Abstract
Spatial distribution of genes within the nucleus contributes to transcriptional control, allowing optimal gene expression as well as constitutive or regulated gene repression. Human immunodeficiency virus type 1 (HIV-1) integrates into host chromatin to transcribe and replicate its genome. Lymphocytes harbouring a quiescent but inducible provirus are a challenge to viral eradication in infected patients undergoing antiviral therapy. Therefore, our understanding of the contribution of sub-nuclear positioning to viral transcription may also have far-reaching implications in the pathology of the infection. To gain an insight into the conformation of chromatin at the site of HIV-1 integration, we investigated lymphocytes carrying a single latent provirus. In the silenced state, the provirus was consistently found at the nuclear periphery, associated in trans with a pericentromeric region of chromosome 12 in a significant number of quiescent cells. After induction of the transcription, this association was lost, although the location of the transcribing provirus remained peripheral. These results, extended to several other cell clones, unveil a novel mechanism of transcriptional silencing involved in HIV-1 post-transcriptional latency and reinforce the notion that gene transcription may also occur at the nuclear periphery.
Keywords: chromatin, HIV-1, latency, nucleus, transcription
Introduction
The sub-nuclear organization of chromatin in the nucleus is not random with whole chromosomes occupying discrete territories in preferred positions (Cremer et al, 2006). The position of a specific gene within the nucleus is correlated with its transcriptional status and specialized sub-compartments for gene expression or repression have been described (Sexton et al, 2007). Silent gene loci are often found in close proximity to centromeric heterochromatin, and centromeres are known to be preferentially associated with the nuclear periphery or the nucleolus (Brown et al, 1997, 1999, 2001; Kim et al, 2004; Merkenschlager et al, 2004). The nuclear periphery has been functionally associated with transcriptional silencing in yeast (Andrulis et al, 1998) and Drosophila melanogaster (Marshall et al, 1996; Pickersgill et al, 2006), although this view has been challenged by the demonstration of regions of active transcription associated with nuclear pores (Casolari et al, 2004; Taddei et al, 2006). In addition, in mammals, the nuclear periphery has been associated with transcription repression, particularly through associations with the nuclear lamina< (Guelen et al, 2008). Tethering of transgenes to the periphery of the nucleus causes transcriptional silencing that can be reverted by repositioning towards the interior of the nucleus (Dietzel et al, 2004; Finlan et al, 2008; Reddy et al, 2008). Several endogenous genes have also been shown to behave similarly (Kosak et al, 2002; Zink et al, 2004; Chuang et al, 2006; Williams et al, 2006), although the picture might be more complex with genes being unaffected by their proximity to the periphery (Nielsen et al, 2002; Zhou et al, 2002; Hewitt et al, 2004; Finlan et al, 2008; Kumaran and Spector, 2008; Reddy et al, 2008). In addition to the nuclear periphery, different types of silenced domains containing Polycomb group (PcG) proteins have also been found throughout the nucleoplasm of Drosophila and human nuclei (Saurin et al, 1998; Martinez et al, 2006). Changes in gene expression after cellular differentiation, or after various external stimuli, are often associated with changes in the sub-nuclear positioning of genes and regulatory sequences that may establish long-range interactions (Spilianakis et al, 2005; Lomvardas et al, 2006). In some cases, large chromosome loops containing active genes extend outside of a defined chromosome territory (Volpi et al, 2000; Chambeyron and Bickmore, 2004; Williams et al, 2006; Noordermeer et al, 2008). Sites of active transcription are enriched in RNA polymerase II and are called transcription factories (Jackson et al, 1998). In some cases, more than one gene, also from different chromosomes, have been shown to occupy the same transcription factory on activation (Osborne et al, 2004).
The human immunodeficiency virus type 1 (HIV-1) requires integration into host chromatin to replicate. Transcriptional silencing of integrated HIV-1 is crucial for the establishment of latent reservoirs of infected cells that persist during antiretroviral therapy and pose a constant threat to infected individuals. Discontinuation of therapy induces transcriptional reactivation of latent provirus and a consequent relapse of viremia. As the HIV-1 provirus is found integrated into the host genome, regulation of the viral gene expression depends on the chromatin environment at the site of integration (Marcello, 2006). Clearly, multiple mechanisms could concur in this process. The U3 region of the HIV-1 long terminal repeat (LTR) functions as the viral promoter and contains consensus sequences for several host transcription factors (Pereira et al, 2000). In addition, HIV-1 transcription is boosted by the viral Tat trans-activator that recruits host transcription kinases and acetyltransferases to the integrated promoter (Marcello et al, 2001). Both host transcription factors and the viral Tat trans-activator have been proposed as limiting factors for transcriptional reactivation of latent HIV-1. However, as HIV-1 is found integrated into the genome of resting memory T cells, it has been proposed that the chromatin environment at the viral integration site may have a role in the transcriptional silencing of the HIV-1 genome (Jordan et al, 2001, 2003). Indeed, integrated HIV-1 has nucleosomes positioned in its 5′-LTR that are remodelled by deacetylase inhibitors, cytokines and Tat (Van Lint et al, 1996; Lusic et al, 2003; Marcello et al, 2004). Histones are important in regulating HIV-1 transcription as they integrate signals for repression, like the heterochromatin markers H3K9 trimethylation, Suv39H1 and HP1γ (du Chene et al, 2007), and reactivation, like histone acetylation (Lusic et al, 2003). HIV-1 functions as an ectopic transcription unit that can integrate at various positions within chromatin and as such becomes part of this dynamic framework of chromatin interactions that control transcription. It is possible that by integrating into a region involved in a specific cellular pathway that requires inter- or intra-chromosomal interactions to regulate transcription, HIV-1 becomes regulated in the same manner as has been proposed for cellular genes (Fraser and Bickmore, 2007; Misteli, 2007). At the molecular level, such dynamic genomic reorganizations correlate with the formation of inter- and intra-chromosomal associations that can be revealed by the chromosome conformation capture (3C) technique (Dekker et al, 2002).
To explore this possibility, we took advantage of a lymphoid cell line that carries a single silenced provirus (Jordan et al, 2003). To clone putative sequences that were spatially proximal to the integrated provirus, we developed a circular 3C (4C) technique (Lomvardas et al, 2006; Zhao et al, 2006). The 4C method allowed the identification of a region of chromosome 12 (position Ch12q12) that physically interacted with the provirus that was confirmed by subsequent 3C analysis. It is interesting that, however, the interaction was lost in cells where transcriptional activation of the provirus occurred. To confirm the spatial proximity of these two regions, we performed fluorescent in situ hybridization (FISH) and found that the provirus and Ch12q12 were associated in a significant number of quiescent cells. On activation, the association between these two regions was again lost, although the transcribing provirus remained localized at the nuclear periphery. We conclude that HIV-1 transcriptional repression correlates with nuclear positioning of the provirus to the periphery and with its interaction with pericentromeric heterochromatin. However, both inactive and actively transcribing provirus did not move towards the interior of the nucleus but remained peripheral.
Results
The J-lat A1 cellular model
J-lat A1 cells are a convenient model to study HIV-1 post-integrative latency. J-lat A1 cells were generated by transduction of the lymphocytic cell line Jurkat with an HIV-1 vector containing the Tat and the green fluorescent protein (GFP) open reading frames under the control of the viral 5′-LTR (Jordan et al, 2003). Cells that were negative for GFP have been isolated by sorting and then stimulated with phorbol esters (TPA) to isolate GFP-positive cells. Cells that had the latent phenotype and that could be reactivated by TPA treatment were cloned and further characterized. These cells carry a single integrated HIV-1 vector at position ChXp21.1 (Figure 1A). The vector was derived from the prototype pHR′ series where flag-tagged Tat101 (corresponding to the two-exon form of the HIV-1 Tat gene with a C-terminal flag tag) and GFP were under the control of the HIV-1 5′-LTR by using an internal ribosome entry site (LTR-Tat-IRES-EGFP) (Jordan et al, 2003).
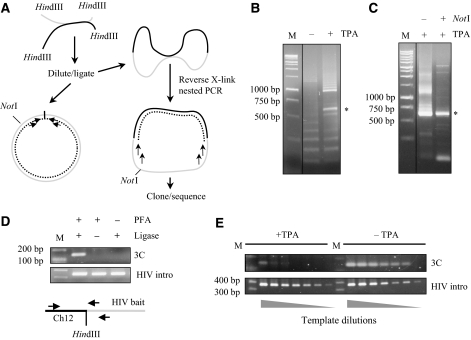
Figure 1.
The J-lat A1 cellular model. (A) Diagram of the HIV-1 construct integrated in position ChXp21.1 of J-lat A1. The 5′-LTR is followed by the packaging signal, the major splice donor site (SD) and a portion of the gag gene fused to the Rev-responsive element (RRE). The Tat acceptor site SA7 precedes a cassette containing the Tat101 gene fused to a flag tag (f-Tat101), an internal ribosome entry site (IRES) and the GFP reporter. Sites of cleavage by HindIII (H) and NotI (N) are also indicated. The position of the primers used in the 4C protocol is indicated by black arrows above the bait indicated by a double grey arrow. Below, the pre-mRNA and spliced RNA are shown together with the dimensions of the fragment amplified by RT–PCR with the indicated primers. (B) Activation of the J-lat A1 cell line. Cytofluorimetric analysis of cells was conducted in non-induced cells (left panel) and after induction with TPA for 15 h (right panel). More than 85% of cells were activated as shown by the expression of the GFP reporter. (C) Assembly of Cyclin T1 with Tat on induction of J-lat A1 cells. To show that induction by TPA induces f-Tat101 expression and the formation of the Tat∷Cyclin T1 complex, cells were lysed and immunoprecipitated with a flag antibody. Top panel, western blot analysis probing with an anti-flag antibody: IgL immunoglobulin light chains. Bottom panel, western blot probing with an antibody against Cyclin T1. (D) Time course of provirus expression in J-lat A1 cells. RT–PCR analysis was conducted for the spliced RNA (exon, top panels), the pre-mRNA (intron, middle panels) and the β-actin control (bottom panels) before (left panels) and after induction with TPA (right panels). Samples were analyzed at the indicated time points.
On induction with TPA, ∼86% of cells express GFP after 24 h (Figure 1B), Flag-tagged Tat is produced and the cellular cofactor Cyclin T1 can be immunoprecipitated with anti-flag antibodies (Figure 1C). Induction of viral mRNA is already visible at 4 h post-induction with TPA and accumulates further on (Figure 1D). A time course of nascent viral RNA was measured with primers for the intronic region, and in this case, the precursor RNA was also produced within 4 h and peaked 8 h after induction (Figure 1D). We conclude that the J-lat A1 cell line may be a convenient model to study HIV-1 latency with a defined silenced state (off state) and an activated state (on state). We also set 8 h after treatment with TPA as the maximal activation of the integrated provirus.
Identification of genomic sequences that interacted with the HIV-1 provirus by the 4C technique
On the basis of formaldehyde cross-linking of living cells followed by enzyme digestion, intra-molecular ligation and quantitative PCR analysis, the 3C method provides the opportunity to study chromosomal folding in the nucleus only if previous knowledge of the bait and interacting sequence exists (Dekker et al, 2002). This limitation has been overcome by exploiting a circular intermediate by the strategic positioning of primers within the bait (Lomvardas et al, 2006; Zhao et al, 2006). Therefore, this latter method enables the identification of unknown interacting sequences, which form part of the circular DNA, rendering it very useful for genome-wide studies. The 4C method is based on the proximity ligation principle in which DNA–protein/protein–DNA complexes generate circular DNA molecules under prolonged incubation with high concentrations of ligase (Gondor et al, 2008).
J-lat A1 cells were either left untreated or incubated with TPA for 8 h and then cross-linked with formaldehyde as described in detail in Material and Methods. Cross-linked material was digested with HindIII, diluted and ligated. After reversal of cross-links, primers pointing outwards from a HindIII fragment (position 1088–1672 according to the sequence of the provirus) within the HIV-1 provirus were used as a bait to amplify DNA sequences without any previous knowledge of their identities (Figure 2A). As shown in Figure 2B, a smear of several bands was evident in agarose gels after nested PCR. The major difference consisted in the appearance of bands at ∼600 and 1200 bp only in the induced cells. The bands were excised from the gel, cloned and sequenced. Identity was confirmed to be the fragment of the provirus used as bait for the 4C (584 bp) and multimers of this sequence. We hypothesized that this could be due to multiple runs of PCR over the junction on religated templates (Figure 2A). To confirm this hypothesis, we introduced an additional step in the 4C procedure by cutting with NotI (position 1145 of the provirus), a unique restriction site within the bait fragment, before the nested PCR. As shown in Figure 2C, treatment with NotI reduces the products of the religated template. However, despite this modification of the protocol, we were unable to detect host chromatin sequences associated with the provirus in activated cells. Both single excised bands or bulk cloning of the PCR products showed only religated templates, primer multimers or boundary sequences of the provirus owing to incomplete HindIII digestion. This could be explained by a technical failure to identify all the fragments involved and may be solved by more in-depth sequencing or the use of tiling arrays (Simonis et al, 2006). Alternatively, the observation that the intra-molecular ligation by-products of the 4C reaction increase on induction may indicate that the chromatin loops out from a more compact conformation and therefore is more accessible to enzymatic digestion. Looping of HIV chromatin on transcriptional activation coupled to high levels of Tat-mediated transcription and local enrichment of nascent RNA, although not formally proven, may well account for a single transcription factory exclusive for HIV transcription (Boireau et al, 2007; Perkins et al, 2008).
Figure 2.
4C analysis at the site of HIV integration in J-lat A1 cells. (A) Diagram of the 4C protocol. The bait (grey line) derived from the HIV-1 provirus (see Figure 1A) is cross-linked to an unknown genomic locus (black line). After digestion with HindIII, the reaction is diluted and ligated to generate both intra- and inter-molecular ligations. Intra-molecular ligations could be reduced by NotI treatment. After reversal of cross-linking, a nested PCR is performed with primers within the bait pointing outwards, as shown also in Figure 1A. (B) 4C amplified fragments from J-lat A1 cells. Products of the nested PCR amplification of cells either not induced or induced with TPA were resolved on agarose gels. Molecular weight marker (1 kb, M) and positions of the major product of intra-ligation (asterisk) are shown. (C) 4C amplified fragments from J-lat A1 cells after NotI digestion. To reduce intra-molecular ligation in the TPA-activated cells, the cross-linked material was treated with NotI before performing the nested PCR. Molecular weight marker (1 kb, M) and positions of the major product of intra-ligation (asterisk) are shown. (D) 3C analysis of the interaction of the provirus and Ch12q12. To confirm 4C data, a 3C analysis was performed by hemi-nested amplification using a primer within the Ch12q12 region (see diagram). Control amplification was performed with primers mapping within the provirus (see Figure 1A). (E) Loss of the interaction between the provirus and Ch12q12 on induction of transcription. 3C analysis of the Ch12q12/provirus interaction was performed both in induced (left) in and non-induced (right) cells. Two-fold serial dilutions of the template show that in non-induced cells there is more cross-linked material for the interaction.
By contrast, in silenced cells, after bulk cloning of all the nested PCR products, we identified a specific fragment from host chromatin that corresponded to the positions 37 017 830–37 017 941 of chromosome 12q12. To confirm this interaction, we proceeded to 3C analysis with a primer mapping within the identified fragment (Supplementary Table 1). As shown in Figure 2D, a hemi-nested PCR on cross-linked material from latent J-lat A1 cells confirmed the interaction of the provirus with Ch12q12 (Figure 2D, top panels). As control of template amplification, we used a set of primers that detected the HIV provirus (HIV intro, bottom panels). However, if the ligation step was omitted or the material was not cross-linked, no amplification was observed (Figure 2D). Hence, in the off state, the provirus interacts physically with Ch12q12. Next, we examined this interaction by 3C after induction with TPA. As shown in Figure 2E, the amount of template that could be amplified by hemi-nested PCR was reduced on activation of HIV transcription. We conclude that in J-lat A1 quiescent cells, the provirus integrated in ChXp21.1 interacts physically with Ch12q12. On activation of proviral transcription with TPA, this interaction is lost.
Mapping of the genomic regions by 3D-FISH
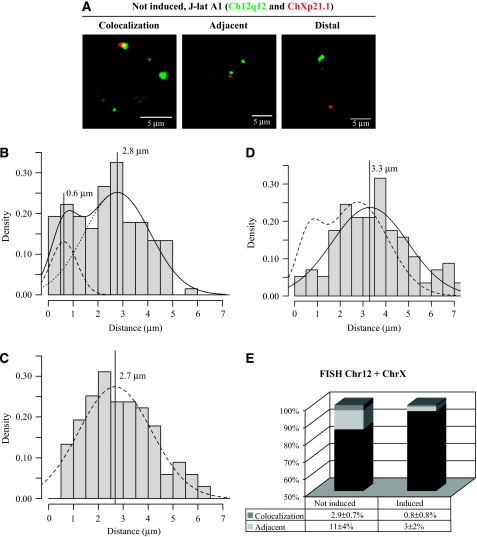
To confirm the interaction between the provirus integrated into ChXp21.1 and Ch12q12, we performed FISH on J-lat A1 cells that were fixed by a method previously shown to preserve the 3D structure of the nucleus (Solovei et al, 2006). To mark ChXp21.1, we used BAC RP11-77013 and to mark Ch12q12 we used BAC RP11-379CZ4. These probes were selected to cover the region where the provirus is integrated (ChX) and the region that was showed by 3C to be physically associated with it in Ch12. As shown in Figure 3A, single-cell disposition of the two marked regions varied. First, we measured the minimal distance between ChXp21.1 and Ch12q12, to look for the interaction detected in 4C, and between Ch12q12 and Ch12q12 for comparison. More than 100 cells were analyzed and the results are shown in Figure 3B and C. Both samples show a Gaussian distribution around a 3-μm mean of the minimal distances of interaction. However, for the Ch12/ChX analysis, the data could be fitted with two Gaussians, with the smaller representing a subset of close associations centred around 0.6 μm (Figure 3B). Within the population of cells tested in the silenced state, 2.9±0.7% showed colocalization of the two regions and 11±4% showed proximity at a distance of <0.6 μm (Figure 3E). Clearly, this could be the fraction of the interaction that could be detected by 3C; therefore, we wanted to explore what happens when HIV transcription is induced by TPA. As shown in Figure 3D, treatment of cells with TPA induces the disappearance of the cluster of close associations of Ch12q12 with ChXp21.1. Both fractions of cells were significantly reduced to 0.8±0.8% (colocalizing) and 3±2% (adjacent) (Figure 3E).
Figure 3.
Analysis of the interaction of Chr12q12 with ChXp21.1. (A) J-lat A1 cells were analyzed in FISH with a probe for Ch12q12 (BAC RP11-379CZ4, green) and a probe for ChXp21.1 (BAC RP11-77013, red). The position of the two loci varied from colocalizing (left panel) to adjacent at ⩽0.6 μm (central panel) to distal at >0.6 μm (right panel). (B) Quantitative analysis of the distribution of the distances between Ch12q12 and ChXp21.1 in non-induced J-lat A1 cells (n=135). The density is intended as the frequency of distances between the two loci that fall within a discrete interval divided for the interval amplitude. The data were fitted and showed a bi-Gaussian distribution (solid line with the two Gaussians shown as dotted lines) with a subset of nuclei where the two genomic loci were closely associated. The mean values of the two Gaussians are also indicated by vertical lines (0.6 and 2.8 μm, respectively). (C) As a control, the same analysis as in panel B was conducted for the distances between chromosomes 12. This analysis showed a Gaussian distribution (dotted line). (D) The distribution of minimal distances between chromosome X and 12 is compared between non-induced (dotted line, n=135) and induced cells (solid line, n=114). Histogram shows the distribution of the induced cells. (E) Quantitative analysis of the frequency of nuclei showing colocalization (dark grey), proximity at ⩽0.6 μm (light grey) or distance at >0.6 μm (black). Numeric values are indicated below. Decrease of the colocalization is significant (χ2, P=0.016).
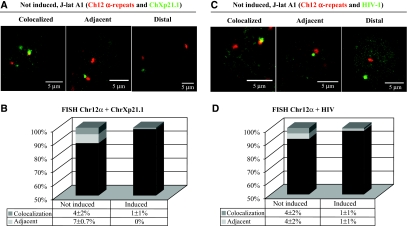
The region of interaction at position Ch12q12 is close to the centromer of chromosome 12, a heterochromatic region that might be involved in the silencing of HIV-1 in trans. Indeed, pericentromeric satellite alphoid DNA repeats are present and can be used as a marker for heterochromatin at that location. Therefore, we exploited a DNA probe specific for these α-satellite repeats present in Ch12 (Baldini et al, 1990). As expected, the distances between the two marked regions varied (Figure 4A). A fraction of the non-induced cells showed colocalization of signals (4±2%), whereas 7±0.7% of cells showed proximity at a distance of <0.6 μm (Figure 4B). When cells were induced, the proximity was lost (Figure 4B). We next explored the localization of the provirus with respect to Ch12 α-satellites (Figure 4C). In this case, 4±2% of cells showed colocalization of the provirus with the centromer and 4±2% showed proximity at a distance of <0.6 μm (Figure 4D). Again, when cells were induced, the colocalization was reduced to 1±1% and the fraction showing proximity at a distance of <0.6 μm was also reduced to 1±1% (Figure 4D).
Figure 4.
Interaction of the Chr12 centromere with ChXp21.1 and the provirus. (A) J-lat A1 cells were analyzed in FISH with a probe for Ch12 α-repeats (red) and BAC RP11-77013 (green) mapping at ChXp21.1. Similar to Figure 3, the position of the two loci varied from colocalizing (left panel) to adjacent at ⩽0.6 μm (middle panel) to distal at >0.6 μm (right panel). (B) Quantitative analysis performed as in Figure 3E (non-induced, n=105; induced, n=103). Decrease of the colocalization is significant (χ2, P=0.016). (C) J-lat A1 cells were analyzed in FISH with a probe for Ch12alpha repeats (red) and the provirus (green). Variation in the position of the two loci is shown as in panel A. (D) Quantitative analysis (non-induced, n=104; induced, n=78). Decrease of the colocalization is significant (χ2, P=0.04).
These results confirm the 3C data since we could demonstrate that a fraction of the population showed interaction between the provirus and the pericentromeric region of chromosome 12. It is possible that this sub-population of cells corresponds to those that are not activated by TPA (Figure 1B). However, we show in Supplementary Figure 1A that there is no correlation between the inability of being reactivated by TPA and the association of the provirus with centromeric heterochromatin in different cell clones. Most likely, the timing of transcriptional activation differs among different cells in the population, also possibly reflecting the chromatin interactions of the provirus.
Interestingly, one can also observe that on TPA treatment, the interaction between provirus and Ch12 is disrupted both in 3C and in FISH. How this disruption occurs remains to be established. An increase in the cell volume in J-lat A1 was not observed on TPA stimulation as it occurs, for example, in resting T cells (Branco et al, 2008). Furthermore, although the TPA treatment induces cell-cycle arrest, it remains possible that in the period after TPA stimulation a certain proportion of cells are still capable of completing mitosis, thus justifying the disruption of the interaction observed in J-lat A1. Alternatively, active rearrangements of chromatin domains during interphase may be required (Louvet and Percipalle, 2009).
Sub-nuclear positioning of the HIV-1 provirus
As in our case the proximity between the loci correlates with silencing and it has been shown that silencing correlates with positioning near the nuclear periphery in some cases, we measured the distance from the nuclear periphery of the interactions. It is noted that the distribution of pericentromeric heterochromatin in J-lat A1 is characteristic of cycling lymphocytes (see Supplementary Figure 1B and C) with a high, but not exclusive, prevalence at the nuclear periphery. The distribution appears similar to what was observed in interphase lymphocytes (Weierich et al, 2003; Solovei et al, 2004) and corresponds to the so-called ‘conventional architecture' (Solovei et al, 2009). Evidence that the nuclear lamina forms an important repressive compartment also comes from the recent observations that this domain appears to be discontinuous with discrete lamin-associated chromatin domains (Guelen et al, 2008).
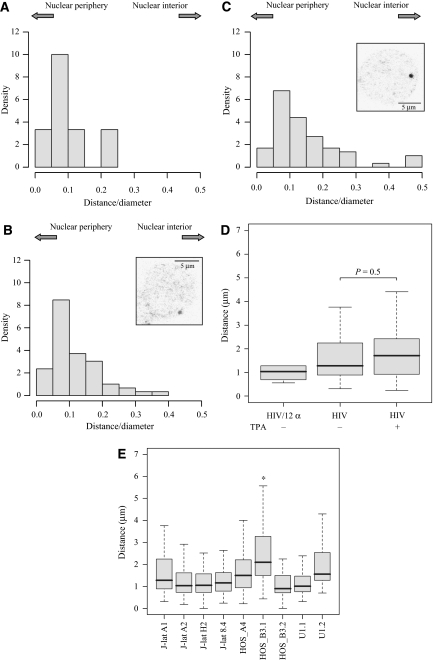
A subset of cells was chosen where the signals for Ch12 pericentromeric α-satellites and the provirus were at a distance of <0.6 μm (the mean value of the first Gaussian of Figure 3B compatible with the mean radius of confinement of each genomic locus) and measured their distance from the periphery of the nucleus. In Figure 5A, the distribution of these distances is shown as values normalized to the diameter of the nucleus where the value 0 represents the periphery and 0.5 the centre of the nucleus. It is interesting that the position of the interaction is consistently associated with the nuclear periphery (median of the absolute values=1 μm, Figure 5D). These results show that in a certain proportion of cells, the silent provirus is found in close proximity of Ch12 pericentromeric α-satellites close to the nuclear periphery. Next, we repeated the analysis looking at the position of the provirus. As shown in Figure 5B, the provirus associates with the periphery irrespective of its association with chromosome 12. On induction with TPA, the position of the provirus seems to move slightly to the interior of the nucleus (Figure 5C). The shift is from a median absolute distance of 1.3 μm in non-induced cells to a distance of 1.7 μm in induced cells and was not significant (K–S test, P=0.5) (Figure 5D). As more than 86% of cells are activated on induction (Figure 1A), it appears that activation of the provirus does not correlate with a significant move towards the interior of the nucleus. However, as shown in the box plot of Figure 5D, the interactions between Ch12q12 and ChXp21.1, as well as between the provirus and the α-satellite repeats showed a significant sharp clustering at the nuclear periphery, whether the provirus remained more loosely associated both in the non-induced and in induced states.
Figure 5.
Localization of the J-lat A1 provirus to the nuclear periphery. (A) The sub-population of J-lat A1 non-induced cells carrying the interaction between the provirus and Ch12 centromeric α-repeats was analyzed for distance of the interaction from the nuclear periphery (n=25). The abscissa represents the radius of the cell where 0 is the periphery and 0.5 is the centre. Values are normalized for the nuclear diameter. The density is intended as the frequency of distances between the two loci that fall within a discrete interval divided for the interval amplitude. (B) J-lat A1 cells were also analyzed for the localization of the provirus with respect to the nuclear periphery in the non-induced state (n=60) (B) and on induction (n=60) (C). Analysis was conducted as in Figure 5A. Inset: example of provirus detection. (D) Box plot analysis of the distribution of absolute distances from the periphery of the subset of interactions of provirus and Ch12 and centromeric α-repeats (median=1 μm). In addition, the distance of the provirus from the periphery, independent of its association with chromosome 12, is shown both in the inactive (median=1.3 μm) and in active (median=1.7 μm) states. Status of induction with TPA is indicated below. The differences in the distribution of the distances to the periphery of the provirus in the inactive or active state were not statistically significant (K–S test, P=0.5). (E) Box plot analysis of the distribution of distances from the periphery of the integrated provirus in different cell clones. When two integrations are present, as for U1 and HOS_B3, each of them is shown. Distances are presented as absolute values (values normalized to the diameter of the cell are shown in Supplementary Figure 1D). Values that differ significantly from those obtained from J-lat A1 are indicated by an asterisk (K–S test, P<0.01).
These observations are conducted on one cell clone, J-lat A1, and may be of limited general significance. Therefore, the analysis of the localization of the provirus was extended with respect to nuclear periphery to: (i) two additional Jurkat cell clones derived similarly to J-lat A1 (J-lat A2 and J-lat H2) (Jordan et al, 2003); (ii) a Jurkat cell clone carrying a complete provirus (J-lat 8.4) (Jordan et al, 2003); (iii) two HOS cell clones obtained with a different protocol carrying a Tat-inducible provirus whose RNA can be visualized in single living cells (HOS_A4 and HOS_B3) (De Marco et al, 2008); and (iv) a pro-monocytic cell line carrying wild-type integrated HIV-1 (U1) (Folks et al, 1987). These cell lines represent a wide range of models of proviral latency, from virus/vector integration within alphoid repeats in -cis (J-lat H2) (Jordan et al, 2003) to integration within expressed genes (J-lat 8.4, HOS_A4 and HOS_B3) (De Marco et al, 2008; Lenasi et al, 2008) to latency resulting from cells surviving a wild-type infection (U1) (Folks et al, 1987). For all these cells, the distance of the provirus from the nuclear periphery was measured. As shown in Figure 5E, most latent proviruses localize closely at the periphery of the nucleus and do not differ significantly from J-lat A1, with one exception. HOS_B3 cells harbour two integrated proviruses, both within active genes and transcriptionally competent (data not shown). One of the two integrants appears more distal from the periphery compared with J-lat A1 (K–S test, P=0.001). This cell line requires further investigation to explain how the internal provirus may be kept silenced.
We show that the majority of the silent provirus is found at the nuclear periphery, but as we could establish only indirectly in J-lat A1 that transcriptional activation does not result in changes of sub-nuclear positioning of the gene, we decided to visualize HIV-1 RNA at the site of transcription. We took advantage of the HOS_A4 cell line that allows visualization of viral RNA in single living cells by the MS2 tagging method as described extensively in De Marco et al (2008). First, we confirmed that nascent HIV-1 RNA remains at the nuclear periphery upon transcriptional activation by in situ hybridization of RNA (Figure 6A). Interestingly, the distribution of distances from the periphery measured for the provirus in HOS_A4 in the silenced state (median of the absolute values=1.5 μm) did not differ significantly from those of nascent RNA in the activated state (median of the absolute values=1.2 μm), indicating that the transcribing provirus did not move from its position (Supplementary Figure 1E). To further confirm that HIV-1 RNA is transcribed at the nuclear periphery, a time course of RNA biogenesis in living cells expressing both Tat and EYFP-MS2nls was measured. As clearly shown in Figure 6B, active transcription from an integrated provirus is also continuously associated with the nuclear periphery in living cells.
Figure 6.
Localization of the HIV-1 nascent RNA to the nuclear periphery in single living cells. (A) HOS_A4 cells were analyzed for the localization of the HIV-1 nascent RNA with respect to the nuclear periphery in Tat-activated cells (n=62). Analysis of distances was conducted as in Figure 5. (A) Inset: example of the RNA in situ hybridization with an intronic probe that shows localization of nascent HIV-1 RNA at the site of transcription close to the nuclear periphery. (B) Localization of transcription in single living cells. HOS_A4 cells expressing Tat and EYFP_MS2nls were monitored in time for the localization of nascent RNA. Distances from periphery for 13 cells (±s.d.) are plotted at 5 min intervals for 30 min. A single nucleus is also shown at the various time points to show the position of the transcribing locus (white arrowhead).
Discussion
Regulation of gene expression is profoundly involved in HIV-1 pathogenesis. Failure of anti-retroviral therapy to eradicate the virus is determined by long-term post-integration transcriptional latency. In this work, the role of spatial positioning on proviral function in a well-established model of HIV-1 post-integrative latency was explored. J-lat A1 cells carry a transcriptionally silent HIV-1 proviral construct that could be induced by phorbol esters or other stimuli. We found that the provirus is located at the nuclear periphery and that it is associated with a pericentromeric region of chromosome 12 in ∼10% of cells. In this calculation, we include both loci that colocalized and loci that were within a distance of 0.6 μm, considering that the mean radius of confinement of a genomic locus in a mammalian nucleus is between 0.5 μm (Spector, 2003; Lanctot et al, 2007) and 0.3 μm for more constrained peripheral loci (Chubb et al, 2002). Interestingly, upon transcriptional activation of the provirus, the interaction of the two loci dropped significantly, although the localization of the provirus remained peripheral.
In the original work by Jordan et al (2003), HIV-1 post-integrative latency correlated with integration in close proximity to alphoid repetitive DNA in cis. However, the J-lat A1 cell line differed because the integrated provirus was not associated with these repetitive sequences (Jordan et al, 2003). Heterochromatin is associated with specialized chromosome structures, such as centromeres and telomeres, which define transcriptionally inactive nuclear domains. The silencing effect of heterochromatin is not restricted to the region packaged into heterochromatin itself but also to neighbouring DNA sequences providing an environment of condensed chromatin enriched in specific histone modifications and heterochromatin proteins (Dillon and Festenstein, 2002). Therefore, heterochromatin centred at the chromosome 12 centromere might extend its repression also in trans involving closely associated chromatin loci such as ChXp21 harbouring the integrated provirus. Accordingly, recruitment of chromatin domains to centromeric heterochromatin in trans imposes silencing in D. melanogaster and differentiating mouse lymphocytes (Csink and Henikoff, 1996; Dernburg et al, 1996; Brown et al, 1997, 1999, 2001; Merkenschlager et al, 2004). Heterochromatin markers, such as Suv39H1, HP1 and histone H3Lys9 trimethylation, have an important function in chromatin-mediated repression of integrated HIV-1 gene expression being reversibly associated with HIV-1 in a transcription-dependent manner (Marban et al, 2007; du Chene et al, 2007; Mateescu et al, 2008). Interestingly, reactivation of J-lat A1 cells by siRNA knockdown of HP1α and HP1γ has been shown (du Chene et al, 2007). All these observations point to an involvement of heterochromatin in the silencing of HIV-1. However, a paradox emerges from our observations as in only ∼10% of cells the provirus was found in the proximity of the centromere of chromosome 12 when all cells were in fact silenced. Two explanations could be proposed: either within the same clonal population, the provirus associates with different heterochromatin environments that went undetected in our 4C approach, or two or more states of silencing exist, ranging from one closely associated with heterochromatin to one possibly more poised for transcription. The former hypothesis is unlikely, as the fraction of proximity of the provirus with a pan-α-satellite probe did not differ significantly from that with the specific Ch12 pericentromeric region (see Supplementary Figure 1A). The latter state could involve the assembly of a pre-initiation complex with a stalled RNAPII that requires further signals to elongate (Figure 7). Indeed, it has been shown that RNAPII elongation complexes initiating from the viral LTR prematurely terminate transcription in the absence of Tat, also in quiescent lymphocytes from patients (Kao et al, 1987; Lassen et al, 2004). Furthermore, experiments involving bulk analysis of cells, such as, for example, chromatin immunoprecipitation, detected both HP1β and RNA polymerase on the viral LTR in the latent state (Mateescu et al, 2008). Here, we show by single-cell analysis that these two states could be distinct within the same population.
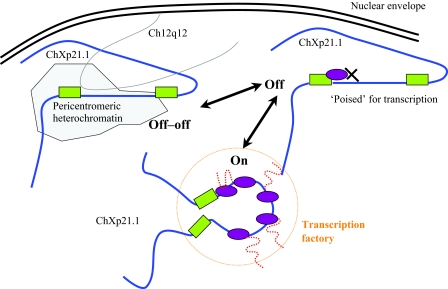
Figure 7.
Schematic drawing that summarizes the concepts emerging from the experimental data. In J-lat A1 cells, the provirus is found integrated in the X chromosome and localized at the nuclear periphery. Although all cells harbour a transcriptionally silent provirus, this could be found spatially associated close to the centromere of Chromosome 12 in a fraction of the nuclei. We identify two states of the silenced provirus: one associated with pericentromeric heterochromatin of chromosome 12 at the nuclear periphery (off–off) and one not associated with chromosome 12 that may be poised for transcription (off). On activation, this interaction is lost and HIV RNA is transcribed, possibly within a transcription factory, without changing its localization to the nuclear periphery (on).
Regardless of the status of chromatin at the site of integration, the provirus is also found associated with the nuclear periphery in induced cells. To note that peripheral localization of the region of interaction of chromosome territories corresponding to Ch12 and ChX has been observed in both human primary fibroblasts (Bolzer et al, 2005; Guelen et al, 2008) and lymphocytes (Kim et al, 2004; Parada et al, 2004). Recent studies that addressed gene expression by locus targeting at the nuclear periphery consistently showed that transcriptional competence is also maintained at this location (Finlan et al, 2008; Kumaran and Spector, 2008; Reddy et al, 2008). Indeed, our analysis of several additional cell clones confirmed localization of the silent provirus at the nuclear periphery in most cases (Figure 5E). Furthermore, we could also show continuous transcription at that location in single living cells where nascent RNA could be visualized (Figure 6).
Local changes profoundly modify chromatin at the site of HIV integration, including histone acetylation, the removal of negative factors, the recruitment of positive factors and the phosphorylation of RNA polymerase (Lusic et al, 2003; Marcello et al, 2004). In addition, HIV-1 DNA adopts a circular conformation associated with transcriptional activation (Perkins et al, 2008). However, on activation, such local conformational changes appear to occur at the integrated provirus without a significant modification of the spatial position of the locus.
HIV is a retrovirus that crosses the nuclear pore to integrate into cellular chromatin. The pre-integration complex is found preferentially at the nuclear periphery in decondensed chromatin (Albanese et al, 2008). In most cells, the virus is transcriptionally active, but in some cells it may be subjected to reversible silencing. This latter feature, although infrequent, is critical for the establishment of long-term viral reservoirs in patients undergoing antiviral therapy. From our work, it appears that certain portions of the periphery provide such an environment required for reversible silencing. Analysis of the integration site in latently infected cells show that the provirus is found both in gene-poor and in gene-rich regions (reviewed in Marcello, 2006) and also within transcribed genes as recently described (De Marco et al, 2008; Han et al, 2008; Lenasi et al, 2008). Therefore, it appears that positioning at the nuclear periphery is dominant over the site of integration in determining the latent/inducible state.
To conclude, we propose that HIV-1 integrated close to pericentromeric heterochromatin at the nuclear periphery in quiescent lymphocytes finds a convenient environment to maintain a silent but inducible state. However, it would be important to extend these observations to a wider population of latent cells from infected patients undergoing antiviral therapy to get a general picture of the phenomenon.
Materials and methods
Cells and their characterization
The J-lat cell lines, derivatives of the Jurkat cell line originally developed in Eric Verdin's laboratory, were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. The pro-monocytic U937 cell line U1, which contains two copies of the integrated provirus (Folks et al, 1987; Lusic et al, 2003), and the HOS 143b cells (ECACC n. 91112502) carrying an integrated HIV-1 vector (De Marco et al, 2008) were described earlier. Cells were grown in RPMI 1640 or DMEM medium, 10% fetal calf serum and antibiotics. As expected for tumour cell lines in culture, which are frequently subject to chromosomal abnormalities and polyploidy, we observed that J-lat A1 cells were often polyploid with loss of the original Y chromosome (Schneider et al, 1977). Cytofluorimetric analysis was carried out on untreated cells or on cells treated with tetradecanoyl phorbol acetate (TPA) and analyzed after 15 h. For RT–PCR, total RNA was extracted at the indicated time points as described by the RNeasy Kit (Qiagen). Reverse transcription was performed with M-MLV RT (Moloney Murine Leukemia Virus Reverse Transcriptase) (Invitrogen) using random primers. Amplification was conducted with the indicated primers (Supplementary Table 1).
For the immunoprecipitation, activated cells were lysed and treated with agarose beads conjugated to the anti-flag antibody (Sigma). Membranes were then blotted with the same anti-flag antibody to detect f-Tat and with anti-Cyclin T1 (Santa Cruz Biotechnology).
4C technique
The circular chromatin conformation capture (4C) protocol was established as per published procedures (Zhao et al, 2006).
J-lat A1 cells were maintained in warm fresh medium at 0.5 × 106 cells per ml. After 1 h, TPA (Fluka) was added to the final concentration of 1.6 μM. After 5–8 h, 108 cells were cross-linked on a rocker platform in 40 ml of phosphate-buffered saline (PBS) with 1% formaldehyde at room temperature for 10 min. The reaction was then quenched with 0.125 M of glycine and kept on ice. After two washes with cold PBS, the pellet was resuspended in lysis buffer (10 mM Tris pH 8.0, 10 mM NaCl, 0.2% NP40 and freshly added protease inhibitors) and kept on ice for 10 min. Cell lysis was completed with 10 strokes of Dounce homogenizer (pestle B, Wheaton). Nuclei were pelletized at 600 g for 8 min and washed with 10 ml of 1 × restriction buffer (NEB2, New England Biolabs). Nuclei were resuspended in 712.8 μl of restriction buffer including 0.1% SDS and incubated by shaking for 1 h at 37°C. Triton X-100 was then added to a final concentration of 1.8% and further incubated at 37°C for 1 h before adding 4000 U of HindIII. The reaction was incubated at 37°C overnight by shaking. The restriction enzyme was inactivated by the addition of 2% SDS (final concentration) and incubated at 65°C for 20 min. The digestion mixture was diluted in 8 ml of 1 × T4 ligase buffer (New England Biolabs) including 1% Triton X-100 and incubated at 37°C for 1 h. The digested sample was ligated using 4000 U of T4 DNA ligase (New England Biolabs) for 3 days at 4°C, followed by incubation at 16°C for 5 h and for 1 h at room temperature with additional enzyme and ATP. The reaction was stopped with 10 mM EDTA. RNase (1 μg/ml) of was then added and incubated at 37°C for 30 min, followed by 0.1 mg/ml protein kinase and incubated at 56°C for 30 min. Finally, cross-links were reversed by incubation at 65°C overnight. The DNA was purified by 2 × phenol/chloroform, 2 × chloroform extraction and ethanol precipitation.
In some experiments, the purified DNA was digested with a restriction enzyme (NotI) that cuts between the inverse PCR primers to eliminate products from self-ligated templates. The DNA samples were then amplified by nested PCR. The high-fidelity AccuPrime Pfx (Invitrogen) was used for first and second round amplification. The PCR conditions were as follows: 94°C for 2 min followed by 35 cycles at 94°C for 30 s, 58°C for 30 s and 72°C for 30 s, followed by one cycle at 72°C for 2 min. For the second round, the PCR conditions were as follows: 94°C for 2 min followed by 35 cycles at 94°C for 30 s, 62°C for 30 s and 72°C for 30 s, followed by one cycle at 72°C for 2 min. The primers used are shown in Supplementary Table 1.
The PCR products were either resolved on agarose gels and the fragments cloned individually or bulk cloned into Zero Blunt TOPO PCR Cloning Kit (Invitrogen) and analyzed by DNA sequencing.
To confirm the interaction, we performed 3C assays following the protocol of 4C technique described previously. With a primer on Ch12q12 (found by 4C) and the primers on the provirus, a hemi-nested PCR was performed following the same conditions as in 4C PCR. The primers used are shown in Supplementary Table 1. As a positive control, we used internal amplification of the provirus using Nuc1-B177 and splice-Intro primers (see sequence in Supplementary Table 1).
FISH
Three-dimensional FISH (3D-FISH) was performed essentially as described in Solovei et al (2006). Briefly, freshly grown J-lat A1 cells were resuspended at 3 × 106 cells per ml in 50% serum and seeded for 20 min on glass coverslips that were previously treated with polylysine. The cells were then fixed in 4% paraformaldehyde in 0.3 × hypotonic PBS for 10 min, permeabilized with Triton X-100 for 10 min and left in PBS with 20% glycerol for 1 h overnight. The next day, the coverslips with the cells were subjected to three cycles of liquid nitrogen in glycerol, treated with 0.1 N HCl for 10 min and incubated in 0.002% of Pepsin in 0.01 N HCL at 37°C for 8 min, and then washed in PBS followed by RNase A (200 μg/ml) treatment. The coverslips were then equilibrated in 50% formamide, 2 × SSC for 1–2 h.
Bacterial artificial chromosome (BAC) DNA containing the regions under study, ChXp21.1 and Ch12q12 (RP11-77013 and RP11-379CZ4, respectively), and the DNA probe specific for Ch12 α-satellite repeats (Baldini et al, 1990) were obtained from Dr Mario Rocchi (University of Bari, Italy). The pan-satellite repeats probe p82H, which is specific for the centromeric regions of all human chromosomes (Mitchell et al, 1985), was obtained from Wendy Bickmore (MRC Human Genetics Unit, Edinburgh, Scotland). Probes were conjugated to digoxigen-11-dUTP and labelled with α-digoxigenin fluorescein (Roche) or tetramethyl-rhodamine-5-dUTP (Roche). The provirus was conjugated to biotin-16-dUTP (Roche) and labelled with the TSA Cyanine 5 System (Perkin-Elmer). The incorporation of modified dUTPs for all samples was obtained by nick translation (Roche), and unincorporated nucleotides were removed by gel filtration using a MicroSpin G-25 Column (GE Healthcare). The probes were then resuspended into the hybridization mix and applied to the slides. The coverslips were immobilized with rubber cement and left to hybridize at 37°C in a humid chamber for at least 2 days. Washes and antibody staining were performed using standard techniques. The coverslips were then mounted in Vectashield (Vector Labs).
Fluorescent images of fixed cells were captured on a Zeiss LSM 510 META confocal microscope (Carl Zeiss Microimaging) with a × 63 NA 1.4 Plan-Apochromat oil objective. The pinhole of the microscope was adjusted to obtain an optical slice of <1.0 μm for any wavelength acquired. Samples labelled with digoxigenin-11-dUTP were excited with the 488 nm line of the Ar laser and its emission was monitored using a custom-made Meta band pass filter between 510 and 563 nm. Biotin/TSA-Cy5 System and tetramethyl-rhodamine-5-dUTP fluorophores were excited using the 543 nm HeNe laser and their emission collected using a custom-made Meta band pass filter between 552 and 670 nm.
Z-Stacks of images were analyzed with a Zeiss LSM Image Examiner. The shortest distance from the centres of signal intensity between FISH spots (gene locus) or from the spot to the nuclear periphery (defined by a sharp drop in stain) was measured using the Ortho tool of the LSM510 software with the 3D distance display mode. The periphery data were normalized using the maximum diameter of each cell.
RNA FISH on HOS_A4 cells has been performed as described (De Marco et al, 2008). For live cell experiments, cells were plated on glass-bottom plates (MatTek), transiently transfected with Lipofectamine with plasmids expressing the Tat transactivator and EYFP-MS2nls and analyzed next day at 37°C in a 5% CO2 humidified atmosphere in a non-fluorescent complete DMEM medium. Stacks of 21 planes were acquired at bin=1 with steps of 0.5 μm in the z axis using a wide-field Leica DMRI inverted microscope ( × 63 objective, NA 1.3) controlled by Metamorph (Universal Imaging) and equipped with a conventional light source (Hg, 100 W), a filter cube for YFP detection (Leica Microsystems) and an automated shutter control to minimize exposure of samples to light (Sutter). Digital images were collected using a CoolSnap K CCD camera (Roper Scientific). The 3D deconvolution and reconstruction was performed with the ImageJ plug-in ‘Iterative Deconvolve 3D' (http://rsb.info.nih.gov/ij/).
Cells treated as described for 3D-FISH at the 20% glycerol step were blocked with 5% horse serum in PBS for 1 h at 37°C for immunofluorescence. As a primary antibody, we used the rabbit antiserum for trimethyl-histone H4 Lys20 (H4K20me3) (cat. no. 07-463, Upstate). The antibody was diluted 1:500 in 5% horse serum and left for 2 h at 37 °C. Anti-rabbit Alexa Fluor 594 (Invitrogen) was used as the secondary antibody. The proteins were cross-linked with EGS of 0.5 M (diluted 1:500; Sigma, E-3257) for 10 min at room temperature. Optical sectioning of fixed cells were captured on a Zeiss LSM 510 META confocal microscope as described above.
Statistical analysis
The statistical analysis of the data was performed with R software (http://www.r-project.org/).
The distribution of distances between chromosomes (Figure 3B–D) was analyzed with the MCLUST package (http://www.stat.washington.edu/mclust/). The parametric method, Bayesian Information Criterion (BIC), was used as a useful statistical criterion for model selection to determine the number of components in a Gaussian mixture model. The value of BIC allowed the distinction between a single Gaussian (Figure 3C and D) and the sum of two Gaussians (Figure 3B). The two Gaussians in Figure 3B were centred at 0.6 and 2.8 μm, with a standard deviation of 0.5 and 1 μm, respectively. The mean and standard deviation of the curve in Figure 3C were 2.7 and 1 μm and in Figure 3D were 3.3 and 2 μm.
The differences in spatial associations measured in Figures 3E and 4B and D were analyzed with the χ2-test and were always significative (significance level set at χ2, P<0.05).
The differences between the distribution of distances from the provirus to the periphery in induced and not induced cells (Figure 5D) were analyzed using the Kolmogorov–Smirnov test (K–S test) and were not significative (significance level set at P<0.01). The K–S test is a non-parametric and distribution-free statistical test that determines whether two data sets differ significantly.
The distributions of distances of the provirus from the periphery for different cell lines compared with the J-lat A1 distribution (Figure 5E) were also analyzed with a K–S test (significance level set at P<0.01).
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Edouard Bertrand (IGMM, Montpellier, France) for the gift of reagents and useful suggestions; Eric Verdin (Gladstone Institute, San Francisco, USA) and Albert Jordan (Centre for Genomic Regulation, Barcelona, Spain) for plasmid pEV731; Vanna Pecile (IRCCS Burlo Garofolo, Trieste, Italy) for cytogenetic analysis of J-lat A1 cells; Mario Rocchi (University of Bari, Italy) for providing BACs; Wendy Bickmore (MRC Human Genetics Unit, Edinburgh, Scotland) for reading the paper and for the pan-α-satellite probe; and Luis Parada (CIC bioGUNE, Derio, Spain) for useful hints on FISH. This work was supported in part by an HFSP Young Investigators Grant, by the Italian FIRB program of the ‘Ministero dell'Istruzione, Università e Ricerca' of Italy and by the AIDS Program of the ‘Istituto Superiore di Sanità' of Italy.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albanese A, Arosio D, Terreni M, Cereseto A (2008) HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS ONE 3: e2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 [DOI] [PubMed] [Google Scholar]
- Baldini A, Rocchi M, Archidiacono N, Miller OJ, Miller DA (1990) A human alpha satellite DNA subset specific for chromosome 12. Am J Hum Genet 46: 784–788 [PMC free article] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E (2007) The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T (2005) Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Branco T, Ramirez F, Pombo A (2008) Changes in chromosome organization during PHA-activation of resting human lymphocytes measured by cryo-FISH. Chromosome Res 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG (2001) Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol 3: 602–606 [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG (1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell 3: 207–217 [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91: 845–854 [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18: 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS (2006) Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA (2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S (2006) Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol 18: 307–316 [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S (1996) Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381: 529–531 [DOI] [PubMed] [Google Scholar]
- De Marco A, Biancotto C, Knezevich A, Maiuri P, Vardabasso C, Marcello A (2008) Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 5: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW (1996) Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85: 745–759 [DOI] [PubMed] [Google Scholar]
- Dietzel S, Zolghadr K, Hepperger C, Belmont AS (2004) Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing beta-globin regulatory sequences. J Cell Sci 117: 4603–4614 [DOI] [PubMed] [Google Scholar]
- Dillon N, Festenstein R (2002) Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet 18: 252–258 [DOI] [PubMed] [Google Scholar]
- du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M (2007) Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 26: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS (1987) Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238: 800–802 [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W (2007) Nuclear organization of the genome and the potential for gene regulation. Nature 447: 413–417 [DOI] [PubMed] [Google Scholar]
- Gondor A, Rougier C, Ohlsson R (2008) High-resolution circular chromosome conformation capture assay. Nat Protoc 3: 303–313 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Han Y, Lin YB, An W, Xu J, Yang HC, O'Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF (2008) Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M (2004) Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur J Immunol 34: 3604–3613 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR (1998) Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell 9: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20: 1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM (1987) Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330: 489–493 [DOI] [PubMed] [Google Scholar]
- Kim SH, McQueen PG, Lichtman MK, Shevach EM, Parada LA, Misteli T (2004) Spatial genome organization during T-cell differentiation. Cytogenet Genome Res 105: 292–301 [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H (2002) Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL (2008) A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115 [DOI] [PubMed] [Google Scholar]
- Lassen KG, Bailey JR, Siliciano RF (2004) Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J Virol 78: 9105–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, Contreras X, Peterlin BM (2008) Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R (2006) Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413 [DOI] [PubMed] [Google Scholar]
- Louvet E, Percipalle P (2009) Transcriptional control of gene expression by actin and myosin. Int Rev Cell Mol Biol 272: 107–147 [DOI] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M (2003) Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22: 6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O (2007) Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 26: 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A (2006) Latency: the hidden HIV-1 challenge. Retrovirology 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A, Lusic M, Pegoraro G, Pellegrini V, Beltram F, Giacca M (2004) Nuclear organization and the control of HIV-1 transcription. Gene 326: 1–11 [DOI] [PubMed] [Google Scholar]
- Marcello A, Zoppe M, Giacca M (2001) Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 51: 175–181 [DOI] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW (1996) Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell 7: 825–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM, Colomb S, Dejardin J, Bantignies F, Cavalli G (2006) Polycomb group-dependent Cyclin A repression in Drosophila. Genes Dev 20: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Bourachot B, Rachez C, Ogryzko V, Muchardt C (2008) Regulation of an inducible promoter by an HP1beta–HP1gamma switch. EMBO Rep 9: 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Amoils S, Roldan E, Rahemtulla A, O'Connor E, Fisher AG, Brown KE (2004) Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J Exp Med 200: 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Mitchell AR, Gosden JR, Miller DA (1985) A cloned sequence, p82H, of the alphoid repeated DNA family found at the centromeres of all human chromosomes. Chromosoma 92: 369–377 [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Hudson LD, Armstrong RC (2002) Nuclear organization in differentiating oligodendrocytes. J Cell Sci 115: 4071–4079 [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W (2008) Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet 4: e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T (2004) Tissue-specific spatial organization of genomes. Genome Biol 5: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ (2000) A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res 28: 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ (2008) Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell 29: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS (1998) The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol 142: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Schwenk HU, Bornkamm G (1977) Characterization of EBV-genome negative ‘null' and ‘T' cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer 19: 621–626 [DOI] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM (2007) Gene regulation through nuclear organization. Nat Struct Mol Biol 14: 1049–1055 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368 [DOI] [PubMed] [Google Scholar]
- Solovei I, Schermelleh L, During K, Engelhardt A, Stein S, Cremer C, Cremer T (2004) Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma 112: 410–423 [DOI] [PubMed] [Google Scholar]
- Solovei I, Walter J, Cremer M, Schermelleh L, Cremer T (2006) FISH on three-dimensional preserved nuclei. In FISH, A Practical Approach, Beatty B, Mai S, Squire J (eds), pp 119–157. Oxford, UK: Oxford University Press. [Google Scholar]
- Spector D.L. (2003) The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72: 573–608 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA (2005) Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM (2006) Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778 [DOI] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Ott M, Verdin E (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 15: 1112–1120 [PMC free article] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D (2000) Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci 113(Part 9): 1565–1576 [DOI] [PubMed] [Google Scholar]
- Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I (2003) Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res 11: 485–502 [DOI] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, Fisher AG (2006) Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci 119: 132–140 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL (2002) Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol Cell Biol 22: 4876–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D (2004) Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol 166: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File