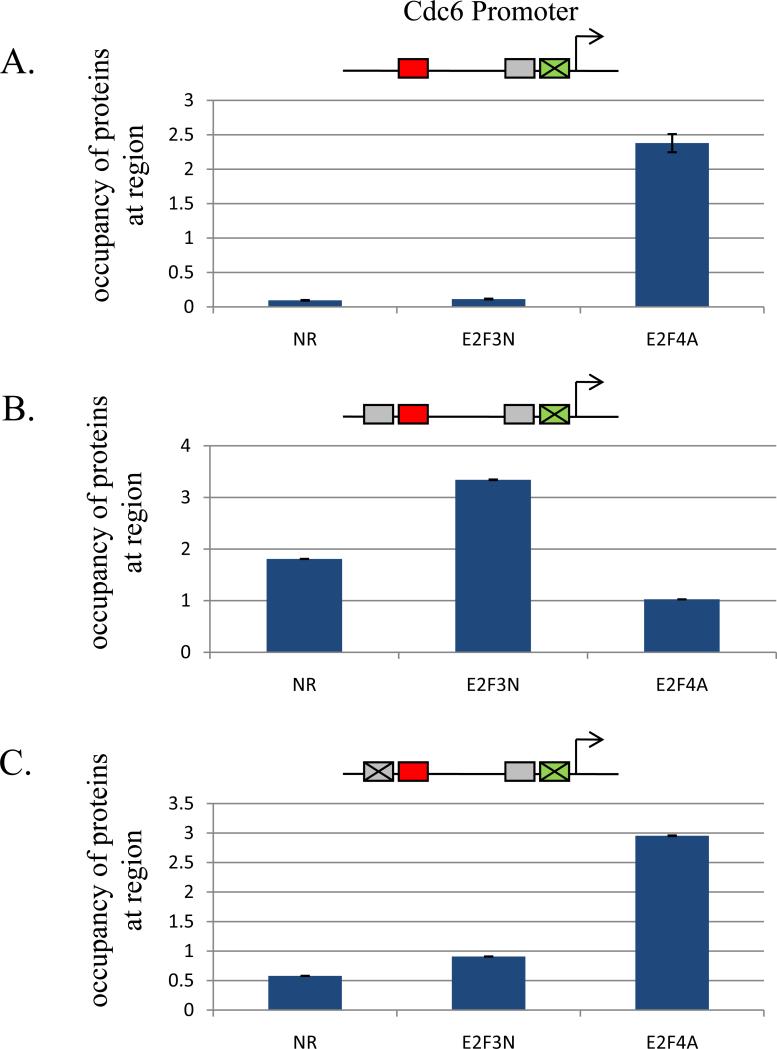
Figure 2. Role of promoter context in determining specificity of interaction of E2F proteins with the Cdc6 promoter.
T98G cells were co-transfected with mutant promoter-reporter constructs and an internal control plasmid containing the β-galactosidase gene. Cells were harvested 18 hours after HU block and reporter ChIP experiments were done as described in Materials and Methods. Promoter constructs assayed are indicated above each graph. Positive-acting and negative-acting E2F sites are depicted by green and red boxes, respectively. YY1 sites are depicted by grey boxes. Antibodies used in the reporter ChIP experiments are as indicated, NR=normal rabbit, E2F3N=anti-E2F3, E2F4A=anti-E2F4. A. Interaction of E2F4 with the −36 E2F site within the Cdc6 promoter during G1/S phase of the cell cycle. B. Interaction of E2F3a with the −36 E2F site within the Cdc6 promoter in the presence of an YY1 element adjacent to the −36 E2F site during G1/S phase of the cell cycle. C. Interaction of E2F4 with the −36 E2F site within the Cdc6 promoter in the presence of a mutated YY1 element adjacent to the −36 E2F site during G1/S phase of the cell cycle.
As seen in Figure 2B, insertion of a YY1 binding site adjacent to the −36 E2F site within the Cdc6 promoter does influence the member of the E2F family that interacts with the −36 E2F site during the G1/S phase of the cell cycle, with E2F3a now preferentially interacting with the −36 E2F site within the Cdc6 promoter instead of E2F4. As shown in Figure 2C, mutation of the inserted YY1 binding site adjacent to the −36 E2F site within the Cdc6 promoter confirmed that in the absence of an YY1 binding site E2F4 preferentially interacts with the −36 E2F site within the Cdc6 promoter during the G1/S phase of the cell cycle. These data suggest that transcription factors that participate in combinatorial gene regulation with E2Fs direct specific E2F family members to particular E2F sites within E2F target genes.
As a second example of E2F specificity, we have focused on the control of the G2/M regulated Cdc2 gene where previous work has shown distinct binding of activator E2Fs to a positive-acting distal E2F site and binding of a repressor E2F to a negative-acting proximal E2F element (Zhu et al., 2004). We have again utilized ChIP assays to examine the interactions of E2Fs on transfected plasmid-borne promoter constructs, in this case utilizing the Cdc2 promoter.
As seen in Figure 3A, and as previously shown (Zhu et al., 2004), E2F4 interacts with the negative-acting proximal E2F site within the Cdc2 promoter during the G1/S phase of the cell cycle. As seen in Figure 3B, insertion of an YY1 binding site adjacent to the proximal E2F site within the Cdc2 promoter does influence the member of the E2F family that interacts with the proximal E2F site during the G1/S phase of the cell cycle, with E2F3a now preferentially interacting with the proximal E2F site within the Cdc2 promoter instead of E2F4. As shown in Figure 3C, mutation of the inserted YY1 binding site adjacent to the proximal E2F site within the Cdc2 promoter confirmed that in the absence of an YY1 binding site E2F4 preferentially interacts with the proximal E2F site within the Cdc2 promoter during the G1/S phase of the cell cycle.
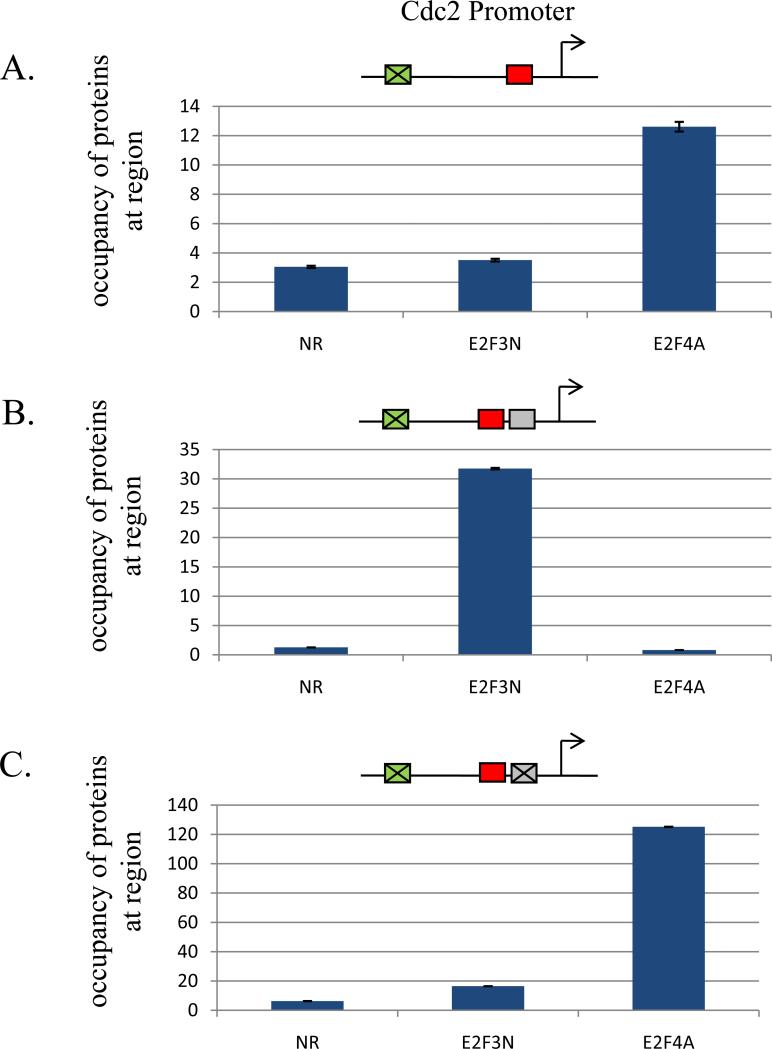
Figure 3. Role of promoter context in determining specificity of interaction of E2F proteins with the Cdc2 promoter.
T98G cells were co-transfected with mutant promoter-reporter constructs and an internal control plasmid containing the β-galactosidase gene. Cells were harvested 18 hours after HU block and reporter ChIP experiments were done as described in Materials and Methods. Promoter constructs assayed are indicated above each graph. Positive-acting and negative-acting E2F sites are depicted by green and red boxes, respectively. YY1 sites are depicted by grey boxes. Antibodies used in the reporter ChIP experiments are as indicated, NR=normal rabbit, E2F3N=anti-E2F3, E2F4A=anti-E2F4. A. Interaction of E2F4 with the proximal E2F site within the Cdc2 promoter during G1/S phase of the cell cycle.
B. Interaction of E2F3a with the proximal E2F site within the Cdc2 promoter in the presence of an YY1 element adjacent to the proximal E2F site during G1/S phase of the cell cycle.
C. Interaction of E2F4 with the proximal E2F site within the Cdc2 promoter in the presence of a mutated YY1 element adjacent to the proximal E2F site during G1/S phase of the cell cycle.
Taken together, these data provide direct evidence that the specificity for interaction of a particular E2F with a promoter element is dictated by the promoter context involving adjacent transcription factor binding elements.
Extending the concept of combinatorial E2F control
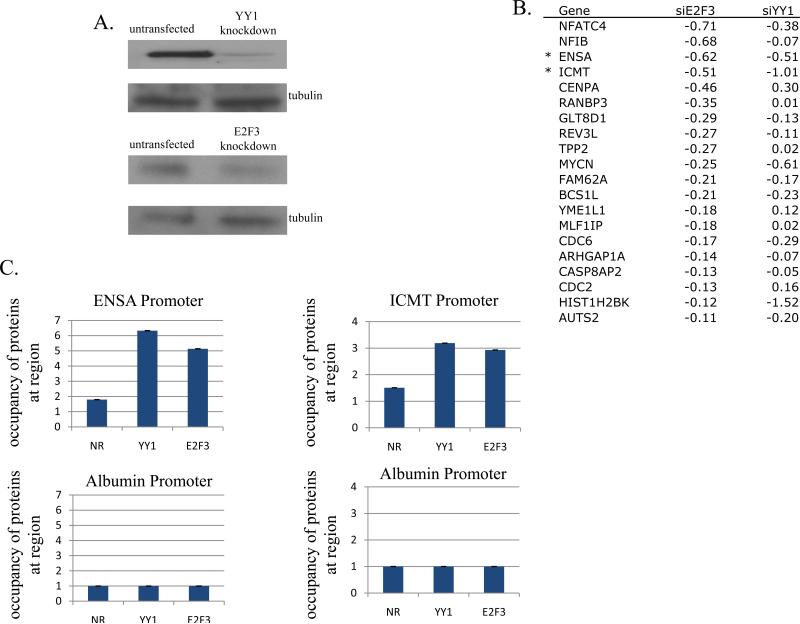
Given the role of combinatorial interactions in defining the specificity of E2F function, we would predict that sets of genes within the human genome exist which have promoters containing co-occurring binding sites for E2F and the binding partner that determines the specificity of the E2F activator or repressor that binds to the E2F element. We have sought to address this in the context of the role of YY1 as a partner for the E2F3 activator. To do so, we examined the effect of knockdown of either E2F3 or YY1 on the expression level of the predicted target genes. As shown in Figure 4A, siRNA duplexes targeting E2F3 and YY1 were effective in reducing E2F3 and YY1 protein levels, respectively. An analysis of the genes predicted to be regulated jointly by E2F3 and YY1 revealed a strong association with replication and/or cell cycle functions, based on reduced expression observed in cells transfected with both E2F3 and YY1 siRNAs (Figure 4B).
Figure 4. Effect of E2F3 and YY1 knockdown on global gene expression.
A. Western blot analysis using cell extracts from asynchronously growing HEK293 cells with untransfected as a control, or transfected with effective YY1 RNAi duplexes, or transfected with effective E2F3 RNAi duplexes. Protein levels were detected by Western blotting using antibodies against YY1 or E2F3, respectively. Levels of tubulin protein were detected by Western blotting using an antibody against α-tubulin as a control.
B. Effect of E2F3 and YY1 knockdown on gene expression. Microarray analysis was done to identify genes exhibiting changes in regulation in response to transfection of siRNAs targeting YY1 or E2F3. Fold changes in expression of the twenty genes most effected in E2F3 knockdowns relative to untransfected cells are given (fold changes in expression of the same genes in YY1 knockdown cells relative to untransfected cells are also given). Asterisks indicate genes examined in Figure 4C.
C. Asynchronously growing HEK293 cells were harvested and endogenous ChIP experiments were done as described in Materials and Methods. Promoters assayed and antibodies used in the endogenous ChIP experiments are as indicated, NR=normal rabbit.
To verify that genes predicted to be jointly regulated by the combined action of E2F3 and YY1 were indeed direct targets, we made use of ChIP assays to measure the interaction of E2F3 and YY1 with the promoters of genes most affected by the joint knockdown (ENSA and ICMT). As shown in Figure 4C, ChIP analyses reveal that E2F3 and YY1 interact with the ENSA and ICMT promoters. As a control, no binding of either E2F3 or YY1 was observed with the albumin promoter. Taken together, the results demonstrate a broad array of genes whose expression is dependent on functional E2F3 and YY1 coinciding with the presence of E2F and YY1 elements in the promoters and joint binding of the two proteins to these elements. This supports the concept of selective activation of E2F target genes via combinatorial gene control across the human genome.
Discussion
Although the recognition of specific DNA sequences is critical to the specificity of action of transcription factors, a number of observations suggest that this is not sufficient. In particular, most transcription factors recognize DNA sequences of five to eight base pairs in length, which is not sufficient information to identify unique promoter sequences within a three billion base pair genome. Various studies point to a role for protein-protein interactions as one mechanism to increase the specificity of promoter recognition (Pilpel et al., 2001; Smale, 2001). This mechanism of combinatorial gene regulation involving cooperative binding of two transcription factors results in the potential to increase the complexity of the DNA recognition, and thus add specificity of promoter selection (Yamamoto et al., 1998). One of the first such examples of combinatorial gene regulation can be seen with respect to the control of herpes simplex virus transcription in which an interaction of the viral VP16 protein with the Oct-1 and HCF-1 cellular transcription factors creates a complex that recognizes a unique nine base pair DNA sequence (TAATGARAT) found in herpes virus immediate early promoters (Wysocka and Herr, 2003). In essence, the functional transcription factor is the complex of these proteins, recognizing not just the individual elements, but the combined elements in the form of a transcription module. Importantly, this concept provides a further opportunity for combinatorial action in which the elements of the complex can interact with other partners to achieve additional specificities of transcription control.
The evolution of the E2F family can be seen as an example of complexity of transcriptional control in which individual members of the E2F family participate in the activation of distinct sets of genes with distinct functional outputs (Muller et al., 2001). This includes both positive and negative control of these genes by distinct E2F family members (Schlisio et al., 2002; Zhu et al., 2004). As an example, E2F1 and E2F3 are critical for activation of genes at the G1/S phase of the cell cycle to generate DNA replication activities, but then also at the G2/M phase of the cell cycle to produce the mitotic gene functions (Dyson, 1998; Ishida et al., 2001a; Nevins, 1998; Polager et al., 2002; Ren et al., 2002; Zhu et al., 2004). In addition, while E2F1 activates a collection of genes important for cellular proliferation, E2F1 also signals an apoptotic program (Bates et al., 1998; Croxton et al., 2002; DeGregori et al., 1997; Hallstrom and Nevins, 2006; Irwin et al., 2000; Lissy et al., 2000; Moroni et al., 2001; Nahle et al., 2002; Rogoff et al., 2004; Stiewe and Putzer, 2000). In each case, distinct sets of genes are activated by the individual E2Fs under different circumstances. The Cdc6 gene is an example of an E2F target gene regulated at G1/S and critical for initiation of DNA replication (Yan et al., 1998). Our previous work has described a model for control of Cdc6 in which RYBP acts as a bridging protein within a ternary complex, involving E2F2/3, RYBP, and YY1, which is required for activation of the promoter (Schlisio et al., 2002). Thus, YY1 is one of the distinct partner proteins that play a role in imparting specificity of transcription control within the E2F family.
In most instances, the genes positively regulated by the activator E2Fs are also subject to negative control by the repressor E2F proteins. In particular, the negative control is an active process whereby distinct E2F family members actively repress the transcription of target genes. Previous work has suggested that distinct promoter elements are involved in the positive or negative control. For example, as shown previously, the Cdc2 promoter contains two E2F sites, a distal E2F site, which is involved in positively controlling the promoter and to which the activator E2Fs (E2F1−3) bind, and a proximal E2F site, which is involved in negatively controlling the promoter and to which the repressor E2F (E2F4) binds (Zhu et al., 2004).
The work we present here now demonstrates that these distinctions are dictated not by the E2F elements, but rather by the context of the E2F elements, with the presence of elements for interacting proteins determining which form of an E2F forms the complex and thus the consequence of the interaction. Thus, at least part of the specificity of transcription control within the E2F family appears to reflect a mechanism similar to that described for VP16, with interaction with distinct partner proteins imparting specificity of transcription control. Such a mechanism for specificity of E2F function implies that protein-protein interactions direct particular E2Fs to specific subsets of E2F target gene promoters allowing particular E2Fs to regulate different target genes, thus resulting in distinct functional roles for E2Fs. In addition, this mechanism for specificity of E2F function also implies that proteins that participate in combinatorial gene regulation with E2Fs appropriately direct activator or repressor E2Fs to particular E2F sites involved in positive or negative control of E2F target genes, respectively.
Taken together, we believe this study provides strong evidence for the role of transcription factor interactions in the determination of specificity of function, particularly with respect to the role of individual E2F family members to impart positive or negative control. When combined with previous work that points to a role for such interactions in determining the specificity of individual E2F family members in the activation of distinct sets of genes, we suggest that this represents a more general mechanism for providing the specificity of transcription factor function.
Materials and Methods
Cell culture
Human embryonic kidney cells (HEK293 cells) and human ganglioblastoma cells (T98G cells) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS).
Plasmid constructions
To generate the Cdc6 promoter construct containing an inserted YY1 binding site adjacent to the −36 E2F binding site, site-directed mutagenesis was performed using the QuikChange Site-Directed Mutagenesis Kit from Stratagene. The Cdc6 promoter construct containing a mutated −1 E2F site (Yan et al., 1998) was mutagenized using the following mutagenic primers 5′TCAGAATCGAGGCCGGGCCATTCGGCTTTGGCGGGAGG3′ and 5′AGTCTTAGCTCCGGCCCGGTAAGCCGAAACCGCCCTCC3′. To generate the Cdc6 promoter construct containing an inserted mutated YY1 binding site adjacent to the −36 E2F binding site, the aforementioned plasmid was mutagenized using the following mutagenic primers 5′TCAGAATCGAGGCCGGGCGGTTCGGCTTTGGCGGGAGG3′ and 5′AGTCTTAGCTCCGGCCCGCCAAGCCGAAACCGCCCTCC3′.
To generate the Cdc2 promoter construct containing an inserted YY1 binding site adjacent to the proximal E2F site, the hcdc2-dE2Fm promoter construct (Zhu et al., 2004) was mutagenized using the following mutagenic primers 5′GCCCTTTAGCGCGGTGAGGCCATTCCTGCTCGCACTTGGC3′ and 5′GCCAAGTGCGAGCAGGAATGGCCTCACCGCGCTAAAGGGC3′. To generate the Cdc2 promoter construct containing an inserted mutated YY1 binding site adjacent to the proximal E2F site, the aforementioned plasmid was mutagenized using the following mutagenic primers 5′GCCCTTTAGCGCGGTGAGGCGGTTCCTGCTCGCACTTGGC3′ and 5′GCCAAGTGCGAGCAGGAACCGCCTCACCGCGCTAAAGGGC3′.
All constructs were confirmed by DNA sequencing.
Electrophoretic mobility shift assays
HA-E2F3 and HA-E2F4 Schlisio et al. (2002) were in vitro transcribed and translated using the TNT Quick Coupled Transcription/Translation System from Promega. DNA probes were generated by PCR amplification. For the Cdc6 promoter, the template used was the minimal Cdc6 promoter region (Schlisio et al., 2002) and the primers used were as follows: to generate the −1 E2F site probe 5′CCGGAATTCGGTGGGAACGCTGTGGCC3′ and 5′CCCAAGCTTACAGCGGCAGCAGCAAAC3′ and to generate the −36 E2F site probe 5′CCGGAATTCGTGACTACAGCCAATCAG3′ and 5′CCCAAGCTTCGAATGGCCACAGCGTTC3′. For the Cdc2 promoter, to generate the distal site probe the template used was hcdc2-pE2Fm (Zhu et al., 2004) and the primers used were 5′CCGGAATTCTGCTTTTTCTCTAGCCGC3′ and 5′CCCAAGCTTTTGAAGCCAAGTGCGAGC3′. To generate the proximal site probe the template used was hcdc2-dE2Fm (Zhu et al., 2004) and the primers used were 5′CCGGAATTCTGCTTTTTCTCTAGCCGC3′ and 5′CCCAAGCTTTTGAAGCCAAGTGCGAGC3′. PCR products were digested with EcoRI and HindIII, radiolabeled, and purified using Quick Spin Columns (TE) for Radiolabeled DNA Purification from Roche. EMSAs were performed as previously described (Ikeda et al., 1996). The polyclonal antibody α-HA (Y-11) from Santa Cruz Biotechnology was used in the EMSAs.
Reporter chromatin immunoprecipitation assays
T98G cells were co-transfected with mutant promoter-reporter constructs and an internal control plasmid containing the β-galactosidase gene, pCMV-βgal, using SuperFect Transfection Reagent from Qiagen. Transfections were performed using 5μg of promoter-reporter plasmid, 5 μg of internal control plasmid, 60 μl of SuperFect, and 700 μl of Optimem. After incubation for 20 minutes the transfection mix was added to the cells and incubated for six hours at 37°. Cells recovered overnight at 37° in DMEM containing 10% FBS. Cells were then split 1 to 3 into starvation medium (DMEM containing 0.1% FBS) and starved for 48 hours. Following starvation, cells were brought to the G1/S phase of the cell cycle by growth in DMEM containing 10% FBS and 1mM hydroxyurea for 18 hours. Cells were harvested 18 hours after HU addition and reporter ChIP experiments were done as previously described (Zhu et al., 2004), but with the following modification. Immunoprecipitated chromatin was detected by quantitative PCR using the QuantiTect SYBR Green PCR Kit from Qiagen and the 7900HT Fast Real-Time PCR System from Applied Biosystems. Primers used in the quantitative PCR were as follows: for the Cdc6 promoter 5′GTGACTACAGCCAATCAG3′ and GLprimer2 from Promega, for the Cdc2 promoter 5′GCTTGCGCTCGCACTCAGTTGGCG3′ and GLprimer2 from Promega, and for the β-galactosidase promoter 5′ACTGGCAGATGCACGGTTACGATG3′ and 5′CACATCTGAACTTCAGCCTCCAG3′. Antibodies used in the reporter ChIP experiments were as follows: anti-E2F3 sc879 from Santa Cruz Biotechnology, anti-E2F4 sc1082 from Santa Cruz Biotechnology, and ImmunoPure Rabbit Gamma Globulin from Pierce. ChIP data was analyzed as described by Aparicio et al. (Aparicio 2005).
Endogenous chromatin immunoprecipitation assays
Asynchronously growing HEK293 cells were harvested and endogenous ChIP experiments were done as previously described (Zhu et al., 2004), but with the following modification. Immunoprecipitated chromatin was detected by quantitative PCR using the QuantiTect SYBR Green PCR Kit from Qiagen and the 7900HT Fast Real-Time PCR System from Applied Biosystems. Primers used in the quantitative PCR were as follows: for the ENSA promoter 5′GGTCCTTGTGGCTCACTCTC3′ and 5′GGGCAATGACGTAACGATCT3′, for the ICMT promoter 5′GGGACTAAGTTTGGACAGACG3′ and 5′GGGAAGTGGTGGGAGAAGTC3′, and for the albumin promoter 5′TGGGGTTGACAGAAGAGAAAAGC3′ and 5′TACATTGACAAGGTCTTGTGGAG3′. Antibodies used in the endogenous ChIP experiments were as follows: anti-E2F3 sc879 from Santa Cruz Biotechnology, anti-YY1 sc7341x from Santa Cruz Biotechnology, and ImmunoPure Rabbit Gamma Globulin from Pierce. ChIP data was analyzed as described by Aparicio et al. (Aparicio et al., 2005).
siRNA assays
Asynchronously growing HEK293 cells were transfected using DharmaFECT Reagent 2 from Dharmacon with the following siRNAs from Dharmacon: human YY1 siGENOME duplex D-011796−06 and D-011796−07, or human E2F3 siGENOME duplex D-003261−05, D-003261−06, D-003261−07, and D-003261−09. We performed three replicates of each knockdown. Cells were harvested after forty-eight hours. Nuclear extracts were prepared from half of the cells and used in Western Blot Assays to confirm knockdown of the respective proteins. Antibodies used in the Western Blot Assays were as follows: anti-YY1 sc-7341 from Santa Cruz Biotechnology, anti-E2F3 sc-878 from Santa Cruz Biotechnology, and anti-α-tubulin T5168 from Sigma. Total RNA was prepared from the other half of the cells using the Qiashredder and the RNeasy Mini Kit from Qiagen. RNA quality was confirmed by an Agilent 2100 Bioanalyser.
DNA microarray analysis
Affymetrix DNA microarray analysis was prepared according to the manufacturer's instructions, and targets were hybridized to the Human U133A GeneChip (Affymetrix, Santa Clara, CA). To process the microarrays, we normalized the CEL files using the MAS5 implementation in Bioconductor (Gentleman et al., 2004). We mapped the genes to Enztrez Gene records using the annotations provided by NetAFFX. We discarded unannotated genes as well as genes expressed at signal levels lower than 100. Then, we averaged the three replicates in each condition and calculated fold enrichment for a gene as the ratio of the average expression of the gene in either the E2F or YY1 knockdown to the control. Next, we downloaded the sequence of the human genome from the UCSC database and used version hg18 (Kuhn et al., 2009). From the complete assembly, we extracted the proximal promoter of each gene, using the transcription start site (TSS) annotations from both the known Gene and refFlat files. We determined the proximal promoter to be 250 base pairs upstream of the TSS to 50 base pairs downstream due to enrichment of predicted E2F sites in that region (data not shown). Using the promoter sequence for all annotated genes in the genome, we searched for the presence of binding sites for E2F and YY1. We defined E2F binding sites as the sequences that match the position-specific scoring matrix (PSSM) MA0024 from the JASPAR database with a p-value<0.0001 using the patser tool (Bryne et al., 2008). For YY1, we used M00793 from TRANSFAC with a cutoff of p<0.005 (Matys et al., 2006). To choose the matrices and cutoffs, we used the known binding sites of E2F and YY1 in the CDC6 promoter as a model. We predicted E2F-YY1 modules by identifying predicted binding sites for E2F and YY1 with fewer than 15 bases in between the predicted sites. We did not allow overlapping binding sites. Using the CDC6 promoter as a model, we only accepted modules where the binding sites occurred on the same strand.
Acknowledgments
We thank members of the Nevins laboratory for valuable input throughout the course of this work and for comments on the manuscript. The project described was supported by Award Number R01CA104663 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- Aparicio O, Geisberg JV, Sekinger EA, Yang A, Moqtaderi Z, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol, Chapter 21: Unit 21.3. 2005 doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- Araki K, Nakajima Y, Eto K, Ikeda MA. Distinct recruitment of E2F family members to specific E2F-binding sites mediates activation and repression of the E2F1 promoter. Oncogene. 2003;22:7632–41. doi: 10.1038/sj.onc.1206840. [DOI] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, et al. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucl. Acids Res. 2008;36:D102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cloos P, Toftegaard U, Klinkenberg D, Bracken AP, Trinh E, et al. Characterization of E2F8, a novel E2F-like cell cycle regulated repressor of E2F activated transcription. Nucl. Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–1369. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2003;278:42041–42049. doi: 10.1074/jbc.M308105200. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad BM, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande P, Zhu W, Schlisio S, Sun XH, Mori S, Gaubatz S, et al. A role for E2F6 in distinguishing G1/S and G2/M specific transcription. Genes & Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande PH, Hallstrom TC, Tunyaplin C, Calame K, Nevins JR. Identification of the E box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 2003;23:3707–3720. doi: 10.1128/MCB.23.11.3707-3720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2007;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci USA. 2003;100:10848–10853. doi: 10.1073/pnas.1831408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes & Dev. 2006;20:613–623. doi: 10.1101/gad.1345006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M-A, Jakoi L, Nevins JR. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci U S A. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Martin MC, Phillips AC, Seelan RS, Smith DI, Liu W, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001a;21:4684–99. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in the control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001b;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L-J, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Leone G, Nevins JR. E2F1-specific induction of apoptosis and p53 accumulation is modulated by mdm2. Cell Growth & Diff. 1998;9:113–118. [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J. Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–61. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–644. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- Logan N, Graham A, Zhao XD, Fisher RI, Maiti B, Leone G, et al. E2F-8: an E2F family member with a similar organization of DNA binding domains to E2F-7. Oncogene. 2005;24:5000–5004. doi: 10.1038/sj.onc.1208703. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its moduel TRANSCompel: transcriptional gene regulation in eukaryotes. Nucl. Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Denchi EL, Caprara G, Colli E, Cecconi F, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nature Cell Biology. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- Nevins JR. Toward an understanding of the functional complexity of the E2F and Retinoblastoma families. Cell Growth & Diff. 1998;9:585–593. [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nature Genetics. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoff HA, Pickering MT, Frame FM, Debatis ME, Sanchez Y, Jones S, et al. Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol. 2004;24:2968–2977. doi: 10.1128/MCB.24.7.2968-2977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RC, Nevins JR. Signaling networks that link cell proliferation and cell fate. J. Biol. Chem. 2002;277:11617–11620. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes & Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Stiewe T, Putzer BM. Role of the p53 homologue p73 in E2F1-induced apoptosis. Nature Genetics. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and Rb families in vivo: distinct E2F proteins mediate activation and repression. Genes & Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Kassatly RF, Cress WD, Horowitz JM. Subunit composition determines E2F DNA-binding site specificity. Mol. Cell. Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi S, Pfeifer GP. In vivo structure of the human cdc2 promoter: release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Wen J, Lees JA. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Nat'l. Acad. Sci. 2001;98:1519–1524. doi: 10.1073/pnas.041597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, et al. CDC25A phosphatase is a target of E2F and is required for efficient E2f-induced S phase. Mol. Cell. Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J, Boyd KE, Fry CJ, Bartley SM, Farnham PJ. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka A, Herr W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci. 2003;28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- Yamamoto KR, Darimont BD, Wagner RL, Iniguez-Lluhi JA. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harbor Symp Quant Biol. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet RV, Leone G, Stillman B, Nevins JR, et al. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci U S A. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Fraenkel E, Pabo CO, Pavletich NP. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes & Dev. 1999;13:666–674. doi: 10.1101/gad.13.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande P, Nevins JR. E2Fs link the control of G1/S and G2/M. EMBO J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes & Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]