Abstract
While the contribution of specific tumor suppressor networks to cancer development has been the subject of considerable recent study, it remains unclear how alterations in these networks are integrated to influence the response of tumors to anti-cancer treatments. Here, we show that mechanisms commonly used by tumors to bypass early neoplastic checkpoints ultimately determine chemotherapeutic response and generate tumor-specific vulnerabilities that can be exploited with targeted therapies. Specifically, evaluation of the combined status of ATM and p53, two commonly mutated tumor suppressor genes, can help to predict the clinical response to genotoxic chemotherapies. We show that in p53-deficient settings, suppression of ATM dramatically sensitizes tumors to DNA-damaging chemotherapy, whereas, conversely, in the presence of functional p53, suppression of ATM or its downstream target Chk2 actually protects tumors from being killed by genotoxic agents. Furthermore, ATM-deficient cancer cells display strong nononcogene addiction to DNA-PKcs for survival after DNA damage, such that suppression of DNA-PKcs in vivo resensitizes inherently chemoresistant ATM-deficient tumors to genotoxic chemotherapy. Thus, the specific set of alterations induced during tumor development plays a dominant role in determining both the tumor response to conventional chemotherapy and specific susceptibilities to targeted therapies in a given malignancy.
Keywords: Cancer, ATM, p53, DNA-PK, chemotherapy, mouse models
Following DNA damage, cells activate distinct signaling networks that mediate cell cycle checkpoints, DNA repair, and apoptosis. These pathways and networks are highly interconnected through complex nonlinear relationships (Sancar et al. 2004), but how they function together at the systems level is poorly understood. The selective sensitivity of cancer cells to DNA-damaging chemotherapy suggests that interconnections between cell cycle checkpoint and survival pathways may be altered in tumors. These network-level differences could be used to specifically enhance the killing of tumor cells.
One of the master regulators of the DNA damage response is the protein kinase ATM, which recruits and phosphorylates a variety of proteins involved in all three of these DNA damage responses—DNA repair, cell cycle regulation, and programmed cell death—through downstream targets such as H2AX, MDC1, Rad50, Nbs1, Chk2, p53, and MDM2 (Matsuoka et al. 1998, 2007; Khosravi et al. 1999; Rotman and Shiloh 1999; Hirao et al. 2000; Lim et al. 2000; Schultz et al. 2000; Burma et al. 2001; Kim et al. 2002; Goldberg et al. 2003; Verdun et al. 2005; Harrison and Haber 2006). Primary cells from ataxia telangiectasia (A-T) patients and ATM knockout mice are hypersensitive to ionizing radiation (IR) and chemotherapy-induced DNA double-strand breaks (DSBs) (Shiloh et al. 1983; Barlow et al. 1996; Chun and Gatti 2004). Consistent with this idea, ATM-deficient tumors have been shown to be sensitive to DNA DSB-inducing cancer treatments (Tribius et al. 2001). Thus, it has been proposed that ATM inhibition might provide a general strategy to sensitize tumors to the cytotoxic effects of DNA-damaging therapies by preventing the execution of critical survival programs, such as activation and maintenance of cell cycle checkpoints and initiation of DNA repair (Hickson et al. 2004; Helleday et al. 2008). Small molecule-based ATM inhibitors have been shown to sensitize cancer cells to DNA-damaging agents and are currently under investigation for their use as DSB-sensitizing agents for anti-cancer therapy.
Given this preclinical data, it is surprising that, while some reports show loss of ATM in tumors correlates with beneficial clinical outcomes, a large number of studies show the exact opposite result. In fact, certain studies suggest that loss of ATM may correlate with actual resistance to DNA-damaging chemotherapy and poor patient survival (Haidar et al. 2000; Ripolles et al. 2006; Austen et al. 2007). Similarly, loss of p53, a known ATM target, has also been shown to correlate with both favorable and poor prognoses (Rahko et al. 2003; Bertheau et al. 2007). Thus, the status of either of these tumor suppressor genes alone appears to be insufficient to predict therapeutic outcome. Furthermore, there are significant differences between the p53 and ATM loss-of-function phenotypes in cells, precluding any simple epistatic relationship between these two prominent tumor suppressor genes.
p53 signaling contributes to two major cellular responses following DNA damage—cell cycle arrest and apoptosis (Vogelstein et al. 2000). However, it remains largely elusive which cues dictate the choice between p53-mediated cell cycle arrest and apoptosis. Here we provide genetic and biochemical evidence using a diverse collection of cell lines in culture, two murine cancer models, and human clinical data that ATM functions to direct the p53 response toward a dominant apoptotic outcome in tumor cells after genotoxic stress. In cells and tumors that lack a functional p53 pathway, ATM inactivation is sufficient to globally sensitize them to killing by genotoxic chemotherapy, demonstrating a synthetic lethal interaction between these two tumor suppressor genes.
Further investigation into the binary switch-like role of ATM in modulating p53-directed proapoptotic versus procytostatic signaling led to the discovery of an additional synthetic lethal interaction between ATM and DNA-PKcs, the two kinases that control the major DSB repair pathways—homologous recombination (HR) and nonhomologous end-joining (NHEJ), respectively. We show that depletion of DNA-PKcs can resensitize inherently chemoresistant ATM-deficient p53-containing tumors to DNA DSB-inducing anti-cancer drugs in vitro and in vivo. These findings imply that ATM-deficient cancer cells become addicted to DNA repair via the NHEJ pathway to survive DNA DSBs. Thus, by correlating ATM, Chk2, p53, and DNA-PKcs status in human tumors, our study establishes a framework for predicting the effect of ATM mutation or pharmacological inhibition on patient survival, and provides a new therapeutic strategy to sensitize inherently therapy-resistant tumors that show loss of ATM but contain intact p53.
Results
ATM acts as a binary switch by regulating the ability of p53 to induce cell death after chemotherapy
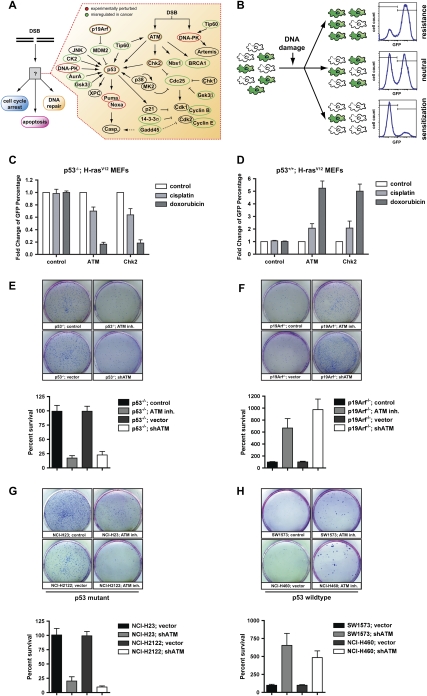
To functionally dissect the contribution of core components of the DNA damage signaling network (Fig. 1A) for cancer cell responses to genotoxic chemotherapy, we used a retroviral RNAi approach in conjunction with 16 different murine and human cell lines and two independent mouse models of cancer. Our retroviral transfer vector encoded for miR30 embedded shRNAs and GFP coexpression to tag shRNA-expressing cells. We assayed cellular survival in a GFP-based competition assay (Fig. 1B), in which the ratio of GFP-positive and GFP-negative cells was scored by flow cytometry before and after exposure to DNA-damaging chemotherapeutics. In this assay, the change in the relative abundance of GFP-positive, shRNA-expressing cells in a partially transduced population after DNA damage is a direct reflection of relative cellular proliferation and survival. Retroviral transduction of shRNAs targeting ATM and Chk2 led to stable suppression of target genes in murine embryonic fibroblasts (MEFs) (Supplemental Fig. 1A), without affecting cellular proliferation and survival (data not shown). We initially analyzed the effects of ATM depletion by comparing p53-proficient and p53-deficient MEFs transformed with H-rasV12. For p53-proficient tumor cells, we chose to use p19ARF−/−;H-rasV12 MEFs, since they allow for bypass of Ras-induced senescence, while retaining an intact p53 response following DNA damage (Stott et al. 1998). When p53-deficient H-rasV12 MEFs were exposed to either the DNA cross-linking agent cisplatin or the topoisomerase II inhibitor doxorubicin, we observed a robust survival defect in cells expressing an ATM-specific shRNA (Fig. 1C). A similar chemosensitizing effect was observed when the canonical ATM substrate Chk2 was depleted in p53-deficient H-rasV12 MEFs (Fig. 1C). In striking contrast, when ATM or Chk2-specific shRNAs were expressed in p53-proficient H-rasV12 transformed MEFs, the exact opposite effect was seen (Fig. 1D). In the context of functional p53, depletion of ATM or Chk2 resulted in a substantial survival benefit in shRNA-expressing cells following cisplatin and an even more pronounced survival benefit after doxorubicin. Similar results were obtained in long-term colony survival assays using either RNAi or the well-characterized ATM inhibitor KU-55933. In these experiments, cells expressing an ATM shRNA, or cells pretreated with KU-55933 for 30 min, were exposed to doxorubicin for 4 h before replating 5 × 103 cells into fresh media. Surviving colonies were counted 2 wk later (Fig. 1E,F). In p53-deficient MEFs, ATM abrogation strongly reduced the number of surviving colonies, while in p53-proficient MEFs, ATM abrogation resulted in substantially more surviving colonies when compared with their respective controls (Fig. 1E,F). In an additional line of experimentation, we analyzed the effects of RNAi-mediated ATM depletion in cell lines derived from two well-established mouse models of lung adenocarcinoma. ATM depletion in tumor cells derived from lung adenocarcinomas that lost both alleles of p53 and expressed oncogenic K-rasG12D from its endogenous promoter (Jackson et al. 2005) resulted in a substantial increase in the sensitivity to doxorubicin (Supplemental Fig. 2A,B). In contrast, ATM depletion in tumor cells derived from K-rasG12D-driven lung adenocarcinomas with retained p53 function (Johnson et al. 2001) resulted in resistance to the cytotoxic effects of doxorubicin treatment (Supplemental Fig. 2C,D). These data suggest that p53-mediated cell death in response to DNA-damaging chemotherapy depends on the presence of functional ATM signaling.
Figure 1.
Functional integration of the ATM–Chk2 and p53 pathways determines the in vitro response to genotoxic chemotherapy. (A) Core components of the DNA damage response examined in this study. (B) Schematic representation of the GFP enrichment assay. Following drug treatments, the surviving population was assayed by flow cytometry to determine the percentage of GFP/shRNA-expressing cells. (C) Suppression of ATM and Chk2 in H-rasv12;p53−/− MEFs sensitizes cells to doxorubicin (1 μM) and cisplatin (1 μM). Bars indicate the mean of three experiments, with error bars indicating standard deviation. (D) Suppression of ATM and Chk2 in H-rasv12;Arf−/− MEFs confers resistance to doxorubicin (1 μM) and cisplatin (1 μM) in vitro (n = 3). Cells were treated as in C. (E–H) Clonogenic survival assays. (E) ATM inhibition (10 μM KU-55933 30 min prior to doxorubicin, top row) or shRNA-mediated ATM depletion (bottom row) results in reduced long-term survival in doxorubicin-treated (1 μM), p53-deficient MEFs. The bottom panel is a quantification of the results shown. (F) ATM inhibition or shRNA-mediated ATM depletion (as in E) confers a long-term survival benefit in doxorubicin-treated (1 μM), p53-proficient MEFs. (G) p53 mutant human cancer cells show increased doxorubicin sensitivity when ATM is inhibited. Cells were pretreated with KU-55933 (10 μM) for 30 min and colony survival assays were performed as in E. (H) p53-proficient human cancer cells show increased doxorubicin resistance when ATM is inhibited. Cells were treated as in G. Assays were performed in triplicate for each condition and representative images are shown.
The depletion of ATM or Chk2 shRNA-expressing populations that we observed in p53-deficient cells could be the result of reduced proliferation, or cell death. To evaluate the difference between p53+/+ and p53−/− cells, we stained MEFs with antibodies against cleaved caspase-3 and analyzed them by flow cytometry. While ATM- and Chk2-depleted p53−/− MEFs showed increased apoptosis after DNA-damaging chemotherapy when compared with control cells (Supplemental Fig. 3A–C,G), the opposite was true for p53-proficient MEFs (Supplemental Fig. 3D–F,H). These cells appeared to be protected from DNA damage-induced apoptosis, as indicated by a significant reduction of apoptotic cells when compared with control cells. Together these data indicate that the combined loss of p53 and ATM/Chk2 is synthetic lethal in the context of DNA-damaging chemotherapy, while the inactivation of each of these genes in isolation confers chemoresistance.
To further rule out any specific effects of ARF deficiency or murine biology on therapeutic response, we examined the effects of ATM inhibition in a diverse panel of human cancer cell lines using colony formation assays (Fig. 1G,H; Supplemental Fig. 4). In MDA-MB-453 (breast cancer) and SW1573 and NCI-H460 (both lung cancer) cell lines, which are p53-proficient, ATM inhibition significantly protected cells from doxorubicin-induced cell death. In contrast, in HT-29 (colon cancer), A431 (epithelial cancer), NCI-H23 (lung cancer), and NCI-2122 (lung cancer) cell lines, all of which carry mutations in p53, ATM inhibition resulted in a dramatically reduced number of surviving colonies (Fig. 1G,H; Supplemental Fig. 4). Given the genetic diversity of these human cancer cell lines, these results suggest a dominant role for ATM/Chk2 in regulating the ability of p53 to modulate cell death after DNA-damaging chemotherapy.
Inhibition of ATM promotes drug resistance in p53-proficient tumor models
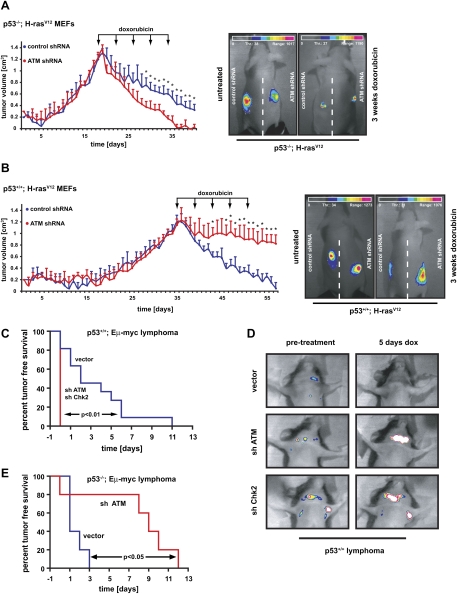
Next, we sought to determine whether ATM similarly affected the impact of p53 on therapeutic response to chemotherapy in vivo. The effect of ATM suppression on chemosensitivity in transformed H-Rasv12 MEFs was examined in vivo using a nude mouse allograft model. Cells (106) were injected into the flanks of NCRnu/nu mice and resulting tumors were allowed to grow to ∼1 cm in diameter before treatment with doxorubicin was administered as indicated in Figure 2A,B. Consistent with the in vitro phenotypes seen in cell culture, suppression of ATM in H-Rasv12;p53−/− tumors strongly sensitized these tumors to the cytotoxic effects of doxorubicin. While tumors arising from cells expressing a control shRNA showed a moderate reduction in volume after five treatment cycles, tumors expressing an ATM-specific shRNA displayed a dramatically increased sensitivity, mirrored by a significant reduction in tumor volume (Fig. 2A). This observation further strengthens the notion of a synthetic lethal interaction between p53 and ATM in the context of DNA-damaging chemotherapy.
Figure 2.
The in vivo effect of ATM and Chk2 pathway abrogation on tumor chemosensitivity is strictly dependent on p53 status. (A) ATM depletion sensitizes p53-deficient tumors to the cytotoxic effects of doxorubicin in vivo. H-RasV12-transformed p53−/− MEFs expressing either a control shRNA (blue) or ATM-specific shRNA (red) were subcutaneously injected into the flanks of NCRnu/nu mice (n = 4 for each experimental group). Arrows indicate the timing of intraperitoneal (i.p.) doxorubicin administrations. Asterisks indicate significant size difference (Student's t-test, two-tailed, P < 0.05). (Right panels) Maintenance of GFP expression in tumors was verified at the termination point. (B) Depletion of ATM in tumors arising from Arf−/−;H-RasV12 MEFs confers resistance to doxorubicin in vivo (n = 4 for each experimental group). Animals were subcutaneously injected with Arf−/−;H-RasV12 MEFs expressing either a control shRNA (blue) or an ATM-specific hairpin (red). Doxorubicin was administered i.p. at the indicated times and tumor growth was monitored as in A. (Right panels) Maintenance of GFP expression in tumors was verified at the termination point. (C) Suppression of ATM or Chk2 in Eμ-Myc;Arf−/− mouse lymphoma confers resistance to doxorubicin in vivo. Lymphoma cells were transduced with vector control, shATM, or shChk2; sorted for GFP; and injected into recipient mice. Resulting lymphomas were treated with 10 mg/kg doxorubicin. Tumor-free survival displayed in Kaplan-Meier format. n = 10 for shATM and shChk2; n = 11 for vector control. (D) Representative images of lymphoma burden before treatment and 5 d post-therapy. (E) ATM depletion sensitizes p53-deficient lymphomas to the cytotoxic effects of cyclophosphamide in vivo. p53-null lymphomas were transduced and injected as in C. n = 8 for both vector and shATM.
Similar to what was observed in vitro, suppression of ATM strongly protected tumors arising from H-Rasv12;p53+/+ MEFs from the cytotoxic effects of doxorubicin in vivo. While control shRNA-expressing tumors responded favorably to treatment with five cycles of doxorubicin, tumors arising from ATM-depleted cells showed only a minimal reduction in volume at the end of the doxorubicin course (Fig. 2B). To eliminate the possibility of tumor/cell-type specificity in the observed chemotherapeutic resistance of p53-proficient tumors, a second mouse model using Eμ-Myc;p53+/+ or Eμ-Myc;p53−/− lymphoma cells expressing an ATM shRNA was examined. Transduced Eμ-Myc;p53+/+ cells were injected into recipient mice, which were treated following lymphoma onset and monitored for tumor-free survival. As shown in Figure 2, C and D, tumors formed from vector-transduced cells responded favorably to doxorubicin, with 90% of mice bearing these tumors showing complete tumor regression after treatment. In contrast, ATM- or Chk2-deficient tumors responded very poorly to treatment. At 5 d after doxorubicin, most ATM- or Chk2-deficient, but p53-proficient, tumors increased in size, and none of the 20 mice bearing these tumors exhibited complete remission (Fig. 2C,D).
As seen in transformed p53−/− MEFs, down-regulation of ATM in Eμ-Myc;p53−/− tumors strongly sensitized these tumors to the cytotoxic effects of genotoxic chemotherapy (Fig. 2E). While tumors arising from cells expressing a control vector showed a transient response to therapy, tumors expressing an ATM-specific shRNA displayed a dramatically increased therapeutic sensitivity with a significant extension of tumor-free survival. These two different murine tumor models directly confirm the central role of ATM as a binary switch that dictates the effect of p53 activation on tumor response to chemotherapy in vivo.
The combined status of p53 and ATM is a key determinant of the human clinical response to chemotherapy
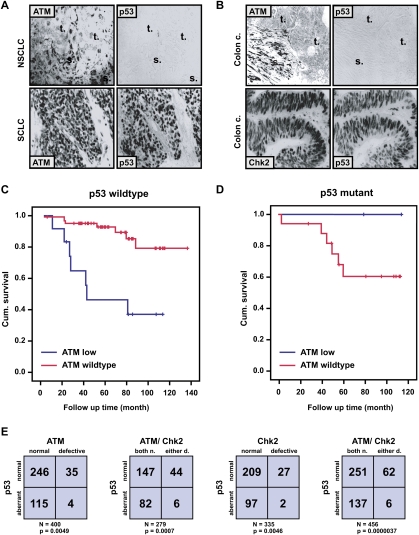
Our cell culture and mouse model data strongly suggest that, despite the molecular complexity of DNA damage response networks, a simple stratification based solely on the combined status of the ATM–Chk2 pathway and the p53 apoptotic network might help determine the response of human cancer patients to genotoxic chemotherapy. Our in vitro and in vivo data suggest that ATM inhibition is likely to benefit only a select group of cancer patients; i.e., those with tumors that have lost p53. In contrast, tumors with preserved ATM and p53 function might become resistant to the effects of DNA-damaging chemotherapy when simultaneously treated with ATM inhibitors. To evaluate this, we examined the 10-yr cumulative survival of 93 breast cancer patients for whom tissue samples and clinical response data were available. The presence of normal levels of both ATM and p53 in tumors (defined as detectable levels of ATM and nonstabilized p53 by immunohistochemistry [IHC]) (; Fig. 3A,B) correlated with favorable prognosis (Fig. 3C; Supplemental Fig. 5). Similarly, the aberrant decrease or absence of detectable ATM staining in the presence of wild-type p53 levels correlated with a significantly reduced cumulative patient survival rate (Fig. 3C; Supplemental Fig. 5). Finally, the presence of both ATM and p53 alterations in single tumors, although quite rare (Fig. 3E), invariably correlated with an excellent prognosis (Fig. 3D). Importantly, the incidence of aberrant ATM and p53 expression detected by IHC in our cohort is very similar to the incidence of ATM and p53 mutation reported recently in a human cancer genome sequencing effort, further validating our IHC-based approach (Ding et al. 2008).
Figure 3.
Combined ATM and p53 status is a key determinant for survival of breast cancer patients treated with DNA-damaging chemotherapy. (A,B) Examples of ATM, p53, and Chk2 aberrations in human tumors. Immunohistochemical staining with antibodies against the indicated proteins, showing parallel sections from a large-cell lung carcinoma (NSCLC, with loss of ATM but normal p53), small-cell lung carcinoma (SCLC, with normal ATM pattern but overabundant, mutant p53), and two colon carcinomas—one with loss of ATM and normal p53, the other with normal Chk2 but aberrant p53. The letters “t” and “s” on the images indicate the positions of tumor nests and stromal cells, respectively, to highlight the selective absence of ATM in the carcinoma cells. (C,D) Kaplan-Meier curves showing overall survival in 93 breast cancer patients (omitting those treated with IR). (C) ATM deficiency in a p53 wild-type background correlates with poor patient survival relative to tumors with wild-type ATM and p53 (P = 0.0059). (D) Relative survival of patients with tumors showing ATM deficiency on a p53 mutant background versus mutant p53 alone (P = 0.32). (E) The combined inactivation of ATM/Chk2 and p53 is underrepresented in human cancers. (Far left) ATM versus p53 status in 400 tumors examined for both proteins. (Middle left) ATM/Chk2 versus p53 status in 279 tumors examined for all three proteins. (Middle right) Chk2 versus p53 status in 335 tumors examined for both proteins. (Far right) ATM/Chk2 versus p53 status in 456 tumors examined for p53 and either ATM or Chk2 status.
These results indicate that the combined evaluation of only two commonly mutated cancer genes, ATM and p53, can facilitate the prediction of therapeutic outcome in cancer patients treated with DNA-damaging chemotherapy, and further implies that the clinical use of ATM inhibitors should be limited to those patients carrying p53-deficient tumors with retained ATM function.
ATM signaling is required specifically for the induction of p53-dependent apoptosis, but not for p53-mediated cell cycle arrest
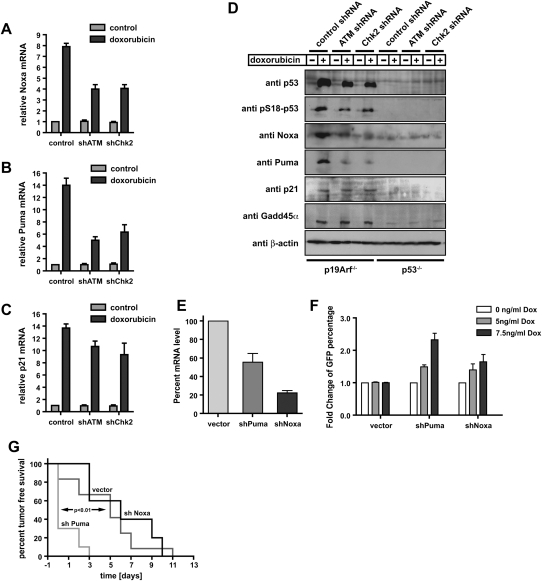
The observation that chemosensitization or chemoresistance following ATM inhibition in cells and tumors was dependent on the functional status of p53 led us to examine the molecular basis for this binary switch-like phenotype. In the presence of p53, we found that ATM signaling is required specifically for the up-regulation of the proapoptotic p53 transcriptional target genes Puma and Noxa. Both ATM and Chk2 knockdown cells showed significant suppression of Puma and Noxa at both the mRNA and protein level after doxorubicin treatment (Fig. 4A–D), indicating that ATM and Chk2 are required for proper p53-mediated activation of an apoptotic program in response to genotoxic chemotherapy. Interestingly, suppression of Puma alone was able to confer chemoresistance both in vitro and in vivo (Fig. 4E–G), suggesting that Puma is a critical effector molecule of the ATM-dependent proapoptotic arm of this p53 response. In marked contrast, the doxorubicin-induced up-regulation of the cell cycle arrest-mediating p53 target genes p21 and Gadd45α was unimpaired in ATM or Chk2 shRNA-expressing cells (Fig. 4C,D). This observation strongly suggests that ATM is specifically required to promote p53-dependent apoptosis, while ATM signaling is dispensable for the induction of a p53-dependent cell cycle arrest.
Figure 4.
Depletion of ATM or Chk2 strongly reduces p53-mediated induction of proapoptotic genes but preserves the induction of p53-dependent cell cycle-regulating genes. (A–D) RNAi-mediated knockdown of ATM or Chk2 strongly reduces doxorubicin-induced Puma and Noxa expression, but only mildly impairs p21 mRNA levels in p53-proficient MEFs. Cells expressing ATM or Chk2-specific shRNAs were treated with 1 μM doxorubicin and mRNA levels of Noxa (A), Puma (B), and p21 (C) were analyzed (n = 3). (D) Depletion of ATM or Chk2 in p53-proficient MEFs abolishes doxorubicin-induced Puma and Noxa protein levels. Protein levels of the p53 target genes p21 and Gadd45α remains largely unchanged in ATM- or Chk2-depleted p53-proficient cells treated with doxorubicin (1 μM). Noxa, Puma, and p21 were not detectable in p53-deficient MEFs, while Gadd45α was present, but at reduced levels. (E) shRNA-mediated suppression of Puma and Noxa. Eμ-Myc cells were transduced with control, shPuma, or shNoxa shRNAs; sorted with for GFP expression; and treated with 15 ng/mL doxorubicin for 12 h. mRNA levels of Puma and Noxa were analyzed by RT–PCR (n = 3). (F) Suppression of Puma and Noxa in Eμ-Myc cells confers resistance to doxorubicin in vitro (n = 3). Cells were analyzed for GFP percentage after doxorubicin treatment. Bars indicate the mean of three experiments ±SEM. (G) Suppression of Puma, but not Noxa, in lymphomas arising from Eμ-Myc cells confers resistance to doxorubicin in vivo. Experiments were carried out as described in Figure 2C, with n = 10 for shPuma, n = 11 for vector control, and n = 5 for shNoxa.
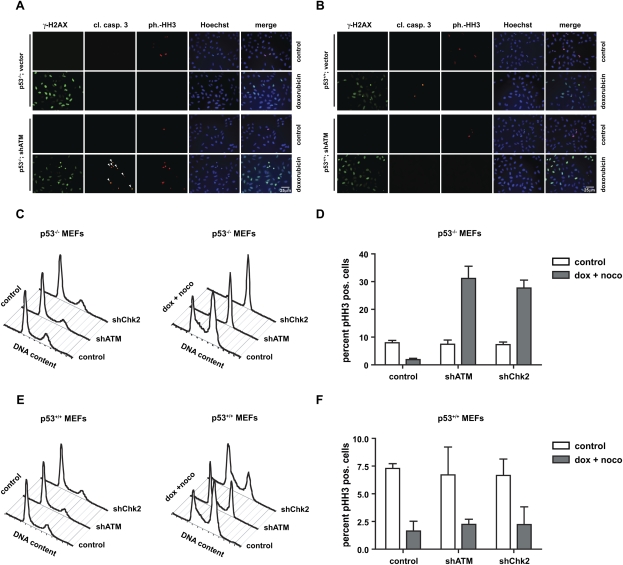
In p53-deficient cells that lack an ATM-dependent p53 apoptotic program, ATM signaling might be redirected to primarily mediate cell cycle arrest; for example, through Chk2/Chk1 modulation of Cdc25 (Bartek et al. 2004). To test this hypothesis, we examined p53-null cells expressing either a control or an ATM-specific shRNA for evidence of mitotic catastrophe, indicated by simultaneous appearance of DNA damage (monitored by γ-H2AX), mitotic markers monitored by phospho-histone H3 (pHH3), and activation of caspase-3. One day following doxorubicin exposure, p53-deficient ATM-expressing cells showed prominent γ-H2AX staining; however, no evidence of mitotic catastrophe was observed as indicated by the absence of cleaved caspase-3 or pHH3 staining. These data suggest that the S/G2/M DNA damage checkpoints remained largely intact in p53-deficient cells (Fig. 5A). In marked contrast, depletion of ATM in p53−/− MEFs resulted in a large fraction of γ-H2AX-positive cells that also stained positive for pHH3 following doxorubicin treatment. Interestingly, these cells appeared to commit to death specifically in mitosis, as no cleaved caspase-3 staining could be observed in cells that did not also stain positively for pHH3 (Fig. 5A). This observation is consistent with mitotic catastrophe, in which cells progress through the cell cycle despite the presence of damaged DNA and activate a mitosis-specific cell death program. Thus, loss of ATM in p53-deficient cells results in fatal DNA damage checkpoint abrogation.
Figure 5.
Depletion of ATM specifically sensitizes p53-deficient MEFs to doxorubicin-induced mitotic catastrophe. p53-deficient (A) and p53-proficient (B) MEFs stably expressing ATM-specific shRNA were either mock-treated or exposed to 1 μM doxorubicin and stained with antibodies detecting γ-H2AX, cleaved caspase-3, pHH3, and Hoechst DNA dye. Costaining with γ-H2AX, cleaved caspase-3, and pHH3 was interpreted as mitotic catastrophe (indicated by arrowheads). (C,D) ATM depletion in p53-deficient MEFs prevents the engagement of a functional G2/M checkpoint following doxorubicin. p53−/− cells expressing control shRNA mounted a robust G2/M arrest in response to nocodazole, as evidenced by an accumulation of 4N cells (monitored by PI staining) and a lack of pHH3 staining. In contrast, ∼30% of ATM-depleted p53-null cells entered mitosis, indicating a bypass of the doxorubicin-induced G2/M arrest in these cells. (E,F) ATM depletion in p53-proficient MEFs does not abrogate the G2/M checkpoint following doxorubicin. Asynchronously growing ATM or control shRNA-expressing p53+/+ MEFs were treated as in C and examined for DNA content by flow cytmoetry. These cells retained a doxorubicin-induced G2/M checkpoint, as evidenced by the accumulation of a largely pHH3-negative 4N population in both the control and ATM shRNA-expressing cells.
When p53+/+;H-rasV12 MEFs expressing either a control vector or ATM-specific hairpins were examined after doxorubicin treatment, we detected positive staining for γ-H2AX in response to doxorubicin (Fig. 5B). None of the cells, however, showed pHH3 staining, indicating the preservation of functional DNA damage despite the absence of ATM. Repression of ATM in these p53-proficient cells strongly reduced the number of cleaved caspase-3-positive apoptotic cells (Fig. 5B; Supplemental Fig. 3D–F). These data further strengthen the notion that p53 retains the ability to mediate a functional cell cycle checkpoint even when the ATM-dependent proapoptotic arm of the p53 response is lost. This could be due to a requirement for ATM to allow proper accumulation of p53, or to promote access of p53 to the promoters of proapoptotic target genes. Alternatively, a parallel pathway that remains intact in the absence of ATM might be responsible for the retained ability of p53 to induce cell cycle regulatory target genes. Additional experiments directly examining cell cycle progression verified that abrogation of ATM/Chk2 signaling in p53−/− cells resulted in cell cycle checkpoint bypass and mitotic entry despite the presence of DNA damage, while cell cycle checkpoints were preserved in ATM- or Chk2-depleted p53-proficient cells (Fig. 5C–F). Thus, p53-dependent cell cycle arrest, but not p53-dependent apoptosis, can occur in the absence of upstream ATM activity. These data suggest that ATM acts as a p53-dependent binary switch, dictating whether the effect of genotoxic chemotherapy is the induction of cell cycle arrest or an apoptotic program.
ATM-deficient tumors can be sensitized to genotoxic chemotherapy by inhibition of NHEJ
Immunostaining of a large panel of different human tumors revealed that ∼10%–20% of tumors show reduced levels of ATM or Chk2 in the presence of normal p53 staining, a value that is in good accordance with ATM–Chk2 mutation frequency determined by recently published tumor genome resequencing efforts (Fig. 3E; Ding et al. 2008). Our data indicate that these tumors will be inherently resistant to DNA-damaging chemotherapy. Thus, we investigated whether ATM deficiency could result in other vulnerabilities in DNA DSB repair that could be therapeutically exploited to enhance the efficacy of targeted chemotherapies.
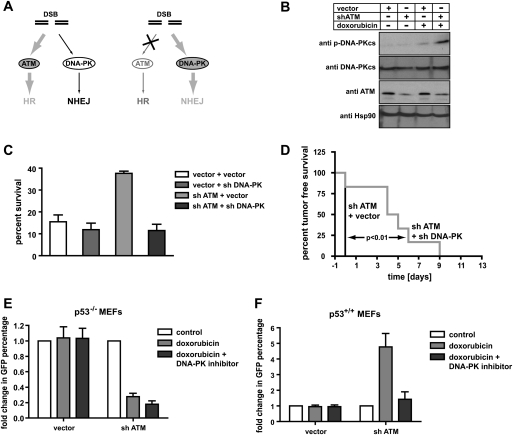
In ATM-deficient p53-proficient cells, doxorubicin-induced DSBs fail to efficiently trigger cell death (Fig. 5; Supplemental Fig. 3D–F). However, repair of these DSBs by either HR or NHEJ is essential for long-term cell survival. It has been suggested that ATM deficiency primarily impairs HR-mediated DSB repair with less effect on NHEJ-mediated DSB repair (Morrison et al. 2000; Yuan et al. 2003; Riballo et al. 2004). Thus, we speculated that NHEJ might become essential for the repair of chemotherapy-induced DSBs in ATM-deficient cancer cells (Fig. 6A). Consistent with compensatory roles for HR and NHEJ in DSB repair, the combined germline deficiency of ATM and DNA-PKcs results in early embryonic lethality (embryonic day 7.5 [E7.5]) (Gurley and Kemp 2001), while single-knockout animals are viable (Xu and Baltimore 1996; Gao et al. 1998). Therefore, we reasoned that drugs that disrupt signaling events important for NHEJ activation should preferentially impact the DSB repair efficiency in ATM-deficient cells and reverse their chemoresistant phenotype. Indeed, doxorubicin-treated ATM-deficient MEFs showed hyperphosphorylation of the essential NHEJ kinase DNA-PKcs on Thr-2606 (corresponding to human Thr-2609) in the autophosphorylation cluster, suggesting enhanced NHEJ activity in the absence of ATM (Fig. 6B).
Figure 6.
A synthetic lethal interaction between ATM and DNA-PKcs in cancer cells. (A) A diagram depicting the DNA repair pathways used for DSB repair. ATM-mediated HR represents a high-fidelity repair mechanism for DSB repair. In the absence of a functional HR pathway, cells rely on the error-prone NHEJ pathway, which requires DNA-PKcs activity. (B) Immunoblot analysis of DNA-PKcs activation in MEFs transduced with an empty vector or ATM-specific shRNA. RNAi-mediated ATM depletion results in increased DNA-PKcs phosphorylation after doxorubicin exposure (1 μM). (C,D) DNA-PKcs suppression resensitizes p53-proficient ATM-depleted Eμ-Myc cells to doxorubicin in vitro (C) and in vivo (D). In C, cells of all four indicated treatments were incubated with 10 ng/mL doxorubicin. The percent cell survival following treatment is shown as mean ± SEM (n = 3). In D, experiments were carried out as described in Figure 2C. n = 8 for both experimental groups. (E,F) Pharmacological inhibition of DNA-PKcs selectively sensitizes p53-proficient ATM shRNA-expressing MEFs to the cytotoxic effects of doxorubicin. MEFs were transduced and treated as indicated and cellular survival was monitored using the flow cytometry-based GFP competition assay. (E) DNA-PKcs inhibition had no significant effect on the survival of p53−/− MEFs or ATM-depleted p53−/− MEFs. (F) DNA-PKcs inhibition selectively sensitized p53+/+ MEFs expressing an ATM shRNA to doxorubicin.
Consistent with a synthetic lethal relationship between DNA-PKcs and ATM in the presence of DSBs, DNA-PKcs suppression sensitized ATM-deficient cells (Supplemental Fig. 1B) to doxorubicin-induced cell death in vitro (Fig. 6C). When tested in vivo, DNA-PKcs and ATM double-knockdown tumors showed a dramatically improved response to doxorubicin when compared with ATM single-knockdown tumors (Fig. 6D). These effects could also be seen in the presence of a pharmacological inhibitor of DNA-PKcs. Using a GFP competition assay, we examined the effect of DNA-PKcs inhibition in the presence or absence of p53 and/or ATM. While ATM suppression sensitized p53−/− MEFs to doxorubicin, DNA-PKcs inhibition had no significant effect on the survival of p53−/− MEFs or p53−/− MEFs expressing an ATM shRNA (Fig. 6E). Conversely, DNA-PKcs inhibition selectively sensitized p53+/+ MEFs expressing an ATM shRNA to doxorubicin (Fig. 6F). This demonstration of a synthetic lethal interaction between ATM and DNA-PKcs in the context of DNA-damaging chemotherapy suggests that the error-prone NHEJ pathway becomes essential for cellular survival after DNA damage in p53 wild-type tumor cells that lack ATM function, and are hence deficient in HR-mediated DSB repair. Thus, while loss of ATM might initially protect cancer cells from the genotoxic effects of oncogenic stress (Halazonetis et al. 2008), by abolishing the proapoptotic p53 response, it ultimately renders these cells susceptible to DNA-PKcs inhibition.
Discussion
We have shown that, in cancer cells retaining functional p53, inhibition of ATM or Chk2 activity promotes a potent chemoresistance phenotype by preventing efficient execution of a p53-dependent apoptotic response. Conversely, ATM suppression directly sensitizes tumors derived from p53−/− MEFs, as well as human p53-deficient cell lines, to chemotherapy. These data indicate that ATM functions as a molecular “binary switch” that channels the effects of p53 signaling toward apoptosis by controlling the relative expression of p53 target genes involved in programmed cell death (Puma and Noxa) relative to those involved in cell cycle arrest (p21 and Gadd45α). In contrast, in p53-deficient tumors, ATM signaling is required for the induction of cell cycle arrest and survival following genotoxic stress. Thus, our data imply that the potential efficacy of ATM inhibitors as chemosensitizing agents is strictly dependent on the status of p53 in target tumors. Similarly, p53 status, as a single factor, fails to predict outcome following chemotherapy in our cohort of patients (P = 0.46) (data not shown). However, in the absence of ATM alterations, p53 status is a significant biomarker of therapeutic outcome (P = 0.05) (Fig. 3D; data not shown), further supporting the notion that the combined status of p53 and ATM allows for a more definitive prediction of the efficacy of genotoxic chemotherapy.
The true incidence of p53 mutations in cancer is likely to have been overestimated based on analysis of cultured tumor cells. We found that most primary tumors, in contrast to cancer cell lines, contain wild-type p53 (Supplemental Fig. 6), suggesting that general use of ATM inhibitors in cancer treatment will generally be contraindicated. Our analysis of publicly available databases revealed that only 32% of primary tumors show p53 mutations (Supplemental Fig. 6; Petitjean et al. 2007)—a number consistent with the 29% seen in human cancers examined in this study. This percentage increased to 69% in tumor cell lines (Wellcome Trust Sanger Institute Cancer Genome Project Web site, http://www.sanger.ac.uk/genetics/CGP), suggesting that p53 mutations are strongly selected in the establishment of cultured tumors. This lower than expected frequency of p53 mutations in primary human cancers argues for caution in the use of Chk2 and ATM inhibitors as adjuncts to standard chemotherapeutic regimens.
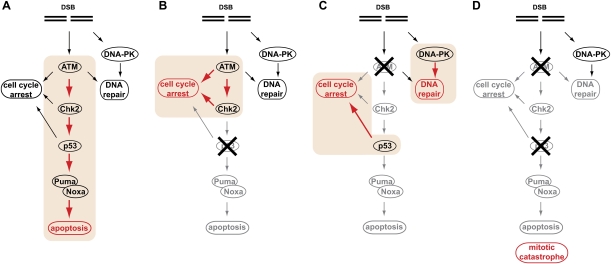
Recent studies have suggested that a DNA damage response similar to that seen following genotoxic chemotherapy may occur in human preneoplastic lesions (Bartkova et al. 2005, 2006; Gorgoulis et al. 2005; Halazonetis et al. 2008). ATM, Chk2, and p53 activation are characteristic of early stages of tumorigenesis, with subsequent checkpoint silencing and escape during tumor progression (Bartkova et al. 2005; Gorgoulis et al. 2005). Our analysis of an extensive set of epithelial malignancies showed an apparent underrepresentation of patients with combined ATM and p53 alterations, suggesting that inactivation of any one of these proteins is sufficient to disable this early tumor checkpoint and render the cells resistant to oncogene-induced DNA damage during malignant development. These data are in line with recent cancer genome sequencing events. Importantly, these data also suggest that tumors may select for DNA damage-resistant genotypes prior to any exposure to anti-cancer therapies, and that the mechanism by which tumor cells evade this early checkpoint may ultimately dictate the most effective treatment regimen for a given tumor (Fig. 7).
Figure 7.
ATM acts as a binary switch to control the contribution of p53 signaling to the DNA damage response. The DNA damage response can be subdivided into three major functional components—cell cycle arrest, DNA repair, and apoptosis. (A) In p53-proficient cancer cells, ATM signaling contributes largely to apoptosis (highlighted in orange). (B) Loss of p53 in tumor cells dramatically reduces apoptosis and redirects ATM signaling to mediate a robust cell cycle (highlighted in orange). In addition, ATM contributes to HR-mediated DSB repair. The net result of this rewired ATM signaling is increased cellular survival in response to DNA damage. (C) Loss of ATM dramatically attenuates apoptotic signaling through p53 and instead promotes a p53-mediated cell cycle arrest (highlighted in orange). ATM-deficient cancer cells expressing functional p53 rely on the DNA-PKcs-mediated NHEJ pathway to repair chemotherapy-induced DSBs (highlighted in orange). Pharmacological abrogation of DNA-PKcs signaling selectively sensitizes ATM-deficient p53-expressing cancer cells to the cytotoxic effects of DNA-damaging chemotherapy. (D) The combined loss of ATM and p53, albeit rare in human tumors, prevents the execution of functional cell cycle checkpoints and promotes death of chemotherapy-treated cancer cells due to mitotic catastrophe. In addition, therapeutic targeting of ATM in p53-deficient cancer cells results in a dramatically increased chemosensitivity of p53-deficient cancer cells.
Our observation of a synthetic lethal interaction between p53 and ATM/Chk2 in the context of DNA-damaging chemotherapy likely also applies to the other two DNA damage effector kinase pathways that establish similar cell cycle checkpoints through inhibiting the Cdc25 family of phosphatases (Busino et al. 2003; Reinhardt et al. 2007). Inhibition of Chk1, for example, has been shown to increase the sensitivity of p53-deficient cancer cells to DNA damage (Mukhopadhyay et al. 2005), while our prior work has shown a similar p53 synthetic lethality with MAPKAP Kinase-2 (MK2) (Reinhardt et al. 2007). Not all of these checkpoint kinases, however, are equivalent drug targets, since disruption of either ATR or Chk1 in mice, for example, results in embryonic lethality (Brown and Baltimore 2000; Liu et al. 2000; Takai et al. 2000). Conditional loss of Chk1 in mammary epithelial tissue is lethal in a homozygous setting and results in progressive DNA damage and uncontrolled mitotic entry in heterozygous animals (Lam et al. 2004), while inhibition of Chk1 by a small-molecule chemical inhibitor in cultured human cells leads to severe stress with catastrophic consequences even in the absence of any exogenous genotoxic insult (Syljuasen et al. 2005). These observations suggest that systemic Chk1 inhibition, in the context of DNA-damaging chemotherapy or radiation therapy, might have severe undesired side effects. In contrast, embryonic lethality was not observed in either the MK2-null or Chk2-null mice (Kotlyarov et al. 1999; Hirao et al. 2002). This observation, in conjunction with the data reported here for ATM/Chk2 and previously for MK2 (Reinhardt et al. 2007), suggests that these kinases are likely to be safer drug targets for chemosensitization of p53-deficient cancer cells than Chk1 or ATR. Whether loss of Chk1 or MK2 in the setting of an intact p53 response results in the profound chemoresistance observed upon loss of ATM/Chk2, however, is not known. Furthermore, there is no data to suggest that MK2 loss is a frequent event in human tumors, in contrast to what is observed with ATM and Chk2.
Our data suggest that analysis of the interplay between the network states of ATM–Chk2-driven cell cycle checkpoints, p53-directed apoptosis, and DNA repair pathways in individual tumors can be used to potentially optimize the personalized treatment of human cancer patients. Despite the complexity of these networks, we made the surprising finding that a simple combinatorial analysis of the DNA damage response, focused on the interface between the ATM–Chk2 pathway and the p53 apoptotic network, can reveal the subset of patients who will likely benefit from an ATM inhibitor, while pharmacological targeting of the error-prone NHEJ pathway is likely to substantially improve the efficacy of DNA-damaging agents in treating inherently chemoresistant ATM-deficient p53 wild-type tumors. Similar DNA damage-based synthetic lethality approaches have been described previously between Brca1/2 and PARP for familial breast cancer (Helleday et al. 2008), which occurs in ∼2% of breast cancer patients, as well as components of the Fanconi anemia pathway and ATM (Kennedy et al. 2007). Importantly, therapies based on ATM mutations have potential efficacy in an estimated 10% of all cancers. Thus, tumor-promoting mutations in ATM underlie both the intrinsic resistance to genotoxic chemotherapy as well as the selective sensitivity to targeted intervention using DNA-PKcs inhibitors in a significant cohort of human cancer patients.
Materials and methods
Cell lines and chemicals
LKR-10 and LKR-13 murine lung adenocarcinoma cell lines were provided by M.S. Kumar. The KP-1A and KPlSIY-5A (Cheung et al. 2008) murine lung adenocarcinoma cell lines were a kind gift from A.F. Cheung. Eμ-Myc;Arf−/− and Eμ-Myc;p53−/− mouse B-cell lymphoma cells were cultured in B-cell medium (50% DMEM and 50% IMDM, supplemented with 10% FBS, L-glutamate, and 5 μM β-mercaptoenthanol). γ-Irradiated NIH 3T3 cells were used as feeder cells. Wild-type, Arf−/−, and p53−/− MEFs, and phoenix, MDA-MB-453, HT-29, A431, NCI-H23, NCI-H2122, SW1573, NCI-H460, LKR-10, LKR-13, KP-1A, and KPlSIY-5A cells were cultured in DMEM plus 10% FBS. Doxorubicin, cyclophosphamide, and the ATM inhibitor KU-55933 were purchased from Calbiochem. For in vivo studies, doxorubicin and cyclophosphamide were dissolved in 0.9% NaCl saline solution.
Antibodies and shRNA constructs
Antibodies against ATM (Genetex), phospho- and total DNA-PKcs (Abcam), β-actin (Sigma), Chk2 (Upstate Biotechnologies and Cell Signaling Technology), p53 (Calbiochem and Cell Signaling Technology), Noxa (Novus), PUMA (Cell Signaling Technology), pHH3 (Upstate Biotechnologies), γ-H2AX (Upstate Biotechnologies), and cleaved caspase-3 (BD Biosciences) were used for immunoblotting and FACS analysis. shRNA constructs targeting mouse ATM, Chk2, Puma, Noxa, and DNA-PKcs were designed and cloned using previously described protocols (Burgess et al. 2008). The hairpin targeting sequences are ATM, 5′-CACGAAGTCCTCAATAATCTA-3′; Chk2, 5′-CAGAAACACATAATCATTAAA-3′; Puma, 5′-CCCAGCCTGTAAGATACTGTA-3′; Noxa, 5′-CAGATTGAATAGTATGTGATA-3′; and DNA-PKcs, 5′-CAGGCCTATACTTACAGTTAA-3′. For ATM, Chk2, and DNA-PKcs, additional shRNAs were constructed and showed similar phenotype (data not shown). An shp53 construct was described earlier (Burgess et al. 2008). After retroviral infection, cells were either directly subjected to GFP competition assays or GFP-sorted for pure population survival assays.
Western blotting
Cells lysates were prepared in 0.5 mL of ice-cold lysis buffer (1% Triton X-100, 25 mM CHAPS, 20 mM Tris-HCl at pH 7.5, 50 mM NaCl, 50 mM NaF, 15 mM Na4P2O7, 2 mM Na3VO4, protease inhibitors) per 10-cm dish for 15 min. Lysates were cleared at 14,000 rpm for 15 min at 4°C, and subsequently subjected to ultracentrifugation (100,000g, 30 min, 4°C). One-hundred microliters of 6× Laemmli buffer were added to the supernatants and protein concentrations were measured using the BCA Protein Assay Kit (Pierce). Alternatively, cell pellets were resuspended in PBS, mixed with 2× SDS sample buffer, and boiled for 15 min at 100°C to generate whole-cell lysates. Each lysate (30∼100 μg of each) was run on 4%–12% SDS-PAGE and transferred to PVDF (Millipore). Membranes were blocked with 5% BSA in 20 mM Tris-HCl (pH 7.5), 137 mM NaCl, and 0.1% Tween-20 and stained with primary antibodies (1:1000 dilution) overnight at 4°C. Membranes were then probed with HRP-conjugated secondary antibody (Amersham or Cell Signaling Technology) at a 1:2000∼1:5000 dilution and visualized by enhanced chemiluminescence (Amersham) on a Kodak Imaging Station (Perkin Elmer). Bands were selected and quantified according to the manufacturer's recommendations.
Competition assays
For MEF experiments, following drug treatment, cells were washed in PBS and trypsinized. Before FACS analysis, the culture supernatant was combined with the trypsinized cells to ensure capture of both floating and adherent cells. Cells were then washed and analyzed on a Becton Dickinson FACScan flow cytometer. We used the FL1 channel for the detection of GFP-labeled retrovirally transduced cells. Dead cells were detected by propidium iodide (PI) (0.05 mg/mL) incorporation. The percentage of GFP was recorded for live cells only. For mouse lymphoma experiments, 106 cells were treated with various drugs at the indicated concentrations. Every 24 h, the cell culture was resuspended by pipetting and half of the culture was replaced with fresh medium. FACS analysis was done at 48 h after the initial treatment.
Clonogenic survival assay
Cells were treated as indicated. After 4 h of treatment, cells were washed three times with growth media and three times with PBS, trypsinized, and replated at a concentration of 5000 cells per 10-cm2 Petri dish. After 14 d, cells were fixed and surviving colonies were stained with 0.1% crystal violet (Sigma-Aldrich). Experiments were performed in triplicate.
In vivo response to chemotherapy
Allograft experiments were performed as described previously (Reinhardt et al. 2007). Eμ-Myc;Arf−/− mouse lymphoma cells were infected with indicated retrovirus and sorted by GFP. Cells (2 × 106) were i.v.-injected into BL/6J mice. Lymphoma burden was monitored by palpation of the axillary and brachial lymph nodes. Upon the appearance of substantial tumor burden (usually 12–13 d after injection), mice were treated with doxorubicin or vincristine. Tumor-free survival was monitored by palpation and in vivo GFP imaging using a NightOwl imaging system (Berthold). For Eμ-Myc;p53−/− lymphoma, 2 × 106 cells were i.v.-injected into BL/6J mice. Due to the aggressiveness of these p53−/− lymphomas, mice were treated 9 d after injection with 300 mg/kg cyclophosphamide. All mouse experiments were performed with the approval of the MIT committee on animal care.
Immunofluorescence
Cells were seeded onto 18-mm2 coverslips and either mock-treated or treated with 1 μM doxorubicin for 30 h. Cells were then fixed in 3% PFA and 2% sucrose for 15 min at room temperature and permeabilized with 20 mM Tris-HCl (pH 7.8), 75 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.5% Triton-X-100 for 15 min at room temperature. Slides were stained with primary antibodies overnight at 4°C. Secondary antibodies were used for 4 h at room temperature. Images were collected on an Axioplan2 (Zeiss) microscope equipped with Openlab software from Improvision.
Flow cytometry and cell cycle analyssis
For flow cytometric analysis of cell cycle and apoptosis, MEFs were treated as indicated. After treatment, cells were washed twice in ice-cold PBS, trypsinized, and fixed in 100% methanol for 3 d at −20°C, blocked with 2% BSA in PBS, and incubated with 1 μg of primary antibody per 106 cells for 3 h at room temperature. Following washing, cells were incubated with Alexa-488-conjugated secondary antibodies (diluted 1:50) (Molecular Probes) for 60 min at room temperature, washed, and resuspended in PBS containing 50 μg/mL PI prior to analysis on a Becton Dickinson FACScan flow cytometer. PI staining was analyzed on FL-3, and Alexa-488 staining was analyzed on FL-1.
Human tissues and immunohistochemical analysis
Formalin-fixed, paraffin-embedded specimens of human normal (n = 65) and tumor (n = 456; including 266 lung, 77 colon, 60 breast, and 53 bladder tumors) tissues were from the tissue bank of the Institute of Cancer Biology, Copenhagen, or provided by M. Sehested (Copenhagen University Hospital) or T. Ørntoft (Aarhus University Hospital) (Bartkova et al. 2005). All biopsy specimens were from sporadic tumors, obtained at surgery before any radiation treatment or chemotherapy of the patient was initiated. The lung tumors were all invasive (including 67 small-cell carcinomas and 199 nonsmall-cell tumors), while the sets of colon, breast, and urinary bladder tumors each included, apart from the bulk of invasive tumors, also a subset of 25%–30% of high-risk preinvasive lesions: grade 3 colon adenomas, breast carcinoma in situ, and bladder Ta lesions, respectively. For IHC, the tissue sections were deparaffinized and processed for sensitive immunoperoxidase staining with the primary monoclonal antibodies against human p53 (DO1, Novocastra Laboratories), ATM (ATML2p) (Tommiska et al. 2008), and rabbit monoclonal Y170 from Abcam), and Chk2 (DCS70 and DCS73) (Lukas et al. 2001) were incubated overnight, followed by detection using the Vectastain Elite kit (Vector Laboratories) and nickel sulphate enhancement without nuclear counterstaining. Staining for p53 was regarded as aberrantly enhanced when >20% of tumor cells showed strong staining, while the criteria for aberrantly reduced expression of ATM and Chk2 were as reported (Vahteristo et al. 2002; Tommiska et al. 2008). Given that well over 90% of all known ATM mutations result in low to undetectable levels of ATM protein (Mitui et al. 2008; http://chromium.liacs.nl/lovd), that the most frequent Chk2 mutations also grossly destabilize the protein (Vahteristo et al. 2002), and the vast majority of p53 mutations stabilize the mutant protein (Bartek et al. 1990; Iggo et al. 1990), immunohistochemical detection of protein loss (ATM and Chk2) or gross overexpression in many cancer cells (p53) is suitable to identify the vast majority of the tumor-associated aberrations of ATM, Chk2, and p53, with only rare false-negative and probably no false-positive cases (given the internal control of normal protein levels in noncancerous cells on each section) to be found. Statistical analysis was performed using the Fisher's exact test, with all P-values being two-sided and considered significant when P < 0.01.
Patient survival analysis
All of the analyses of various subgroups have been made on the total series of 604 tumors on the array stained for both p53 and ATM. For this group of patients (incident cases), the duration of follow-up ranged from 2.0 to 137.0 mo (median: 83.5; mean: 79.9; SD: 30.9). Age at the time of diagnosis ranged from 22.2 to 95.5 yr (median: 54.6; mean: 56.0; SD: 12.6). For non-IR graphs, p53-neg n = 74, P = 0.0002; p53-pos n = 19, P = 0.3236 (altogether, 93 patients). For chemotreated, non-IR, p53-neg, n = 24, P = 0.033. This study was performed with informed consents from the patients as well as with permissions from the Ethics Committee (E8) of the Helsinki University Central Hospital and from the Ministry of Social Affairs and Health in Finland.
Acknowledgments
We are grateful to T. Jacks, A.F. Cheung, and K.A. Janes for critically reading this manuscript. M.T.H. is a Rita Allen Scholar and the Latham Family Career Development Assistant Professor of Biology. This work was supported by the National Institutes of Health (NIH 1 R01 CA128803-01 to M.T.H.; NIH U54 CA112967 and R01 ES15339 to M.B.Y.), the Deutsche Forschungsgemeinschaft (RE2246/1-1 to H.C.R.), the David H. Koch Fund (to H.C.R. and M.B.Y.), the Deutsche Nierenstiftung (to H.C.R), the Danish Cancer Society, the Danish National Research Foundation, the European Community (projects “Active p53,” GENICA, and “Mutant p53”), the Czech Ministry of Education (MSM6198959216), and the Helsinki University Central Hospital Research Fund.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1815309.
Supplemental material is available at http://www.genesdev.org.
References
- Austen B, Skowronska A, Baker C, Powell JE, Gardiner A, Oscier D, Majid A, Dyer M, Siebert R, Taylor AM, et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J Clin Oncol. 2007;25:5448–5457. doi: 10.1200/JCO.2007.11.2649. [DOI] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Bartek J, Iggo R, Gannon J, Lane DP. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990;5:893–899. [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, Plassa LF, Soliman H, Varna M, de Roquancourt A, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4:e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- Cheung AF, Dupage MJ, Dong HK, Chen J, Jacks T. Regulated expression of a tumor-associated antigen reveals multiple levels of T-cell tolerance in a mouse model of lung cancer. Cancer Res. 2008;68:9459–9468. doi: 10.1158/0008-5472.CAN-08-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun HH, Gatti RA. Ataxia-telangiectasia, an evolving phenotype. DNA Repair (Amst) 2004;3:1187–1196. doi: 10.1016/j.dnarep.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Gurley KE, Kemp CJ. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr Biol. 2001;11:191–194. doi: 10.1016/s0960-9822(01)00048-3. [DOI] [PubMed] [Google Scholar]
- Haidar MA, Kantarjian H, Manshouri T, Chang CY, O'Brien S, Freireich E, Keating M, Albitar M. ATM gene deletion in patients with adult acute lymphoblastic leukemia. Cancer. 2000;88:1057–1062. doi: 10.1002/(sici)1097-0142(20000301)88:5<1057::aid-cncr16>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo R, Gatter K, Bartek J, Lane D, Harris AL. Increased expression of mutant forms of p53 oncogene in primary lung cancer. Lancet. 1990;335:675–679. doi: 10.1016/0140-6736(90)90801-b. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, Shimamura A, D'Andrea AD. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–1449. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes & Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nat Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Bartkova J, Latella L, Falck J, Mailand N, Schroeder T, Sehested M, Lukas J, Bartek J. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res. 2001;61:4990–4993. [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mitui M, Nahas SA, Du LT, Yang Z, Lai CH, Nakamura K, Arroyo S, Scott S, Purayidom A, Concannon P, et al. Functional and computational assessment of missense variants in the ataxia-telangiectasia mutated (ATM) gene: Mutations with increased cancer risk. Hum Mutat. 2008;30:12–21. doi: 10.1002/humu.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay UK, Senderowicz AM, Ferbeyre G. RNA silencing of checkpoint regulators sensitizes p53-defective prostate cancer cells to chemotherapy while sparing normal cells. Cancer Res. 2005;65:2872–2881. doi: 10.1158/0008-5472.CAN-04-2502. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Rahko E, Blanco G, Soini Y, Bloigu R, Jukkola A. A mutant TP53 gene status is associated with a poor prognosis and anthracycline-resistance in breast cancer patients. Eur J Cancer. 2003;39:447–453. doi: 10.1016/s0959-8049(02)00499-9. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Ripolles L, Ortega M, Ortuno F, Gonzalez A, Losada J, Ojanguren J, Soler JA, Bergua J, Coll MD, Caballin MR. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;171:57–64. doi: 10.1016/j.cancergencyto.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Rotman G, Shiloh Y. ATM: A mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Tabor E, Becker Y. Abnormal response of ataxia-telangiectasia cells to agents that break the deoxyribose moiety of DNA via a targeted free radical mechanism. Carcinogenesis. 1983;4:1317–1322. doi: 10.1093/carcin/4.10.1317. [DOI] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes & Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, Aittomaki K, Hofstetter B, Lukas J, von Smitten K, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–2506. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- Tribius S, Pidel A, Casper D. ATM protein expression correlates with radioresistance in primary glioblastoma cells in culture. Int J Radiat Oncol Biol Phys. 2001;50:511–523. doi: 10.1016/s0360-3016(01)01489-4. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes & Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- Yuan SS, Chang HL, Lee EY. Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM−/− and c-Abl−/− cells. Mutat Res. 2003;525:85–92. doi: 10.1016/s0027-5107(03)00009-5. [DOI] [PubMed] [Google Scholar]