Abstract
The pseudostratified epithelium of the mouse trachea and human airways contains a population of basal cells expressing Trp-63 (p63) and cytokeratins 5 (Krt5) and Krt14. Using a KRT5-CreERT2 transgenic mouse line for lineage tracing, we show that basal cells generate differentiated cells during postnatal growth and in the adult during both steady state and epithelial repair. We have fractionated mouse basal cells by FACS and identified 627 genes preferentially expressed in a basal subpopulation vs. non-BCs. Analysis reveals potential mechanisms regulating basal cells and allows comparison with other epithelial stem cells. To study basal cell behaviors, we describe a simple in vitro clonal sphere-forming assay in which mouse basal cells self-renew and generate luminal cells, including differentiated ciliated cells, in the absence of stroma. The transcriptional profile identified 2 cell-surface markers, ITGA6 and NGFR, which can be used in combination to purify human lung basal cells by FACS. Like those from the mouse trachea, human airway basal cells both self-renew and generate luminal daughters in the sphere-forming assay.
Keywords: epithelial basal cells, lung, NGFR, stem/progenitor, sphere-forming assay
Basal cells (BCs) make up approximately 30% of the pseudostratified mucociliary epithelium of the lung. They are relatively undifferentiated and characteristically express the transcription factor Trp-63 (p63) and cytokeratins 5 and 14 (Krt5/14) (1–3). In the rodent, BCs are confined to the trachea, where they are interspersed among the ciliated, secretory, and neuroendocrine cells. By contrast, in the human lung, BCs are present throughout the airways, including small bronchioles (1, 4, 5).
Cell turnover in the adult trachea is normally very low (6). However, epithelial injury elicits rapid proliferation of surviving cells, except ciliated cells (7), and the tissue is soon repaired. Several lines of evidence suggest that tracheal BCs function as stem cells for this repair. First, lineage tracing of KRT14-CreER–expressing cells after naphthalene injury, which depletes secretory cells, suggested that some BCs can both self-renew and give rise to ciliated and secretory cells (8). Second, a subset of BCs retains BrdU label over the long term after epithelial damage by SO2 inhalation (9). Third, BCs isolated on the basis of high expression of a KRT5-GFP transgene have a greater capacity to proliferate and give rise to large colonies in vitro than KRT5-GFP− cells (3). Finally, fractionated rat tracheal BCs can restore the entire epithelium of a denuded trachea in a xenograft model (10). Similar xenograft assays have been performed with BCs from human nasal polyps and fetal trachea (11, 12).
Despite the evidence that BCs are stem cells, relatively little is known about their biology and the mechanisms regulating their behavior. To address these problems, we have generated a transgenic mouse line to more efficiently lineage trace and genetically manipulate BCs. We have also isolated mouse BCs and obtained a transcriptional profile. Finally, we have developed a clonal sphere-forming culture system that will greatly facilitate studies on the self-renewal and differentiation of mouse and human BCs.
Results and Discussion
In Vivo Lineage Tracing of Mouse Tracheal BCs.
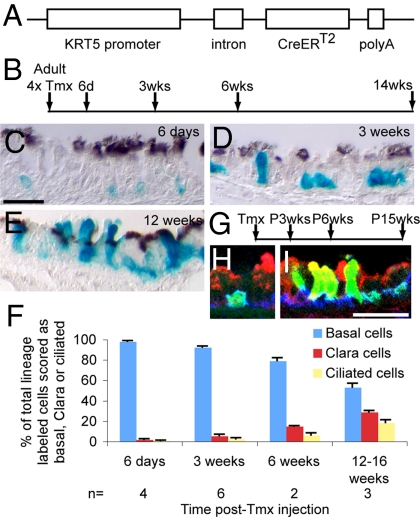
Previous lineage tracing of mouse tracheal BCs after injury used a KRT14-CreER transgene (8). However, this allele is inefficient for labeling BCs in steady state. We therefore made a new line in which a 6-kb human keratin 5 (KRT5) promoter drives CreERT2 (Fig. 1A). We used this allele, in combination with the Rosa26R-lacZ reporter, to lineage trace BCs in adult mice for up to 14 weeks (Fig. 1B). Soon after the last tamoxifen (Tmx) injection, most of the labeled cells (≈98%) were scored as BCs (Fig. 1C). Over time, the percentage of labeled cells scored as BCs declined, whereas that of labeled secretory and ciliated cells increased (Fig. 1 D–F and Table 1). Lineage-labeled BCs give rise to more Clara than ciliated cells over the chase period. There are 2 possible explanations for this result. First, BCs may give rise to Clara cells more frequently than ciliated cells—possibly because the latter have a longer half-life than Clara cells and need to be replaced less often. Alternatively, BCs first give rise to Clara cells, which then slowly transition to ciliated cells. We currently favor the second model because it is supported by tritiated thymidine pulse-chase experiments in the rat (1) and our own pulse-chase lineage labeling studies with Scgb1a1-CreER+ (Clara) cells in the mouse trachea (13).
Fig. 1.
Lineage tracing of tracheal BCs. (A) KRT5-CreERT2 transgene construct. (B) Timeline for steady-state experiments with KRT5-CreERT2;Rosa26R-LacZ adult mice. (C–E) Paraffin sections of X-gal–stained (blue) trachea. Anti-acetylated tubulin (brown, cilia) (C) 6 days, (D) 3 weeks, and (E) 12 weeks after final Tmx injection. (F) Bar graph showing percentage of labeled cells scored as basal, Clara, or ciliated after Tmx exposure and chase. Data shown are mean ± SEM. (G) Timeline for postnatal growth experiments with Rosa26R-eYFP reporter. (H and I) Cryosections stained for GFP (green, lineage label), Scgb1a1 (red, Clara cells), and T1α (blue, BCs). (H) P3 weeks. (I) P15 weeks. [Scale bar in C (for C–E), 20 μm; in H (for H and I), 25 μm.]
Table 1.
BCs of the trachea give rise to ciliated and Clara cells at steady state and in response to epithelial injury
| Time of chase | Basal (%) | Clara (%) | Ciliated (%) | Total no. of lineage-labeled cells scored | Total no. of tracheas |
|---|---|---|---|---|---|
| Tmx + 6 d | 97.8 ± 0.9 | 1.6 ± 0.8 | 0.5 ± 0.3 | 466 | 4 |
| Tmx + 3 wk | 92 ± 1 | 6.6 ± 1.5 | 1.3 ± 0.9 | 1265 | 6 |
| Tmx + 6 wk | 79 ± 2.5 | 14.7 ± 0.6 | 6.2 ± 1.9 | 385 | 2 |
| Tmx + 12–16 wk | 53.1 ± 3.8 | 28.6 ± 1.4 | 18.4 ± 2.6 | 1191 | 3 |
| Tmx + SO2 + 2 wk | 38.5 ± 1 | 21.4 ± 2.6 | 40.1 ± 2.7 | 1265 | 4 |
Percentage of lineage labeled cells, ± SEM, scored as basal, Clara, or ciliated after Tmx exposure and varying times of chase, with or without SO2 injury.
During postnatal growth there is an increase in the length and diameter of the rodent trachea and in the number of epithelial cells, including BCs (1). In previous studies we showed BrdU labeling of tracheal epithelial cells at postnatal (P)6 days, P3 weeks, and P6 months (13). Further analysis of the material demonstrated that 55% of p63+ BCs and 10% of non-BCs are BrdU labeled at P6 days, whereas the corresponding values for P3 weeks were 9% of BCs and 2% of non-BCs (Fig. S1). To follow the behavior of tracheal BCs during postnatal growth a cohort of KRT5-CreERT2;Rosa26R-eYFP mice was given a pulse of Tmx at P10 days or P12 days (Fig. 1G). At P3 weeks, the majority (96%) of lineage labeled cells were BCs (Fig. 1H and Table S1). These represented 7% ± 3% of all BCs in the trachea (Table S2). This percentage remained approximately the same when members of the cohort were examined at P6 weeks and at P15 weeks, suggesting that labeled BCs self-renew and are not diluted by descendants of unlabeled cells. As before, there was an increase over time in the percentage of labeled cells scored as Clara or ciliated cells (Fig. 1I and Table S1). Thus, during postnatal growth BCs both self-renew and give rise to Clara and ciliated cells.
Finally, we followed the fates of labeled BCs in a repair model. Inhalation of SO2 leads to extensive damage of the tracheal epithelium, followed by proliferation of surviving cells and restoration of normal histology by 2 weeks (14). During the repair process, labeled BCs proliferate and give rise to relatively large patches of descendants that include Clara and ciliated cells (Fig. S2). By contrast to the findings at steady state, BCs responding to injury give rise to more ciliated than Clara cells over the same period (Table 1). This suggests that the fates of BC daughters can vary in response to local conditions.
Trancriptional Profile of Tracheal BCs.
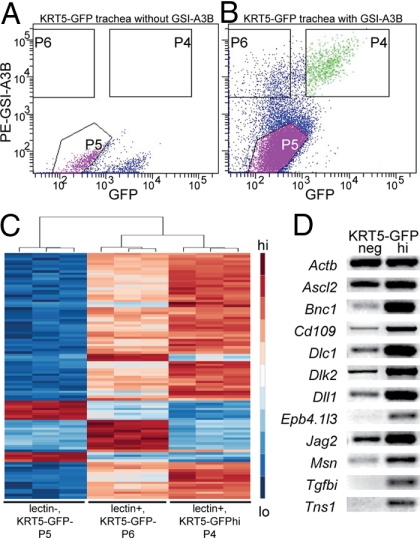
To separate BCs from columnar cells for transcriptional profiling, we first sorted cells on the basis of binding of fluorescently labeled Griffonia simplicifolia isolectin A3B (GSI-A3B), which binds all BCs (3, 10, 15). It also binds a population of dendritic cells present within the epithelium (16). We further purified the lectin+ cells on the basis of expression of GFP using a previously described KRT5-GFP transgenic mouse line in which GFP is expressed at high levels in approximately 60% of p63+ tracheal BCs (3). The lectin+;KRT5-GFP+ cells (P4) were an essentially pure BC subpopulation because >90% express p63. By contrast, the lectin−;KRT5-GFP− (hereafter, non-BC) population (P5) contained very few p63+ cells. The lectin+;KRT5-GFP− cells (P6) contained both basal and dendritic cells (Fig. 2 A and B).
Fig. 2.
Characterization of mouse tracheal epithelial populations. Tracheal epithelial cells were sorted on the basis of labeling with GSI-A3B isolectin and expression of KRT5-GFP. (A) KRT5-GFP tracheal epithelium not labeled with GSI-A3B. (B) Transgenic cells labeled with GSI-A3B. p63+ cells make up 90.5% of cells in P4, 40.4% of cells in P6, and 1.1% of cells in P5. Averaged over 7 experiments, P4 contained 6% of total sorted cells, P5 67%, and P6 12%. (C) Heatmap showing the 100 most differentially expressed genes by Affymetrix microarray of P4, P5, and P6. (D) RT-PCR confirmed the upregulation of 11 genes in KRT5-GFPhi BCs vs. KRT5-GFP− cells.
RNA from all 3 fractions was analyzed by Affymetrix microarray. Clustering of the 100 most differentially regulated genes between the fractions showed good correlation between replicates (Fig. 2C). The genes most highly expressed in the non-BC population included those associated with ciliated and secretory cells (Table S3). As expected, some of the genes upregulated in the lectin+;KRT5-GFP− population are associated with dendritic cells. In the future, new strategies will be used to remove the dendritic cells from this BC fraction. We found 627 genes upregulated at least 21.5-fold in the lectin+;KRT5-GFP+ BCs compared with non-BCs (P ≤ 0.001) (Table S4 and Table S5). The validity of the screen was confirmed in several ways. First, RT-PCR confirmed the enrichment of transcripts in KRT5-GFPhi BCs (Fig. 2D). Second, published reports confirmed expression of several enriched genes in BCs of murine trachea and/or human airways. Examples include genes encoding Trp-63, which is required for BC development in the mouse trachea (2), Snai2, Cd109, claudin 1, Icam1, aquaporin 3, and keratins 5, 14, and 17 (3, 17–21). Finally, immunohistochemistry on sections of mouse trachea and normal human bronchus confirmed BC expression of several genes, including Ngfr (see below).
The genes upregulated in lectin+;KRT5-GFPhi BCs were sorted into functional categories (Table S4). Those associated with cell adhesion include genes encoding components of hemidesmosomes and anchoring fibrils—integrins α6 and β4, laminins α3 and β3, dystonin (BPAG1), and collagen 17a1 (BPAG2) (22). This reflects the abundant hemidesmosomal attachments of BCs to the underlying basal lamina (1). Our results also suggest that BCs produce components of the ECM, including fibulin1, fibrillin2, Tnc, collagen18a1, Sparc, and TGFβ-induced. They are also enriched for cytoskeletal components that influence membrane organization, shape, and motility. Transcription factors that are upregulated in KRT5-GFP+ BCs include Snai2 (Slug) and its corepressor, ajuba (Jub), basonuclin, Lmo1, Tbx2, Barx2, Etv4, Hlf, AP-2 epsilon, Myc, and Ascl2.
Tracheal BCs are normally relatively quiescent but respond rapidly to injury. Both quiescence and activation are likely regulated by reciprocal signaling between BCs and their niche. Transcripts for various signaling ligands (Bmp7, delta-like 1 and 2, jagged2, Ccl20, PdgfC, pleiotrophin, Tgfb1, Ntf3, Wnt3a, Wnt5b, and Wnt9a) and receptors (Notch1, Gfra1, Ngfr, Ptch2, Egfr, Tgfbr3, ephrin receptor B4, and several G protein-coupled receptors) were enriched in this BC subpopulation. In addition, antagonists of intercellular signaling pathways (Socs3, sprouty1, Sfrp1, Dkk3, follistatin-like1, follistatin-like3, Ppp2r2c, and latent TGFβ binding protein 4) were also upregulated.
The rapid proliferative response of BCs to epithelial injury suggests that, although they are normally relatively quiescent, they are poised to respond to activating stimuli. This characteristic is not unique to respiratory BCs. For instance, stem cells in systems that undergo cycles of growth, destruction, rest, and regrowth (e.g., the mammary gland and hair follicle) undergo similar phenotypic switches from quiescence to activation. It is therefore of great interest that a number of genes expressed in stem cells of the hair follicle bulge and mammary gland, including transcription factors, components of the ECM, signaling ligands, receptors, and, importantly, negative regulators of intercellular signaling, are also expressed in tracheal BCs (23, 24). Many of these proteins likely control stem cell–niche interactions and, potentially, the ability of BCs to respond to activating stimuli.
The fact that BCs generate different proportions of ciliated and Clara cells at steady state vs. repair after injury (Table 1) suggests that progeny cell fates are influenced by local tissue conditions. This concept has been proposed for the Drosophila midgut, where varying levels of Notch ligand in the intestinal stem cells regulates the fates of their progeny (25, 26). Significantly, BCs are enriched for transcripts of the Notch pathway (e.g., Notch1, Dll1, and Jag2). Future studies will determine their function in BCs.
Novel Assay for Self-Renewal and Differentiation of BCs.
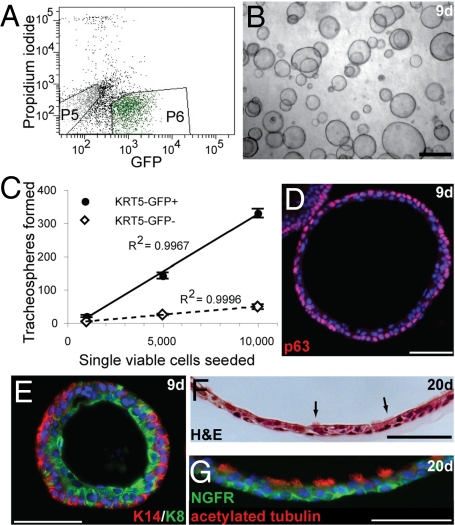
Previous assays for the potential of tracheal BCs to proliferate and differentiate include repopulation of denuded tracheal xenografts and air–liquid interface (ALI) culture. Neither is ideal for quantitative analysis because xenograft models are low-throughput, and we found it difficult to reproducibly differentiate BCs at the ALI in the absence of non-BCs. We therefore adapted 3-dimensional sphere-forming assays from other stem cell populations [e.g., prostate (27)] for the expansion and differentiation of BCs. When single, viable KRT5-GFP+ tracheal BCs were seeded in this assay, “tracheospheres” with a visible lumen formed within 1 week, even in the absence of stroma or non-BCs (Fig. 3 A and B). The sphere-forming efficiency was ≈3% of plated KRT5-GFP+cells. In comparison, the colony-forming efficiency of KRT5-GFP+ cells in 2-dimensional ALI culture in combination with a 500-fold excess of KRT5-GFP− cells was 5% (3). Approximately 0.5% of KRT5-GFP− cells, likely KRT5-GFP− BCs, formed spheres histologically indistinguishable from the KRT5-GFP+ BC-derived spheres (Fig. 3C and Table S6).
Fig. 3.
A novel sphere-forming assay for BC behavior. (A) Single viable KRT5-GFP+ BCs (P6) or KRT5-GFP− (P5) tracheal epithelial cells were seeded. (B) Tracheospheres formed within 9 days of seeding. (C) Approximately 3% of KRT5-GFP+ BCs form tracheospheres, whereas ≈0.5% of KRT5-GFP− cells form spheres. (D and E) 9 days. Tracheospheres stained for (D) p63 (red) and DAPI (blue, nuclei). (E) Basal KRT14 (red) and luminal KRT8 (green). (F and G) Twenty-day differentiated tracheospheres. (F) Hematoxylin and eosin stain. Arrows indicate ciliated cells. (G) Acetylated tubulin (red, cilia) and NGFR (green, BCs). (Scale bars, 1 mm in B; 50 μm in D–G.)
By 9 days of culture, we observed tracheospheres with diameters ranging from <50 μm to >300 μm, suggesting that heterogeneity exists within the sphere-forming population (Fig. S3 and Table S7). Rarely, lobed colonies were observed, but these colonies did not survive over the long term and so have limited capacity for self-renewal in this assay. By day 9, tracheospheres consisted of a pseudostratified epithelium with p63+, KRT14+ BCs peripheral to luminal KRT8+ cells (Fig. 3 D and E). By day 20, surviving spheres had undergone luminal expansion and a thinning of the pseudostratified epithelium, and beating cilia were observed. This was confirmed by antibody staining for acetylated tubulin (Fig. 3G). In 26-day spheres the percentage of KRT8+ columnar cells that were ciliated (≈50%) was approximately the same as in adult wild-type mouse tracheas. Nonciliated KRT8+ cells persisted in spheres but did not express markers of Clara (Scgb1a1), neuroendocrine (CGRP), or mucus-producing (Muc5AC) cells at levels detectable by immunohistochemistry at 9 or 20 days of culture. In addition, very few cells expressed claudin 10. Further experiments are needed to identify this columnar cell type and to optimize conditions for the differentiation of mature secretory cells from BC progenitors. BCs in spheres maintained expression of KRT5-GFP at least up to P21 days. To further demonstrate the self-renewal potential of BCs, we have serially subcultured KRT5-GFP+ cells from spheres twice at the time of submission.
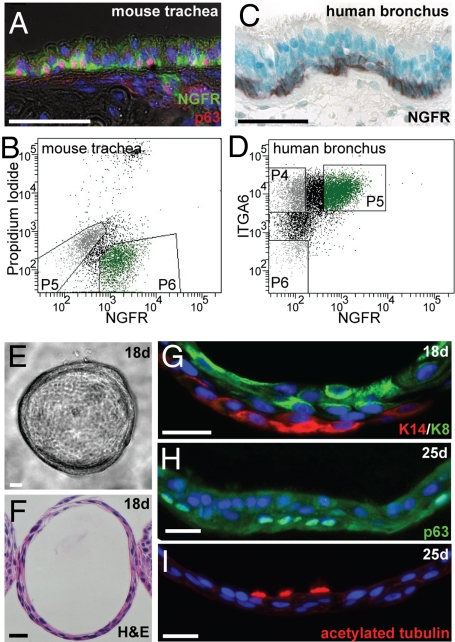
For wide application of the tracheosphere assay, it is important to be able to isolate BCs independently of any transgene. Our transcriptome analysis revealed that Ngfr (p75, Tnfrsf16), a member of the TNF receptor superfamily, is enriched in tracheal BCs (Table S4). Significantly, this gene is also expressed in basal/basal-like cells of other epithelia, including esophagus, corneal limbus, and mammary gland (28–30). Ntf3, encoding a ligand for NGFR, is also enriched in mouse tracheal BCs (Table S5), raising the possibility of autocrine signaling within this population. Immunohistochemistry showed that NGFR is specifically localized to 98% of p63+ mouse BCs (Fig. 4A). We therefore used an NGFR antibody to sort tracheal epithelial cells into basal and nonbasal populations (Fig. 4B). After FACS, 86% of NGFR+ cells were p63+, whereas <1% of the NGFR− cells express p63. The sphere-forming efficiency of NGFR+ BCs was ≈4%, whereas ≈0.05% NGFR− cells gave rise to spheres (Table S6). This finding suggests that although a subset of BCs is able to self-renew and differentiate under these conditions, the ability of BC-depleted populations to do so is significantly reduced. In fact, because 4% of BCs generate spheres and ≈1% of NGFR− cells are p63+ BCs, the 0.05% sphere-forming efficiency in the NGFR− population is potentially attributable to BCs. Spheres derived from NGFR+ cells were histologically the same as those derived from KRT5-GFP+ cells. Furthermore, when NGFR+ BCs from constitutively GFP-expressing or RFP-expressing mice were mixed after sorting, all spheres fluoresced either red or green (Fig. S3D). This finding suggests that spheres are derived from single NGFR+ BCs.
Fig. 4.
NGFR+ BCs of mouse and human airways. (A) NGFR (green) is confined to p63+ (red) BCs in the mouse trachea. (B) FACS using an anti-NGFR antibody to isolate BCs from primary mouse tracheal epithelium. p63+ cells make up >86% of cells in P6. (C) NGFR expression is confined to the BCs of normal human bronchus. (D) FACS using anti-NGFR and anti-ITGA6 antibodies to enrich for BCs from primary human bronchial epithelial cultures. p63+ cells make up 96% of cells in P5, 83% of cells in P4, and only 15% of cells in P6. (E–G) Human spheres at 18 days of culture. (E) Light micrograph. (F) Hematoxylin and eosin–stained section. (G) Basal KRT14 (red) and luminal KRT8 (green). The KRT14+ basal layer appeared somewhat discontinuous. (H and I) Human spheres at 25 days stained for (H) p63 (green, BCs) and (I) acetylated tubulin (red, cilia). (Scale bars, 50 μm in A and C; 25 μm in E–I.)
BCs of the Human Lung.
In humans, p63+ cells with the morphology of BCs are found throughout the lung, although numbers decline distally (1, 4, 5). These cells express a number of genes enriched in mouse BCs, including NGFR (Fig. 4C). To apply our in vitro assay to human BCs, we isolated total epithelial cells from human bronchi and cultured them overnight to enrich for p63+ cells. Our transcriptome analysis of mouse BCs showed that Itga6 is preferentially expressed in tracheal BCs. We therefore sorted human lung epithelial cells by FACS using NGFR and ITGA6 antibodies (Fig. 4D). Under these conditions, ≈96% of the ITGA6+;NGFR+ fraction was p63+, whereas only ≈15% of the ITGA6−;NGFR− cells expressed p63. When seeded in the assay, ITGA6+;NGFR+ BCs gave rise to “bronchospheres” by day 10, with a single lumen that underwent expansion over 20 days of culture (Fig. 4 E–I and Table S8). Very few colonies formed from the ITGA6−;NGFR− cells. Spheres contained KRT14+, p63+ BCs peripheral to KRT8+ luminal cells, and ciliated cells were observed within 25 days of culture. These findings suggest that human BCs are capable of both self-renewal and the generation of differentiated daughters.
In conclusion, we have demonstrated in vivo that BCs of the mouse trachea function as progenitor cells both during postnatal growth and in the adult at steady state and in a repair model. Using a clonal assay, we have shown that BCs of both mouse and human airways can self-renew and differentiate in the absence of stroma or columnar epithelial cells. This assay will facilitate the study of mechanisms regulating the development, maintenance, and repair of the airway epithelium. Finally, we have derived a transcriptional profile of mouse BCs that will inform future investigations on the phenotypic control of this important stem cell population in both normal development and disease.
Materials and Methods
Mice.
The KRT5-GFP transgenic mouse line, Rosa26R-LacZ (Gt(Rosa)26Sortm1Sor), Rosa26R-eYFP (Gt(Rosa)26Sortm1(eYFP)Cos), C57BL/6-Tg(CAG-EGFP)131Osb/LeySopJ, and Gt(ROSA)26Sortm4(ACTB−tdTomato,−EGFP)Luo mice have been described before. The Krt5-GFP transgenic mouse line is maintained by interbreeding on the C57BL/6xC3H background. Rosa26R-LacZ (Gt(Rosa)26Sortm1Sor) and Rosa26R-eYFP (Gt(Rosa)26Sortm1(eYFP)Cos) reporter mice were used at the N3 C57BL/6 backcross generation. To establish the KRT5-CreERT2 line, DNA was injected into fertilized zygotes. The construct is identical to that used to generate the KRT5-GFP line, except that a CreERT2 cassette was inserted in place of GFP. Offspring of 11 founders were screened to obtain 1 that gave robust and inducible recombination in tracheal BCs in response to Tmx. The line is presently on the N3 backcross to C57BL/6.
For adult lineage tracing, 8–10-week-old male and female mice hemizygous for KRT5-CreERT2 and homozygous for Rosa26R-LacZ were injected i.p. 4 times every other day with 0.25 mg/g body weight Tmx in Mazola corn oil (ACH Food Companies). Postnatal mice hemizygous for both KRT5-CreERT2 and Rosa26R-eYFP received i.p. injections of 0.25 mg/g body weight Tmx in corn oil on P10 or P12. Mice injected with corn oil only were included in all experiments. For SO2 injury, adult male mice were exposed to 500 ppm SO2 in air for 3 h 1 week after the final Tmx injection (7).
Immunohistochemistry.
For adult lineage tracing, X-gal–stained paraffin sections were stained with antibody to acetylated tubulin (T7451; Sigma) (7). Cells were scored as ciliated (tubulin+), Clara (columnar, tubulin−), or basal (low, rounded morphology).
For lineage tracing during postnatal growth, cryosections were stained with chick anti-GFP (lineage label, 1:500 GFP1020; Aves Labs), goat anti-Scgb1a1 (Clara cells, 1:10,000; kindly provided by Barry Stripp), mouse anti-acetylated-tubulin (ciliated cells, 1:3,000 T7451; Sigma), and hamster anti-T1α (BCs, 1:1000 clone 8.1.1; DSHB). Alexa-Fluor coupled secondary antibodies (Invitrogen) were used at 1:500. Z-stacks of optical sections were captured on a Leica Sp2 laser scanning confocal microscope. Multiple optical sections were scored manually to distinguish cell boundaries.
For NGFR staining, after citrate buffer antigen retrieval paraffin sections were stained with mouse antihuman NGFR (1:100 05–446; Millipore) and Vectastain ABC Kit (Vector Laboratories) or rabbit antimouse NGFR (Ab8875; Abcam) and Alexa-Fluor coupled secondaries. For staining spheres, paraffin sections were incubated with mouse anti-p63 (BCs, 1:200 sc-8431; Santa Cruz Biotechnology), mouse anti-KRT14 (BCs, 1:500 MS-115-P1; Lab Vision), rat anti-KRT8 (luminal, 1:100 TROMA-I; DSHB), and NGFR and acetylated tubulin as above.
Mouse Tracheal Cell Isolation and FACS.
Mouse tracheas were cut into pieces and incubated in Dispase (BD Biosciences, 16 U/mL) in PBS (30 min) at room temperature. Digestion was stopped by addition of DMEM with 5% FBS. Epithelium was peeled off with forceps and incubated in 0.1% trypsin, 1.6 mM EDTA 20 min at 37 °C, followed by gentle pipetting and passage through a 40-μm cell strainer. GSI-A3B isolectin (a kind gift from Dr. Irwin Goldstein, University of Michigan) was fluorescently labeled using Lightning-Link R-Phycoerythrin Conjugation Kit (Innova Biosciences) and made functionally monovalent by incubation with 5 mM N-acetyl-D-galactosamine (Sigma) (10) before labeling of cells for 40 min on ice. For FACS with NGFR antibody, cells were diluted to 1 × 106 cells/mL in 2% FBS, 2% BSA in PBS and incubated in 18 μg/mL rabbit anti-NGFR (Ab8875; Abcam) or IgG isotype control followed by washing and incubation in allophycocyanin-conjugated or Alexa Fluor 488 donkey antirabbit diluted 1:500. Cells were washed with DMEM + 2% BSA and propidium iodide added to a final concentration of 200 ng/mL before sorting. Sorting was performed on FACS Vantage SE and data analyzed with FACS Diva (BD Biosciences). Cells were collected in DMEM with 2% BSA and cultured immediately or frozen for RNA extraction.
Mouse Tracheosphere Culture.
Sorted cells were resuspended in MTEC/Plus (31), mixed 1:1 with growth factor–reduced Matrigel (BD Biosciences), and 200 μL/cm2 pipetted into a 12-well 0.4-μm Transwell insert (Falcon). MTEC/Plus (1 mL) was added to the lower chamber and changed every other day. Cultures were maintained at 37 °C, 5% CO2. On day 7, MTEC/SF (31) was placed in the lower chamber and changed every other day. The number of spheres per insert was counted on days 7–9 using an inverted microscope. Samples were fixed in 4% paraformaldehyde in PBS, embedded in 3% agarose, then paraffin, and sectioned.
Human BC Isolation and Culture.
Bronchi approximately 1 cm in diameter were processed as described previously (32). Briefly, under institutional review board–approved protocols, excess unaffected airways at the time of tumor resection or donor airways unacceptable for transplantation were obtained and cells dissociated using Protease XIV (Sigma). Cells were plated onto type I/III collagen-coated tissue culture dishes in bronchial epithelial growth medium. After 24 h, day 1 passage 1 cells were harvested using trypsin/EDTA and labeled for FACS using mouse anti-NGFR (5 μg/mL Ab8875; Abcam), Alexa Fluor 488 donkey antimouse, and rat anti-ITGA6 (3 μg/mL Ab19765; Abcam). Anti-ITGA6 was labeled using Lightning-Link R-Phycoerythrin Conjugation Kit (Innova Biosciences). Single viable cells were seeded in 1:1 Matrigel/human ALI medium and fed with ALI medium every other day.
Microarray Analysis.
KRT5-GFP tracheal epithelial cells were separated by FACS into 3 populations: lectin+;K5-GFPhi, lectin+;KRT5-GFP−, and lectin−;KRT5-GFP−. At least 5 mice were used per sort, and samples were pooled from 7 sorts to generate triplicates. RNA was extracted from frozen cells using RNeasy Micro kit (Qiagen) and quality checked with a 2100 Bioanalyzer (Agilent Technologies). RNA (20–30 ng per sample) was amplified and labeled using the Ovation RNA Amplification Kit V2 and FL-Ovation Biotin V2 (NuGEN) by the Duke Microarray Facility. Standard Affymetrix protocols and mouse genomic 430 2.0 chips were used to generate .cel files deposited in the Gene Expression Omnibus of the National Center for Biotechnology Information (accession number GSE15724). These were imported into Bioconductor in the R software environment and preprocessed using robust multichip averaging. Calculation of fold change difference and t tests was performed for each probe set between the nonbasal and lectin+;K5-GFPhi groups. Probe sets with a P value <0.001 and fold change >2.8 were annotated. The functions lmFit, contrasts.fit, and eBayes were used to make all pairwise comparisons between the robust multichip average–normalized data for the 3 groups of cells. The functions decideTests and order were used to order significantly different probes, and the top 100 were used to create the heatmap using the function heatmap.
RT-PCR.
RNA was isolated from tracheal epithelium sorted into KRT5-GFPhi and KRT5-GFP− populations from 8 adult KRT5-GFP hemizygous mice and cDNA synthesized from 120 ng RNA using SuperScript III reverse transcriptase. PCR primers are listed in Table S9.
Supplementary Material
Acknowledgments.
We thank Xiaoyan Luo and Tim Oliver for assistance with FACS and imaging, respectively, and members of the Hogan laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grants HL071303 (to B.L.M.H.) and K12 CA100639–03 (to M.O.) and by a Howard Hughes Medical Institute Early Career Grant (to M.O.). J.R. is supported by training grant 2T32HL007538–26. E.R. is a recipient of a Parker B. Francis Fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession number GSE15724).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906850106/DCSupplemental.
References
- 1.Evans MJ, Van Winkle LS, Fanucchi MV, Plopper CG. Cellular and molecular characteristics of basal cells in airway epithelium. Exp Lung Res. 2001;27:401–415. doi: 10.1080/019021401300317125. [DOI] [PubMed] [Google Scholar]
- 2.Daniely Y, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 3.Schoch KG, et al. A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol. 2004;286:L631–L642. doi: 10.1152/ajplung.00112.2003. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, et al. Immunohistochemical and ultrastructural studies of basal cells, Clara cells and bronchiolar cuboidal cells in normal human airways. Pathol Int. 1998;48:944–953. doi: 10.1111/j.1440-1827.1998.tb03865.x. [DOI] [PubMed] [Google Scholar]
- 5.Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am J Respir Crit Care Med. 1998;157(6 Pt 1):2000–2006. doi: 10.1164/ajrccm.157.6.9707011. [DOI] [PubMed] [Google Scholar]
- 6.Kauffman SL. Cell proliferation in the mammalian lung. Int Rev Exp Pathol. 1980;22:131–191. [PubMed] [Google Scholar]
- 7.Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: Contribution of the ciliated lineage. Proc Natl Acad Sci USA. 2007;104:410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: Evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol. 2004;286:L643–649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 9.Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, Randell SH. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir cell Mol Biol. 2001;24:662–670. doi: 10.1165/ajrcmb.24.6.4217. [DOI] [PubMed] [Google Scholar]
- 10.Randell SH, Comment CE, Ramaekers FC, Nettesheim P. Properties of rat tracheal epithelial cells separated based on expression of cell surface alpha-galactosyl end groups. Am J Respir Cell Mol Biol. 1991;4:544–554. doi: 10.1165/ajrcmb/4.6.544. [DOI] [PubMed] [Google Scholar]
- 11.Hajj R, et al. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells. 2007;25:139–148. doi: 10.1634/stemcells.2006-0288. [DOI] [PubMed] [Google Scholar]
- 12.Avril-Delplanque A, et al. Aquaporin-3 expression in human fetal airway epithelial progenitor cells. Stem Cells. 2005;23:992–1001. doi: 10.1634/stemcells.2004-0197. [DOI] [PubMed] [Google Scholar]
- 13.Rawlins EL, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawlins EL, Hogan BL. Ciliated epithelial cell lifespan in the mouse trachea and lung. Am J Physiol Lung Cell Mol Physiol. 2008;295:L231–L234. doi: 10.1152/ajplung.90209.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu T, Nettesheim P, Mahler JF, Randell SH. Cell type-specific lectin staining of the tracheobronchial epithelium of the rat: Quantitative studies with Griffonia simplicifolia I isolectin B4. J Histochem Cytochem. 1991;39:7–14. doi: 10.1177/39.1.1701188. [DOI] [PubMed] [Google Scholar]
- 16.Sertl K, et al. Dendritic cells with antigen-presenting capability reside in airway epithelium, lung parenchyma, and visceral pleura. J Exp Med. 1986;163:436–451. doi: 10.1084/jem.163.2.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parent AE, Choi C, Caudy K, Gridley T, Kusewitt DF. The developmental transcription factor slug is widely expressed in tissues of adult mice. J Histochem Cytochem. 2004;52:959–965. doi: 10.1369/jhc.4A6277.2004. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, et al. High-level expression of CD109 is frequently detected in lung squamous cell carcinomas. Pathol Int. 2007;57:719–724. doi: 10.1111/j.1440-1827.2007.02168.x. [DOI] [PubMed] [Google Scholar]
- 19.Paschoud S, Bongiovanni M, Pache JC, Citi S. Claudin-1 and claudin-5 expression patterns differentiate lung squamous cell carcinomas from adenocarcinomas. Mod Pathol. 2007;20:947–954. doi: 10.1038/modpathol.3800835. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, Rogers AV, Burke-Gaffney A, Hellewell PG, Jeffery PK. Cytokine-induced airway epithelial ICAM-1 upregulation: Quantification by high-resolution scanning and transmission electron microscopy. Eur Respir J. 1999;13:1318–1328. doi: 10.1183/09031936.99.13613299. [DOI] [PubMed] [Google Scholar]
- 21.Liu YL, et al. Expression of aquaporin 3 (AQP3) in normal and neoplastic lung tissues. Hum Pathol. 2007;38:171–178. doi: 10.1016/j.humpath.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Margadant C, Frijns E, Wilhelmsen K, Sonnenberg A. Regulation of hemidesmosome disassembly by growth factor receptors. Curr Opin Cell Biol. 2008;20:589–596. doi: 10.1016/j.ceb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 25.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 26.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 27.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reis-Filho JS, et al. Distribution and significance of nerve growth factor receptor (NGFR/p75NTR) in normal, benign and malignant breast tissue. Mod Pathol. 2006;19:307–319. doi: 10.1038/modpathol.3800542. [DOI] [PubMed] [Google Scholar]
- 29.Daiko H, et al. Molecular profiles of the mouse postnatal development of the esophageal epithelium showing delayed growth start. Int J Mol Med. 2006;18:1057–1066. [PubMed] [Google Scholar]
- 30.Di Girolamo N, et al. Localization of the low-affinity nerve growth factor receptor p75 in human limbal epithelial cells. J Cell Mol Med. 2008;12:2799–2811. doi: 10.1111/j.1582-4934.2008.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: Selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- 32.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.