Abstract
MicroRNAs (miRNAs) are 21 nt RNAs that regulate many biological processes in plants by mediating translational inhibition or cleavage of target transcripts. Arabidopsis mutants defective in miRNA biogenesis have overlapping and highly pleiotropic phenotypes including serrated leaves and ABA hypersensitivity. Recent evidence indicates that miRNA genes are transcribed by RNA polymerase II (Pol II). Since Pol II transcripts are capped, we hypothesized that CBP (cap-binding protein) 20 and 80 may bind to capped primary miRNA (pri-miRNA) transcripts and play a role in their processing. Here, we show that cbp20 and cbp80 mutants have reduced miRNA levels and increased pri-miRNA levels. Co-immunoprecipitation experiments revealed that pri-miRNAs 159, 166, 168 and 172 could be associated with CBP20 and CBP80. We found that CBP20 and CBP80 are stabilized by ABA by a post-translational mechanism, and these proteins are needed for ABA induction of miR159 during seed germination. The lack of miR159 accumulation in ABA-treated seeds of cbp20/80 mutants leads to increased MYB33 and MYB101 transcript levels, and presumably higher levels of these positive regulators result in ABA hypersensitivity. Genetic and molecular analyses show that CBP20 and 80 have overlapping function in the same developmental pathway as SE and HYL1. Our results identify new components in miRNA biogenesis.
Keywords: ABA, Cap-binding protein, MicroRNA, Post-translational modification, Primary microRNA
Introduction
MicroRNAs (miRNAs) are small, endogenous and non-translated RNAs expressed in a wide range of eukaryotic organisms (Bartel et al. 2004). In plants, these small RNAs, which are about 22 nt in length, are processed from stem–loop regions of long primary transcripts by a nuclear RNase III-like enzyme, DCL1 (Reinhart et al. 2002). Available evidence indicates that miRNAs bind to their target transcripts by base-pairing to regulate translation or to mediate transcript degradation (Chen 2004, Guo et al. 2005). The tissue-specific expression and developmental regulation of miRNAs suggest that they are involved in controlling various developmental processes (Carrington and Ambros 2003). Indeed, plant miRNAs are involved in floral development (Chen 2004), auxin signaling (McConnel et al. 2001, Bonnet et al. 2004,Guo et al. 2005), the promotion of flowering (Lauter et al. 2005) and RNA metabolism (Xie et al. 2003, Vaucheret et al. 2004). Recent studies reveal that miRNAs also function in response to abiotic stresses, such as drought, phosphate starvation, stress hormone (ABA) and UV-B (Sunkar and Zhu 2004, Fujii et al. 2005, Bari et al. 2006, Chiou et al. 2006, Reyes and Chua 2007, Zhou et al. 2007).
Biogenesis of miRNA has been investigated using Arabidopsis mutants defective in miRNA accumulation. In Arabidopsis, several regulators of miRNA biogenesis have been characterized including (i) DCL1, a homolog of Dicer, whose function is to cleave primary miRNAs (pri-miRNAs) into mature miRNAs (Park et al. 2002, Kurihara and Watanable 2004); (ii) HYPONASTIC LEAVES 1 (HYL1), a dsRNA-binding protein, which forms a complex with DCL1 for efficient processing of pri-miRNAs (Lu and Fedoroff 2000, Kurihara et al. 2006, Fang and Spector 2007); (iii) HASTY (HST), a exportin-5 homolog, required for exporting mature miRNAs from the nucleus to the cytoplasm (Park et al. 2005); and (iv) HUA ENHANCER1 (HEN1), a methylase, needed for proper accumulation of miRNAs (Boutet et al. 2003, Yu et al. 2005). Recently, SE has also been identified as another component in miRNA biogenesis (Grigg et al. 2005, Lobbes et al. 2006, Yang et al. 2006). SE also associates with HYL1 to form a protein complex which probably acts together with DCL1 in pri-miRNA processing (Fang and Spector 2007, Fujioka et al. 2007). Notably, these mutants (se, hyl1 and dcl1) share characteristic morphological phenotypes. Both se-2 and hyl1-2 display abnormal phyllotaxy and ABA hypersensitivity in seed germination, and se-2 and dcl1-9 produce serrated leaves.
Recent evidence suggests that miRNA genes, like mRNA-encoding genes, are also transcribed by RNA polymerase II (Pol II; Cai et al. 2004, Lee et al. 2004). For Pol II transcripts that encode proteins, it is known that cap-binding proteins (CBPs) 20 and 80 form a complex with the 5′ cap structure of primary transcripts to perform three functions: transcript stability, splicing efficiency and 3′ end formation. Since pri-miRNA transcripts are also capped and at least some of them appeared to be spliced (Cai et al. 2004, Lee et al. 2004), we reasoned that CBP20 and CBP80 may perform similar functions in stabilizing pri-miRNA transcripts and facilitating its splicing during miRNA biogenesis. Previous studies on cbp20 and cbp80 (abh1) show that these mutants display similar morphological phenotypes (Hugouvieux et al. 2001, Papp et al. 2004). These morphological phenotypes are very similar to those displayed by se-2, hyl1-2 and dcl1-9 (Lu and Fedoroff 2000, Prigge et al. 2001, Park et al. 2002), which are defective in miRNA biogenesis. The molecular basis of the morphological alterations in all these mutants is unknown but they are probably related to changes in miRNA biogenesis. Besides morphological changes, cbp20 and cbp80, as well as se-1, hyl1-2 and dcl1-9, are all hypersensitive to ABA. It is not known whether the molecular mechanism of ABA hypersensitivity is similar in all these mutants and which transcripts and proteins are affected to bring about ABA hypersensitivity during seed germination.
Here, we show that CBP20 and CBP80 function in miRNA biogenesis by binding to pri-miRNA transcripts and presumably increasing its processing efficiency. Mutant cbp20 and cbp80 plants have reduced miRNA levels and increased pri-miRNA levels compared with wild-type (WT) plants. Our co-immunoprecipitation results showed that CBP20 and CBP80 are associated with pri-miRNA transcripts, indicating the involvement of these two CBPs in miRNA processing. ABA induction of miR159 levels was delayed in cbp20 and cbp80 mutants with the corresponding increase in its target transcripts encoding MYB33 and MYB101, two positive regulators of ABA responses. These results, along with the post-translational stabilization of CBP20 and CBP80 protein levels by ABA, can explain in part the ABA hypersensitivity of the mutants. Our genetic and molecular analyses suggest that CBP20 and CBP80 appear to function independently of the presently known miRNA biogenesis regulators, SE and HYL1, in a common pathway.
Results
Reduced miRNA levels in cbp20 and cbp80
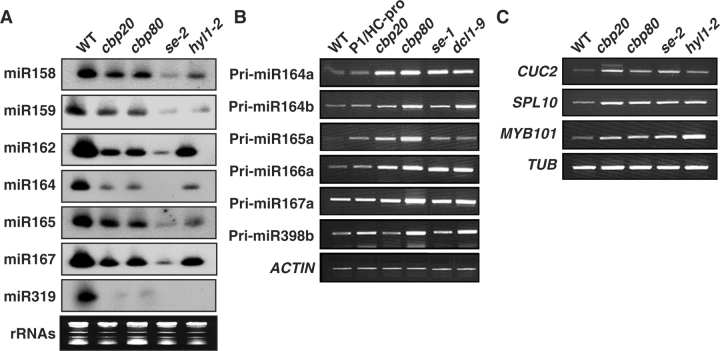
Several mutants (dcl1, se and hyl1) with deficiency in miRNA accumulation produce leaves with serrated margins (Lu and Fedoroff 2000, Prigge et al. 2001, Park et al. 2002). Because cbp20 (Papp et al. 2004) and cbp80 (abh1-2 fri) (Bezzera et al. 2004) mutants also share this morphological phenotype, we investigated whether miRNA levels were also altered in these mutant plants. Fig. 1A shows that the level of several miRNAs was decreased in cbp20 and cbp80 compared with the WT. Consistent with its severe defective developmental phenotype, se-2 showed the most severe reduction of miRNA levels amongst the mutants analyzed.
Fig. 1.
Expression levels of miRNAs and pri-miRNAs in cbp20 and cbp80. (A) Each lane contained 12 μg of total RNA. rRNAs were used as a loading control. (B) Accumulation of pri-miRNA transcripts. The se-1 and dcl1-9 mutants which were affected in miRNA processing were used as positive controls. Pri-miRNA transcripts were detected by RT–PCR using specific primers. ACTIN transcripts were used as a loading control. (C) Levels of transcripts targeted by some miRNAs in cbp20 and cbp80. Target transcript levels were determined by RT–PCR. Results were confirmed with three independent experiments. Tubulin transcripts were used as a loading control.
The level of most miRNAs analyzed was reduced in cbp20 and cbp80 although not to the same severe extent as in se-2. Among them, expression of miR162, miR164 and miR319 was most affected in cbp20 and cbp80 (Fig. 1A). On the other hand, miR159 levels in these two mutants were only slightly reduced compared with those in se-2 and hyl1-2. We found that miR159 levels may be developmentally regulated as a greater reduction in expression level was observed in younger seedlings of cbp20 and cbp80 (Supplementary Fig. S1). This observation suggests that CBP20 and CBP80 may regulate miR159 expression at early developmental stages. The mild reduction of miRNA levels cannot be explained by a partial loss of CBP20 and CBP80 function because cbp20 and cbp80 are null alleles (Hugouvieux et al. 2001, Papp et al. 2004).
To investigate whether the reduction of miRNA expression in cbp20 and cbp80 was due to impaired processing of miRNA from its pri-miRNA precursor, we analyzed levels of several pri-miRNA transcripts in these two mutants using P1/HC-pro transgenic plants and mutant plants of se-1 and dcl1-9 as controls (Fig. 1B) (Prigge et al. 2001, Kasschau et al. 2003, Kurihara and Watanable 2004). All analyzed pri-miRNAs accumulated to higher levels in cbp20 and cbp80 compared with the WT, indicating that CBP20 and CBP80 are required for efficient processing of pri-miRNAs.
Target transcripts of miRNAs accumulate in cbp20 and cbp80
As miRNAs target specific transcripts for degradation, we investigated whether the reduced miRNA level was accompanied by a corresponding increase in their target transcript level. Fig. 1C shows that CUC2 (targeted by miRNA164), SPL10 (targeted by miRNA156/157) and MYB101 (targeted by miRNA159) transcripts indeed accumulated to higher levels in cbp20 and cbp80 compared with the WT.
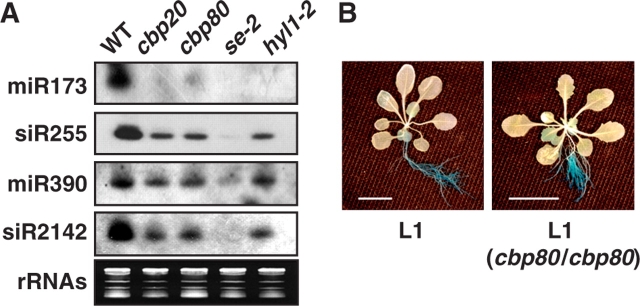
Arabidopsis has three known families of trans-acting small interfering RNA (ta-siRNA)-encoding genes, designated TAS1, TAS2 and TAS3. Biogenesis of ta-siRNAs is initiated by the activity of miR173 and miR390 (Allen et al. 2005, Yoshikawa et al. 2005), which requires SERRATE function (Lobbes et al. 2006). After site-specific cleavage, the processed transcripts are converted to double-stranded RNAs (dsRNAs) through the activities of RDR6 and SGS3. The dsRNAs are then processed by DCL activity to generate mature siRNAs which function in RNA interference (RNAi)-related processes (Papp et al. 2003). Because of the involvement of miRNAs in their biogenesis, we analyzed levels of several ta-siRNAs in cbp20 and cbp80. Fig. 2A shows that miR173 and miR390 levels as well as those of their corresponding siRNAs, ta-siR255 and ta-siR2142, were decreased in cbp20 and cbp80, although the effect was not as severe as that in se-2. Our results indicate that CBP20 and CBP80 also participate in ta-siRNA biogenesis through regulating the biogenesis of miR173 and miR390.
Fig. 2.
CBP20 and CBP80 are needed for ta-siRNA accumulation but not for sense PTGS. (A) siR255 and siR2142 levels in cbp20 and cbp80. Each lane contained 12 μg of total RNA. rRNAs were used as a loading control. (B) GUS staining of seedlings of the L1 line and of the L1 line carrying the cbp80 mutation. The scale bar equals 1 cm.
To see whether the reduction of ta-siRNAs in cbp20 and cbp80 would affect sense post-transcriptional gene silencing (PTGS) activity, we crossed the cbp80 mutant with the L1 line, which carries a silenced 35S::GUS transgene (Mourrain et al. 2000). Silencing of the GUS sense transcript by PTGS requires SGS3 and RDR6 since β-glucuronidase (GUS) activity in L1 was restored in the rdr6 and sgs3 mutant background (Mourrain et al. 2000). GUS activity analysis revealed that L1 plants carrying the cbp80 mutation showed no difference in staining pattern compared with the L1 control line (Fig. 2B). From these results, we conclude that CBP80 is not involved in sense PTGS.
The role of CBP20 and CBP80 in pri-miRNA processing
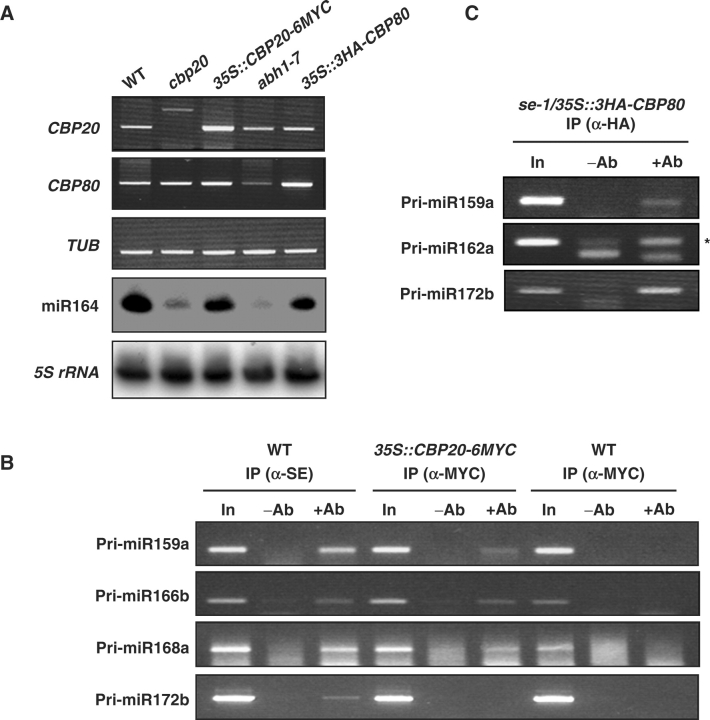
Because miRNA genes are transcribed by Pol II, the resulting pri-miRNA transcripts are expected to be capped (Cai et al. 2004, Lee et al. 2004). It is possible that binding of CBP20 and CBP80 to capped pri-miRNAs is required for their efficient processing. If this is the case, it should be possible to demonstrate an interaction between CBP20/CBP80 and pri-miRNA transcripts by co-immunoprecipitation assays. To this end, we constructed 35S::CBP20–6MYC and 35S::3HA-CBP80, and transferred these constructs into cbp20 and abh1-7, respectively. We found that expression of 35S::CBP20-6MYC or 35S::3HA-CBP80 was able to complement the mutant leaf serration phenotype of cbp20 and abh1-7, respectively, and recover the reduced miRNA levels to those of the WT (data not shown, Fig. 3A)
Fig. 3.
Primary miRNAs are associated with CBP20 and CBP80. (A) cbp20 and abh1-7 mutants were complemented with 35S::CBP20-6MYC and 35S::3HA-CBP80. Transcript levels of CBP20 and CBP80 in 35S::CBP20-6MYC and 35S::3HA-CBP80 lines were determined by RT–PCR. miR164 levels in 35S::CBP20-6MYC and 35S::3HA-CBP80 lines were determined by Northern blot analysis. Each lane contained 12 μg of total RNA, and 5S rRNA was used as a loading control. (B) Polyclonal antibodies to SERRATE and MYC were used to immunoprecipitate SERRATE and MYC-tagged CBP20 in WT and 35S::CBP20–6MYC extracts, respectively. Pri-miRNA levels were determined by RT–PCR using gene-specific primers. Input DNA was diluted 1 : 500. (C) Pri-miR159a, 162a and 172b levels in immunoprecipitates of 3HA–CBP80 using extracts from se-1/35S::3HA-CBP80 plants. HA antibody was used to immunoprecipitate 3HA–CBP80. Pri-miRNA levels in immunoprecipitates were determined by RT–PCR using gene-specific primers. An asterisk indicates the gene-specific band.
Extracts from these complemented plants were processed for immunoprecipitation using antibody against MYC or HA. Fig. 3B shows that pri-miRNA 159a, 166b and 168a could be detected in immunoprecipitates of CBP20–6MYC, suggesting that these pri-miRNAs were bound to CBP20. Pri-miRNA 159a, 166b, 168a and 172b were also found in immunoprecipitates of SERRATE, which was used as a positive control. However, we failed in our first attempts to detect pri-miRNA transcripts in 3HA–CBP80 immunoprecipitates, probably because of its reduced affinity for pri-miRNAs or its low steady-state level. To circumvent this problem, we used 35S::3HA-CBP80 transgenic lines in the se-1 background which accumulated pri-miRNA transcripts. Fig. 3C shows that pri-miR159a, 162a and 172b could indeed be detected in immunoprecipitates of 3HA–CBP80, although miR172 was reported to be largely insensitive to loss of cap-binding complex (CBC) activity (Laubinger et al. 2008). Together, these results show that CBP20 and CBP80 can bind to pri-miRNAs. We were unable to detect accumulation of meaningful levels of other pri-miRNAs in immunoprecipitates of CBP20–6MYC and 3HA–CBP80 even after repeated attempts. This difference may be explained by the differential accumulation levels of pri-miRNAs depending on specific developmental stages.
ABA increases CBP20 and CBP80 protein levels
Mutant plants of abh1 (cbp80) are hypersensitive to ABA and those of cbp20 show enhanced tolerance to drought, although ABH1 transcript levels are not affected by exogenous ABA nor by drought stress (Hugouvieux et al. 2001, Papp et al. 2004). In mammals, on the other hand, CBC activity is increased in response to growth factors, suggesting that CBC activity may be regulated via a post-transcriptional mechanism (Wilson et al. 1999).
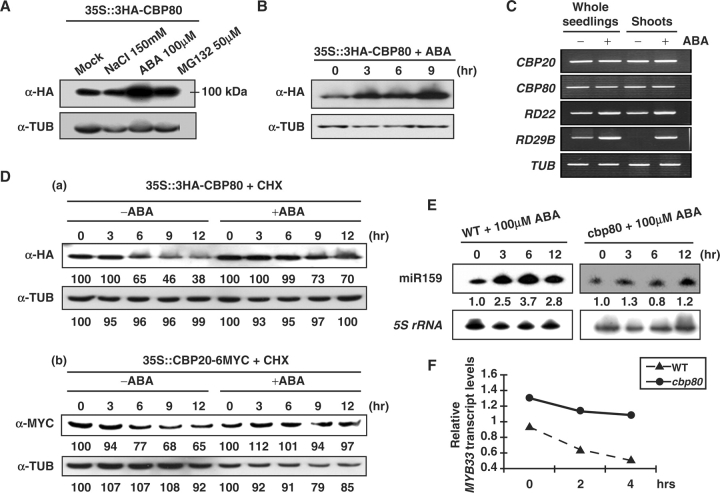
We investigated possible post-transcriptional/post-translational regulation of CBP20 and CBP80. Fig. 4C shows that CBP20 and CBP80 transcript levels were not affected by ABA, although the positive control RD22 and RD29B transcripts were induced in the same treatment (Yamaguchi-Shinozaki and Shinozaki 1993a, Yamaguchi-Shinozaki and Shinozaki 1993b). Next, we analyzed CBP20 and CBP80 protein levels in transgenic seedlings treated with ABA or NaCl. We found that CBP20–6MYC and 3HA–CBP80 levels were increased by ABA but not by NaCl (Fig. 4A, Supplementary Fig. S4). More induction was seen with 3HA–CBP80 upon longer exposure to ABA (Fig. 4B). Upon treating plants with MG132, a 26S proteasome inhibitor, CBP80 levels increased 2-fold compared with mock control (Fig. 4A). To test whether this increase of CBP20 and/or CBP80 was a consequence of enhanced stability by ABA treatment, we performed a protein decay experiment in the presence of cycloheximide (CHX) which blocks new protein synthesis.
Fig. 4.
CBP20 and CBP80 are stabilized and required for increased miR159 levels in response to ABA. (A) 3HA–CBP80 levels in seedlings treated with NaCl (150 mM), ABA (100 μM), MG132 (50 μM) or buffer (mock-treatment) for 2 h. Tubulin levels were used as a loading control. (B) Time course of 3HA–CBP80 accumulation in seedlings treated with ABA. Five-day-old seedlings were incubated in liquid MS medium with ABA (100 μM) for various time periods. (C) CBP20 and CBP80 transcript levels after ABA treatment. Transcripts were detected by RT–PCR using specific primers. RD22 and RD29B transcript levels were used as positive controls. Tubulin transcript levels were used as a loading control. (D) Stabilization of 3HA–CBP80 and CBP20–6MYC by ABA. (a) Five-day-old seedlings expressing 3HA–CBP80 were incubated in liquid MS medium with or without ABA (100 μM) after CHX (100 μM) treatment for 30 min. Proteins were extracted at the indicated times. (b) Seedlings expressing CBP20–6MYC were treated using the same conditions as those in (a). Relative expression levels were calculated using the program Image Gauge V3.12. The value 100 was given for the expression level at 0 time. (E) One-day-old seedlings were treated with 100 μM ABA for various time periods. Each lane contained 12 μg of total RNA. 5S rRNA was used as a loading control. The expression level of miR159 was normalized to that of 5S rRNA. The miRNA level at 0 time was set as 1. Numbers between the panels indicate the relative expression levels compared with 0 time. Signals were measured by Phosphorimager. (F) qRT–PCR analysis of MYB33 transcript levels. One-day-old seedlings were incubated in liquid MS medium with 100 μM ABA for various time periods. MYB33 transcript levels normalized to ACTIN transcript levels.
We determined the time course of CBP20 and CBP80 levels after CHX treatment. CBP80 showed a half-life of about 6 h, which was prolonged to >12 h in the presence of ABA (Fig. 4D). CBP20 showed a longer half-life of about 12 h and was also stabilized by ABA (Fig. 4D). This result showed that CBP20 and CBP80 are stabilized by ABA by a post-translational mechanism.
Because expression levels of certain miRNAs are increased by abiotic stress (Sunkar et al. 2004) and CBP20 and CBP80 appear to regulate miRNA levels, we hypothesized that the increase in CBP80 protein levels by abiotic stress such as ABA treatment may, in turn, facilitate miRNA accumulation. To test this hypothesis, we investigated miR159 whose expression level is controlled by ABA (Reyes and Chua 2007). Fig. 4E confirms a previous report that miR159 levels were indeed induced by ABA. In contrast, this ABA induction of miR159 levels was largely suppressed in the cbp80 mutant (Fig. 4E). One target of miR159 is MYB33 which encodes a positive regulator of ABA response. We performed quantitiative reverse transcription–PCR (qRT–PCR) using RNA samples from 1-day-old seedlings after ABA treatment to determine MYB33 transcript levels. Compared with the WT, the decline of MYB33 transcript levels was much weaker in cbp80, consistent with the decreased miR159 which targets MYB33 transcripts (Fig. 4F). The higher MYB33 transcript levels could account, in part, for the ABA hypersensitivity of the cbp80 mutant as compared with the WT.
Genetic interaction of CBP20/80 and other miRNA regulators
Mutant plants of hyl1, dcl1 and se have serrated leaves, and seedlings of these mutants are hypersensitive to ABA. These shared morphological phenotypes are paralleled by similar alterations in miRNAs. This is not surprising since it has been reported that HYL1, DCL1 and SE together function as a macromolecular complex (Yang et al. 2006, Fang and Spector 2007, Fujioka et al. 2007, Song et al. 2007).
Similar to se-2 and hyl1-2, mutant plants of cbp20 and cbp80 also have serrated leaves, display ABA hypersensitivity and, as shown here, are involved in miRNA biogenesis. To examine possible genetic interactions amongst these loci, we generated several combinations of double mutants by genetic crosses.
CBP20 and CBP80 have been reported to form a heterodimeric complex which functions in diverse aspects of RNA metabolism. Because miRNA expression levels still remained high in each single mutant of cbp20 and cbp80 compared with se, which shows embryonic lethality (Lobbes et al. 2006), we suspected that CBP20 and CBP80 might be functionally redundant. Using a transient expression system in tobacco, we detected a strong interaction between CBP20 and CBP80, confirming their heterodimeric association (data not shown). Therefore, each mutation may not show severe development defects as seen in se-2 or dcl1-9.
However, the phenotype of the homozygous double mutant cbp20/cbp80 was almost identical to that of each single mutant [Fig. 5A(c)]. Consistent with this morphological phenotype, reduction of miRNA levels in the cbp20/cbp80 double mutant was similar to that in each single mutant (Fig. 5C). The miRNA phenotype of cbp20 can be rescued by expression of a 35S::CBP20 transgene (cbp20+35S::CBP20, Papp et al. 2004). This complementation was specific, as the same transgene could not complement the miRNA-deficient phenotype of cbp80 [Fig. 5A(l)]. Notwithstanding their non-redundant functions, CBP20 and CBP80 appear to be non-essential genes in Arabidopsis. This observation is consistent with previous reports that yeast mutants deficient in homologous cap-binding proteins CBC1p (CBP80) and CBC2 (CBP20) are still viable (Chen and Dieckmann 1994, Izaurralde et al. 1994).
Fig. 5.
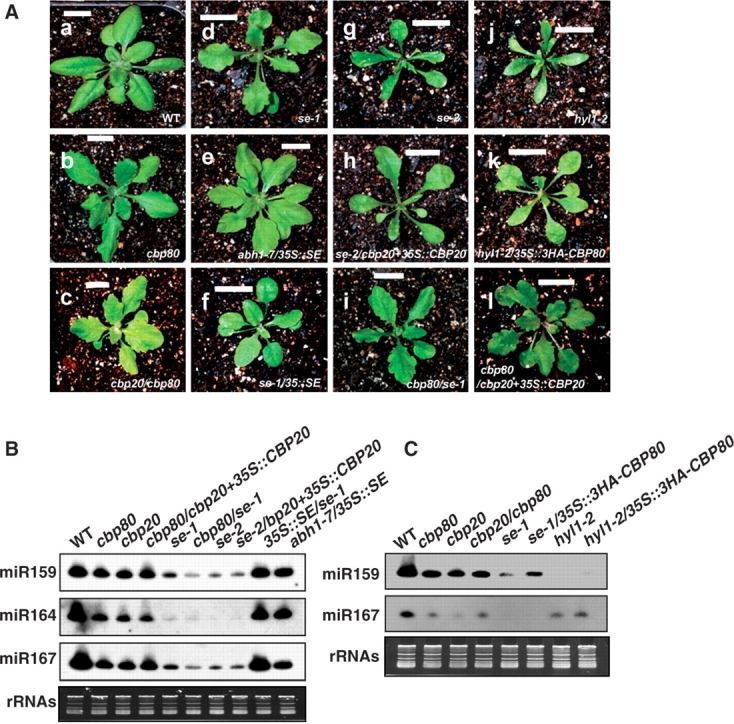
Morphological and molecular analyses. (A) Comparison of leaf phenotype in single and double mutants. Images were taken with 45-day-old plants. Scale bars equal 1 cm. (B) Analyses of miR159, miR164 and miR167 levels in single and double mutants and overexpression plants. Each lane contained 12 μg of total RNA. Consistent results were obtained from three independent experiments. (C) The effect of CBP80 overexpression on miRNA levels in se-1 and hyl1-2. Each lane contained 12 μg of total RNA, and rRNAs were used as a loading control.
To assess the epistasis relationships of cbp20, cbp80 and se, cbp20+35S::CBP20 was crossed to se-2 (Papp et al. 2004, Grigg et al. 2005). Although the overall se-2 phenotype was not altered by cbp20+35S::CBP20, the frequency of upward curling leaves was reduced and WT leaf size was recovered in se-2/cbp20+35S::CBP20 [Fig. 5A(h)]. The presence of the same antibiotic resistance marker in plants overexpressing 3HA-CBP80 and in the se-2 mutant prevented us from generateing se-2/35S::3HA-CBP80. We therefore introduced 35S::3HA-CBP80 into mutant plants of se-1, a weak se allele, by direct transformation. Plants of se-1/35S::3HA-CBP80 showed a similar phenotype to that of se-1 (data not shown).
Next, we introduced 35S::SE into abh1-7 in Col (Prigge et al. 2001, Kuhn et al. 2007). The two alleles of abh1 (abh1-2 and abh1-7) are both null and have the same morphological and molecular phenotype (data not shown). Notably, 35S::SE significantly recovered the leaf serration phenotype of abh1-7 [Fig. 5A(e)]. Along with this phenotypic complementation of abh1-7/35S::SE, expression levels of miR164 were higher than those in cbp80, although not as high as WT levels (Fig. 5B). These results suggest that 35S::SE alters the expression of miR164 target transcripts whose expression is also affected by cbp80, resulting in the partial recovery of the cbp80 phenotype. In addition, miR159 and miR167 expression decreased more in the cbp80/se-1 double mutant than that in the cbp80 single mutant (Fig. 5B), although no apparent difference was seen in their morphological phenotype [Fig. 5A(i)]. Our results suggest that CBP20 and CBP80 also function in a pathway mediated by SE; however, these two proteins have a function in miRNA biogenesis independent of SE.
Yang et al. (2006) have reported that hyl1-2/se-1 double mutants are embryonic lethal, suggesting that HYL1 and SERRATE may act synergistically to control the same genetic pathway. To test for epistasis between hyl1 and cbp80, we crossed hyl1-2 to cbp80 to generate the cbp80/hyl1-2 double mutant. We found that the cbp80/hyl1-2 double mutant had a similar phenotype to the hyl1-2 single mutant (data not shown), although the double mutant plants showed weak fertility, producing only a few seed at maturation. Interestingly, the cup-shaped leaf phenotype of hyl1-2 was rescued by 35S::3HA-CBP80, and hyl1-2/35S::3HA-CBP80 plants showed only mild alteration in leaf shape compared with hyl1-2 plants [Fig. 5A(k)]. This phenotypic compensation was paralleled by a slight elevation of miR159 and miR167 expression levels in hyl1-2/35S::3HA-CBP80 (Fig. 5C).
Collectively, our results on morphological and molecular analyses suggest that CBP20, CBP80, SE and HYL1 are required at the same step but with overlapping function in the same pathway.
Discussion
The nuclear CBPs CBP20 and CBP80 form a heterodimeric complex to bind to the 5′ cap structure of nascent mRNAs transcribed by Pol II, stimulating pre-mRNA splicing (Ohno et al. 1987, Izaurralde et al. 1994, Lewis et al. 1996), polyadenylation (Flaherty et al. 1997) and nuclear RNA export (Izaurralde et al. 2002). In this study, we found that CBP20 and CBP80 are also required for miRNA biogenesis since in cbp20 and cbp80 mutants miRNA expression levels were reduced along with an accumulation of pri-miRNAs, and levels of the corresponding target transcripts of miRNAs were elevated.
The simplest explanation is that CBC binds to the 5′ cap structure of pri-miRNA transcripts and facilitates their processing to generate mature miRNAs. To date, Arabidopsis has been found to have several CBPs such as CUM1 (eIF4E1), eIFiso4E, eIFiso4E-like and a novel CBP identified by their sequence similarity and affinity to the cap structure (Browning 1996, Ruud et al. 1998, Yoshii et al. 2004). As pri-miRNA transcripts are spliced and capped like mRNA transcripts, these other CBPs, like CBP20 and CBP80, also could bind to pri-miRNA transcripts and have a general effect on miRNA biogenesis. To address this issue, we investigated miRNA transcript levels in cum1 and cum2 (eIF4G) (Supplementary Fig. S2). No change of miRNA expression levels was detected in these mutants, confirming that the miRNA decrease in cbp20 and cbp80 does not reflect an indirect consequence of the same effect from the structural location. However, the absence of an obvious mutant phenotype of cum1 and eIF(iso)4E may be due to their overlapping functions in a pathway. It would be interesting to analyze triple or quadruple mutants in future studies to elucidate further the role of CBPs in miRNA biogenesis.
To uncover the biochemical role of CBP20 and CBP80 in miRNA biogenesis, we proposed several hypotheses based on their known function in other organisms, e.g. yeast and mammals. As these proteins have been shown to function in mRNA turnover by external factors, we first suspected that deficiencies in the function of these genes may destabilize mRNA and possibly pri-miRNA transcripts, resulting in their rapid turnover (Chen et al. 1994, Das et al. 2000). However, there was no apparent difference in the decay rate of Expansin-Like1 (EXPL1) transcripts between cbp20/80 mutants and the WT, excluding this possibility of altered mRNA stability in the mutants (Supplementary Fig. S3). In fact, this is consistent with our finding that pri-miRNA levels increased, rather than decreased, in cbp20 and cbp80.
It has also been suggested that CBP20 and CBP80 may modulate another(s) protein, leading to changes in the level of transcripts encoding these proteins (Hugouvieux et al. 2001). CBP20 expression is highly reduced in a yeast strain carrying the null allele of CBP80 (Shen et al. 2000). This observation suggests that protein interaction with CBP80 may modulate the stability of its partners. In addition to CBP20, some RNA-binding proteins that function in mRNA splicing may also interact with CBP80. It has been previously suggested that the large Drosha-containing complex has a function in other RNA processing pathways, because this complex included proteins known to be involved in splicing (Gregory et al. 2004). Also, it has been suggested that CBC has a role in co-transcriptional spliceosome assembly and splicing (Gornemann et al. 2005). Recently, two groups also reported that CBP20 and CBP80 function in miRNA processing and pre-mRNA splicing (Gregory et al. 2008, Laubinger et al. 2008). Laubinger et al. showed that cbp20/80 reduced splicing efficiency in intron-containing pri-miRNAs by tiling assay, suggesting that CBC facilitates the loading of the miRNA processing machinery onto pri-miRNA, in analogy with its role in recruiting the splicing commitment complex onto pre-mRNAs. Interestingly, we found that in cbp20 and cbp80 mutants, the splicing pattern of transcripts for one of the SR proteins which was required for splicing was changed (data not shown). Further experiments should address the role of splicing factor in miRNA generation.
Our co-immunoprecipitation results provide evidence for the participation of CBP20 and CBP80 in pri-miRNA processing. Interestingly, CBP20 and CBP80 expression levels were increased after ABA treatment, although their transcript levels remained unaffected. In contrast to the WT, miR159 which negatively regulates ABA responses during seed germination, failed to accumulate in cbp20 and cbp80 upon ABA treatment. Consequently, transcripts encoding two positive regulators of ABA responses (Reyes and Chua 2007), MYB33 and MYB101, were not down-regulated by miR159 in these two mutants. The elevated MYB33 and MYB101 transcripts and therefore presumably protein levels may explain in part the ABA hypersensitivity of cbp20 and cbp80 mutants. When plants are subject to stress conditions, ABA accumulates and levels of some proteins such as CBP20 and CBP80 are increased, generating more miR159, which mediates degradation of transcripts encoding positive regulators of ABA. This is supported by the notion that plants expressing 35S::miR159a-1 and 35S::miR159a-3 show hyposensitivity to ABA (Reyes and Chua 2007). However, we cannot exclude that the two CBP proteins can also function in additional sites of the ABA signaling cascade since in cbp80, the AtPP2C transcript, which encodes an ABA-negative regulator, was also reduced (Hugouvieux et al. 2001).
It has been proposed that miRNAs are regulated in a developmental and spatial manner and that different regulators of miRNA are needed at different stages. Probably, CBP20 and CBP80 activities are needed as a negative regulator of ABA through miR159 accumulation in certain developmental stages such as germination. However, it should be noted that CBP20 and CBP80 are not the only regulators in ABA responses to generate miRNAs because miR159 levels eventually increased in these mutants (Supplementart Fig. S1). Based on our genetic analysis, SERRATE and HYL1 are strong candidates because mutants deficient in these proteins also display ABA hypersensitivity.
Because miRNA generation appears to follow a similar regulatory mechanism of mRNA transcription, there is increasing evidence that miRNA genes and the corresponding protein-coding genes are similarly regulated through shared motifs in their promoter regions under stress conditions such as UV-B treatment (Zhou et al. 2007). Along with other reports, our findings identify new components in miRNA biogenesis. Our work may provide new directions to understanding how miRNAs are generated and which factors are implicated in their regulation to mediate diverse biological processes such as development, cell proliferation and differentiation.
Materials and Methods
Plant material and growth conditions
All Arabidopsis thaliana lines used were in the Columbia-0 (Col) ecotype. abh1-2, referred to as cbp80 herein, was obtained from Dr. R. Amasino (Bezzera et al. 2004). cbp20 and cbp20+35S::CBP20 seeds were obtained from Dr. C. Koncz (Papp et al. 2004). abh1-7 was an allele of abh1 originally isolated in the C24 background. This mutant abh1-7 was crossed twice to Col and the homozygous line was used for this work (Kuhn et al. 2007). Seeds were stratified on 0.65% phytoagar containing half-strength Murashige and Skoog (MS) medium for 18 d before being transferred to a greenhouse under similar conditions (22°C, 16/8 h photoperiod cycle). For genetic crosses, se-1, se-2, 35S::SE, cbp20, cbp20+35S::CBP20, abh1-2 (cbp80) and hyl1-2 were used (Prigge et al. 2001, Lu and Fedoroff 2004, Papp et al. 2004, Grigg et al. 2005). Homozygous F2 progeny from crosses were obtained using appropriate selective agents: hygromycin (Sigma, St Louis, MO, USA), Basta and/or kanamycin in combination. To generate complementary lines, 35S::CBP20-6MYC and 35S::3HA-CBP80 were transformed into cbp20 and abh1-7 mutant plants, respectively, with Agrobacterium tumefaciens using the floral dip method (Clough and Bent 1998). Transgenic plants of abh1-7/35S::SE, se-1/35S::3HA-CBP80 and hyl1-2/35S::3HA-CBP80 were obtained in a similar manner.
For ABA treatment, 5-day-old seedlings were incubated in liquid MS medium with 100 μM ABA (Sigma) in ethanol or with the same volume of ethanol as controls. These transgenic lines are referred to as 35S::CBP20–6MYC and 35S::3HA-CBP80 in this paper.
Plasmid construction
Full-length cDNAs of CBP20 and CBP80 were amplified by PCR and cloned into pENTR TOPO vector (Invitrogen, Carlsbad, CA, USA). After verification by DNA sequencing, the cDNA sequences were transferred into destination vector, pBA-DC-3HA or pBA-DC-6MYC carrying a cauliflower mosaic virus (CaMV) 35S promoter and a nopaline synthase poly(A) addition sequences, by LR reaction according to the Gateway system (Invitrogen).
Analysis of gene expression
Total RNA was extracted from Arabidopsis seedlings by Trizol reagent (Invitrogen). Low molecular weight RNAs were analyzed according to Wang et al. (2004). Blots were hybridized to 32P-radiolabeled oligonucleotide probes complementary to the miRNAs. Specific primers for pri-miRNAs were described previously (Yang et al. 2006). All experiments were repeated three times. GUS expression was analyzed according to Jefferson et al. (1987). Plants were incubated in 90% acetone at room temperature for 10 min and transferred to staining solution for 3 h at 37°C.
Six-day-old seedlings were incubated in MS medium with 500 μM cordycepin (3′-deoxyadenosine; Sigma-Aldrich). Total RNA was extracted from samples harvested at various time points using Trizol reagent (Invitrogen). For RNA gel blot analysis, 15 μg of total RNA was fractionated on a 1.2% (w/v) agarose gel and then transferred to a Hybond-N+ membrane. DNA probes were labeled with [α-32P]dCTP using the random prime labeling system (GE Biosciences, Milwaukee, WI, USA).
Cycloheximide treatments
Six-day-old seedlings of transgenic Arabidopsis lines (35S::CBP20-6MYC and 35S::3HA-CBP80) were transferred to liquid MS medium and treated with 100 μM CHX for 30 min before addition of 100 μM ABA (Sigma) or ethanol (mock treatment). Treated seedlings were harvested at different time points. Proteins were extracted for Western blot analyses using anti-MYC or anti-HA antibodies [c-Myc (A-14) sc-789; HA probe sc-7392, Santa Cruz Biotechnology, Santa Cruz, CA, USA].
Co-immunoprecipitation assay
Ten-day-old seedlings were used for co-immunoprecipitation assay to detect CBP20/CBP80-associated pri-miRNAs. Protocols provided by the manufacturer's guide (Upstate) were used except that 0.5 U of SUPERaseIN RNase inhibitor (Ambion) was included in all buffers. Antibodies against MYC, HA and SERRATE were used for immunoprecipitation. RNAs associated with immunoprecipitates were recovered using Trizol (Invitrogen). Total RNA was used for reverse transcription after treatment with DNase (TURBO DNA-free, Ambion, Austin, TX, USA). Pri-miRNA transcripts were amplified by RT–PCR; 40 cycles for pri-miR159a and 45 cycles for pri-miR162a, 166b, 168a and 172b. PCR products were resolved by gel electrophoresis and gels were stained with ethidium bromide.
Supplementary data
Supplementary data are available at PCP online.
Funding
The Korea Research Foundation post-doctoral fellowship (to S.K.); Human Frontier Science Program post-doctoral fellowship (LT00385/2007-L to S.K.); National Institutes of Health (NIH) (grant GM44640 to N-H.C.)
Acknowledgments
We thank Dr. Kalyna for providing seeds of atSRp30, Dr. Amasino for providing seeds of abh1-2 fri, Dr. Koncz for providing seeds of cbp20 and 35S::CBP20, and Dr. Reyes for discussion.
Glossary
Abbreviations:
- CaMV
cauliflower mosaic virus
- CBC
cap-binding complex
- CBP
cap-binding protein
- CHX
cycloheximide
- GUS
β-glucuronidase
- miRNA
microRNA
- MS
Murashige and Skoog
- Pol II
RNA polymerase II
- pri-miRNA
primary microRNA
- PTGS
post-transcriptional gene silencing
- qRT–PCR
quantitative reverse transcription–PCR
- RNAi
RNA interference
- siRNA
small interfering RNA
- ta-siRNA
trans-acting small interfering RNA
- WT
wild-type.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bari R, Pant BD, Stitt M, Scheible W. PHO2, microRNA399 and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 2004;40:112–119. doi: 10.1111/j.1365-313X.2004.02194.x. [DOI] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl Acad. Sci. USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S, Vasquez F, Liu J, Beclin C, Fagard M, Gratias A, et al. Arabidopsis HEN1: a genetic link between endougenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS. The plant translational apparatus. Plant Mol. Biol. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen W, Dieckmann CL. Cbplp is required for message stability following of COB mRNA. J. Biol. Chem. 1994;269:16574–16578. [PubMed] [Google Scholar]
- Chen X. A microRNA as translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Das B, Guo Z, Russo P, Chartrand P, Sherman F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 2000;20:2827–2838. doi: 10.1128/mcb.20.8.2827-2838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl Acad. Sci. USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou T, Zhu J. A miRNA involved in phosphate starvation response in Arabidopsis. Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis E, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 2002;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Breton G, Schroeder JI. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 2007;50:1049–1062. doi: 10.1111/j.1365-313X.2007.03110.x. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl Acad. Sci. USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli D, Schmidt DD, Martin C, Clark J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin and cytokinin. Plant Cell. 2000;12:2351–2365. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J, Emery J, Eshed Y, Bao N, Bowman J, Barton M. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Ohno M, Sakamoto H, Shimura Y. Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc. Natl Acad. Sci. USA. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Ruud KA, Kuhlow C, Goss DJ, Browning KS. Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J. Biol. Chem. 1998;273:10325–10330. doi: 10.1074/jbc.273.17.10325. [DOI] [PubMed] [Google Scholar]
- Shen EC, Stage-Zimmermann T, Chui P, Silver PA. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl Acad. Sci. USA. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu J. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel D. The action of ARGONAUT 1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reyes J, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KF, Fortes P, Singh US, Ohno M, Mattaj IW, Cerione RA. The nuclear cap-binding complex is a novel target of growth factor receptor-coupled signal transduction. J. Biol. Chem. 1999;274:4166–4173. doi: 10.1074/jbc.274.7.4166. [DOI] [PubMed] [Google Scholar]
- Xie Z, Kasschau K, Carrington J. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet. 1993a;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993b;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huan H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, et al. The Arabidopsis Cucumovirus Multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.