Abstract
Mice lacking p63 cannot form skin, exhibit craniofacial and skeletal defects, and die soon after birth. The p63 gene regulates a complex network of target genes, and disruption of p63 has been shown to affect the maintenance of epithelial stem cells, the differentiation of keratinocytes, and the preservation of the adhesive properties of stratified epithelium. Here, we show that inactivation of p63 in mice is accompanied by aberrantly increased expression of the Ink4a and Arf tumour suppressor genes. In turn, anomalies of the p63-null mouse affecting the skin and skeleton are partially ameliorated in mice lacking either Ink4a or Arf. Rescue of epithelialization is accompanied by restoration of keratinocyte proliferative capacity both in vivo and in vitro and by expression of markers of squamous differentiation. Thus, in the absence of p63, abnormal upregulation of Ink4a and Arf is incompatible with skin development.
Keywords: adhesion, differentiation, epidermal stem cells, Ink4a–Arf, p63-isoforms
Introduction
p63, a p53 family member, has multiple roles in the development of stratified epithelium (Mills et al, 1999; Yang et al, 1999; Koster and Roop, 2004; Ihrie et al, 2005; Carroll et al, 2006, 2007; Koster et al, 2007; Senoo et al, 2007). Deletion of p63 in the mouse results in a generalized loss of stratified epithelium because of apparent defects in the maintenance of stem cell proliferative capacity, keratinocyte differentiation, and cell–cell adherence (Mills et al, 1999; Yang et al, 1999; Senoo et al, 2007). These functions are controlled by a vast transcriptional network of genes regulated by p63 (Vigano et al, 2006; Yang et al, 2006; Vigano and Mantovani, 2007), but the specific molecular mechanisms governing each of these processes remain largely ill defined.
The Ink4a–Arf locus encodes two tumour suppressor proteins, p16Ink4a and p19Arf, which induce apoptosis, cell-cycle arrest, or senescence by regulating the activities of the retinoblastoma protein and p53, respectively (Sherr and McCormick, 2002). Repression of Ink4a–Arf gene expression helps to maintain haematopoietic stem cell (HSC) and neural stem cell (NSC) function (Jacobs et al, 1999; Molofsky et al, 2003, 2005; Park et al, 2003; Bruggeman et al, 2005; Bracken et al, 2007). The Ink4a–Arf locus is epigenetically silenced by polycomb complexes in adult bone marrow-derived HSCs. Inactivation of the polycomb component, Bmi1, results in the apparent loss of HSC self-renewal capacity and leads to bone marrow aplasia in the first few weeks after birth, but this defect is largely alleviated when Bmi1-null mice are crossed onto an Ink4a–Arf-null background (Jacobs et al, 1999). Recent investigations indicate that inactivation of Ink4a–Arf increases the self-renewal capacity of multiple progenitor cells rather than affecting the frequency of HSCs from which they are derived (Akala et al, 2008). Self-renewal and differentiation of presumptive NSCs in Bmi1-null mice and in young mice lacking the Hmga2 transcriptional regulator are also partially restored with loss of Ink4a or Arf (Molofsky et al, 2003, 2005; Nishino et al, 2008). Conversely, increased expression of Ink4a–Arf as mice age progressively limits the repopulating efficiencies of HSCs and NSCs and the proliferative capacities of B-lymphoid and pancreatic islet cells (Janzen et al 2006; Krishnamurthy et al, 2006; Molofsky et al, 2006; Signer et al, 2008). Although derepression of both products of the Ink4a–Arf locus each contribute to these phenotypes, p16Ink4a and p19Arf exert differential, cell type-specific effects during haematopoiesis, neurogenesis, and lymphopoiesis (Bruggeman et al, 2005; Molofsky et al, 2005, 2006; Miyazaki et al, 2008; Signer et al, 2008).
Negative regulation of Ink4a–Arf gene expression may be essential for tissue stem cell renewal and lineage commitment, but the putative repressors that might serve this function in many lineages have yet to be identified. We reasoned that p63, which is expressed at high levels in the basal layer of the epidermis (Senoo et al, 2007; Fuchs, 2008), might similarly have a function in suppressing Ink4a–Arf gene expression. Indeed, somatic deletion of p63 in the mouse embryo from E8.5 onward or conditional inactivation of p63 in cultured keratinocytes induces a senescence phenotype characterized in part by upregulation of p16Ink4a (Keyes et al, 2005). However, shRNA-mediated knockdown of p16Ink4a was insufficient to bypass senescence induced by p63 ablation implying that other regulators must contribute to the observed phenotype. We, therefore, interbred p63+/− mice with animals lacking either Ink4a or Arf to generate mice lacking both Ink4a and p63 or Arf and p63. Mice with these compound gene deficiencies form stratified epithelium, and their keratinocytes exhibit a renewed capacity to proliferate and differentiate both in vivo and in vitro. These findings show that p63 negatively regulates the Ink4a–Arf locus and that abnormal upregulation of these genes in the absence of p63 inhibits skin development.
Results
Loss of Arf or Ink4a rescues features of the phenotype of p63−/− mice
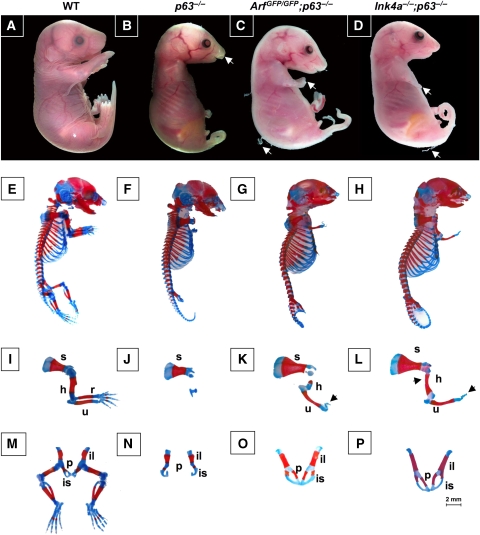
Mice lacking functional p63 are born with defects in the epidermis, have cleft lip and palate, and craniofacial abnormalities (Mills et al, 1999; Yang et al, 1999). These mice die within hours after birth because of desiccation resulting from the lack of a stratified epidermis covering the mouse. We hypothesized that loss of Arf or Ink4a could lead to the formation of a complete epithelium in the p63−/− mice; therefore, ArfGFP/GFP (Zindy et al, 2003) and Ink4a−/− (Sharpless et al, 2001) mice were intercrossed with the p63+/− mice (Yang et al, 1999) to first obtain double heterozygous mice (ArfGFP/+;p63+/− and Ink4a+/−;p63+/−). Subsequent matings by intercrossing compound heterozygous mice gave rise to ArfGFP/GFP;p63−/− and Ink4a−/−; p63−/− mice, respectively. The ArfGFP/GFP knock-in allele is functionally null but yields green fluorescent cells when the Arf promoter is activated (Zindy et al, 2003). Macroscopic examination of 10 neonates of each genotype showed the presence of skin covering 80–95% of the body, but these animals did not survive after birth because of maternal neglect. Day 18.5 embryos (E18.5) of ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− mice had also developed a thin layer of skin covering 85–90% of the body (Table I). Twenty ArfGFP/GFP;p63−/− and 20 Ink4a−/−;p63−/− E18.5 embryos had dull and opaque skin with partially sloughed stratified epithelium (Figure 1C and D), compared with the shiny and translucent appearance of the p63−/− embryos devoid of such epithelium (Figure 1B). In addition, both double mutant embryos exhibited rudimentary limbs and a long tail (Figure 1C and D). Strikingly, 16 of 20 ArfGFP/GFP;p63−/− and 18 of 20 Ink4a−/−;p63−/− mice had normal facial features including a complete palate, lip, and fully developed ears (Figure 1C and D) (Table I).
Table 1.
Quantification of epithelial structures in day 18.5 embryos
| Genotype | Embryos analysed | K5 | K14 | K15 | K10 | Filaggrin | Embryos w/hair follicle | Embryos w/cleft palate | Embryos w/forelimbs |
|---|---|---|---|---|---|---|---|---|---|
| Wild type | 20 | 100% | 100% | 100% | 100% | 100% | 20 | 0 | 20 |
| p63−/− | 20 | 2% | 5% | 0% | 0% | 0% | 0 | 20 | 3 |
| ArfG/G;p63−/− | 20 | 85% | 85% | 90% | 75% | 50% | 0 | 4 | 18 |
| Ink4a−/−;p63−/− | 20 | 85% | 85% | 100% | 85% | 75% | 20 | 2 | 20 |
| p53−/−;p63−/− | 6* | 0% | 0% | 0% | 0% | 0% | 0 | 6 | 0 |
| Shown are the numbers of embryos analysed per genotype. Numbers in columns with % represent the mean from 20 embryos analysed per group. The asterisk (*) indicates that only 6 of 20 p53−/−;p63−/− embryos could be analysed at E18.5; the remaining 14 embryos died in utero at day 15.5. | |||||||||
Figure 1.
Macroscopic analysis of embryos at day E18.5. (A–D) Illustrate images of embryos with their respective genotypes indicated above each panel; WT indicates wild type. The white arrow in (B) points to cleft lip and palate in p63−/− embryo. White arrows in (C, D) designate strings of desquamated epithelium and forelimbs. (E–P) Show skeletal preparations from E18.5 embryos stained with Alcian Blue for cartilage and alizarin red for bone. Whole embryos (E–H) were of the following genotypes: (E), wild type; (F), p63−/−; (G), ArfGFP/GFP;p63−/−; and (H), Ink4a−/−;p63−/−. Analysis of forelimbs (I–L) from (I), wild type; (J), p63−/−; (K), ArfGFP/GFP;p63−/−; and (L), Ink4a−/−;p63−/− embryos. The arrowheads in (K) and (L) point to rudimentary digits. Designated bones include the scapula (s), humerus (h), radius (r), and ulna (u). Analysis of pubic bones and hindlimbs (M–P) from (M), wild type; (N), p63−/−; (O), ArfGFP/GFP;p63−/−; and (P), Ink4a−/−;p63−/− embryos. Designated bones include the ilium (il), ischium (is), and pubic bone (p). A 2 mm scale bar is included in the corner of (P).
A more rigorous analysis of the skeletal structures of 10 embryos of each genotype was therefore undertaken (Figure 1E–P) (Table II). In the ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− embryos, the scapula, humerus, and ulna were well developed and in most cases comparable in size to wild-type embryos (Figure 1I–L) (Table II). In addition, rudimentary digits were apparent in both ArfGFP/GFP; p63−/− and Ink4a−/−;p63−/− embryos (Figure 1K and L), whereas the forelimbs were significantly smaller in the p63−/− embryos (Figure 1J) (Table II). Similarly to the p63−/− embryos, the ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− embryos did not develop hindlimbs; however, their pubic bones were comparable in size to those of wild-type mice and were fused (compare Figure 1O and P with Figure 1M) (Table II), a process that did not occur in the p63−/− mice (Figure 1N). Thus, inactivation of Ink4a and Arf contributes independently to partially reverse cardinal features of the p63-null phenotype.
Table 2.
Quantification of length of limbs in day 18.5 embryos
| Genotype | Scapula | Humerus | Ulna | Pubic bone |
|---|---|---|---|---|
| Wild type | 4.8±0.2 | 4.4±0.1 | 4.8±0.2 | 3.9±0.3 |
| p63−/− | 3.8±0.1 | 0.5±0.1 | 1.5±0.2 | 2.6±0.1 |
| ArfG/G;p63−/− | 5.0±0.1 | 1.5±0.2 | 2.8±0.3 | 3.9±0.1 |
| Ink4a−/−;p63−/− | 5.0±0.1 | 2.8±0.3 | 4.5±0.3 | 3.9±0.2 |
| Shown are the mean±s.d. measured in mm of the indicated skeletal structures. Measurements were taken from 10 embryos per genotype. | ||||
Loss of Arf or Ink4a leads to formation of a uniform layer of skin in the absence of p63
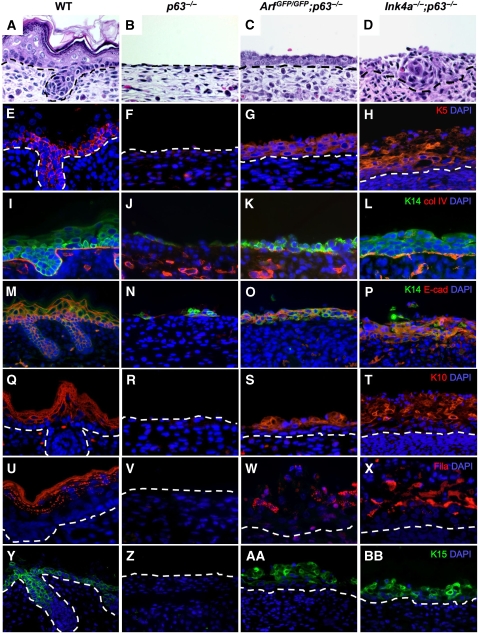
From macroscopic examination of double mutant embryos, we reasoned that loss of Arf or Ink4a alone had rescued epidermal defects detected in the p63−/− embryos. The epidermis of the embryos was characterized by microscopic analysis using haematoxylin and eosin (H&E) staining and immunofluorescence (IF) for the epidermal markers, keratin 5 (K5), keratin 14 (K14), keratin 10 (K10), keratin 15 (K15), and filaggrin (Figure 2; Table I). The H&E-stained sections of E18.5 ArfGFP/GFP;p63−/− embryos showed an intact basal layer of epithelium surrounding the mice (Figure 2C). Twenty ArfGFP/GFP;p63−/− embryos had an average coverage of 85% as determined by H&E staining. The extent of rescue in the 20 Ink4a−/−;p63−/− embryos examined was even more striking with the appearance of structures that resembled hair follicles (Figure 2D; Table I); 10±2 rudimentary hair follicles per centimetre were apparent in all 20 of the Ink4a−/−;p63−/− embryos as compared with 64±4 cm in the wild type and none in the ArfGFP/GFP;p63−/− and p63−/− mice (Table I). These data highlight the fact that inactivation of Ink4a and Arf exerts differential phenotypic effects in the p63-null background.
Figure 2.
Microscopic analysis of the epithelium of embryos at E18.5. Cross sections of skin stained with haematoxylin and eosin (A–D) and immunofluorescence signals using antibodies to keratin 5 (red) (E–H), keratin 14 (green) and collagen IV (red) (I–L), keratin 14 (green) and E-cadherin (red) (M–P), keratin 10 (red) (Q–T), filaggrin (red) (U–X), and keratin 15 (green) (Y–BB) and counterstained with DAPI (blue) are illustrated. The genotypes for each vertical set of panels are indicated at the top of the figure. Broken lines define the border between the dermis and epidermis.
The structure of the epithelium was further analysed by IF using antibodies to K5 and K14, markers of the basal layer, K10, a marker of the spinous layer, and filaggrin, a marker of the granular layer of the epithelium. Immunofuorescence was also performed using markers for the basement membrane, collagen IV (Figure 2I–L), and for cellular adhesion, E-cadherin (Figure 2M–P). All of these markers are completely absent or are only weakly detected in the p63−/− mice (Mills et al, 1999; Yang et al, 1999). Consistent with earlier published data (Yang et al, 1999), 3 of the 20 p63−/− embryos had an average of 2% K5-positive or 5% K14-positive cells in patches above the dermis (Figure 2F, J, and N) (Table I). K5-positive and K14-positive cells in the ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− mice encircled the embryos (Figure 2G, H, K, L, O and P) (Table I) and formed a multi-layer stratum basale. Double IF for K14 and collagen IV further showed a multi-layer stratum basale above the basement membrane marked by collagen IV in the ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− embryos (Figure 2K and L). Immunostaining for K14 and E-cadherin showed cellular disorganization in the stratum basale and somewhat disrupted cell–cell adhesion in the double mutant embryos (Figure 2O and P). In addition, K10 and filaggrin staining were apparent, consistent with further differentiation of suprabasal cells (Figure 2S, T, W and X) (Table I). In the Ink4a−/−;p63−/− mice, the appearance of a multi-layer stratified epithelium, which stained positively for K5, K14, K10, and filaggrin (Figure 2H, L, P, T and X), was even more apparent, and hair follicle-like structures that stained positively for K15, a marker of epithelial stem cells residing in the hair follicle bulge (Liu et al, 2003), were also visible (Figure 2Y, AA, and BB). K15 staining was apparent in the stratum basale of the epidermis of ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− embryos (Figure 2AA and BB) indicating the presence of epidermal precursor cells, as was evident in the developing epidermis of wild-type embryos (Figure 2Y).
To determine whether loss of p53 could likewise rescue the p63−/− phenotype, p53−/−;p63−/− embryos were also analysed at E18.5. Fourteen of 20 p53−/−;p63−/− embryos died in utero at day E15.5. Some had neural tube closure defects as has been reported earlier for p53−/− mice (Sah et al, 1995). However, 6 of 20 embryos were viable at day E18.5 and had a similar phenotype to p63−/− embryos (Table I), raising the possibility that rescue of the p63-null phenotype by Arf inactivation might be mediated, at least in part, through a p53-independent pathway.
Proliferation of p63−/− keratinocytes is restored in the absence of Arf or Ink4a
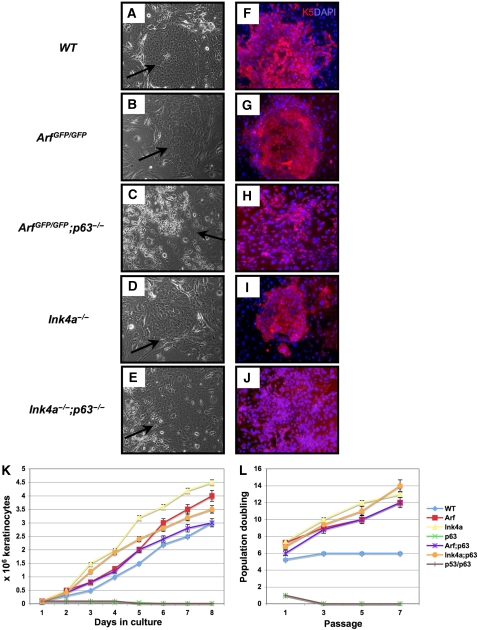
Owing to grossly abnormal epithelialization in p63−/− mice, it is difficult to explant and culture keratinocytes from these embryos. Indeed, when we attempted to culture keratinocytes on J2-3T3 feeder cells under conditions that normally allow expansion of clones from epidermal precursors (Barrandon and Green, 1987; Flores et al, 2000), cells could be extracted from the surface of p63−/− embryos, but they did not proliferate (Koster et al, 2004). In contrast, when epidermis was isolated from ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− embryos at E18.5, recovered keratinocytes formed colonies much similar to those formed by wild-type keratinocytes (Figure 3A–E), and these stained positively with antibodies to the stratum basale marker, K5 (Figure 3F–J). However, the double null colonies were less compact than wild-type colonies (Figure 3C, E, H, and J), and the cells seemed to be less adherent to one another, based on their propensity to be more easily detached from the plate and dispersed by trypsinization. This feature is not surprising, given the discontinous staining of E-cadherin in ArfGFP/GFP;p63−/− and Ink4a−/−; p63−/− embryos and that p63 is known to regulate genes involved in cell adhesion (Ihrie et al, 2005; Carroll et al, 2006).
Figure 3.
Keratinocyte cultures derived from embryos at E18.5. (A–E) Show light micrographs of keratinocytes cultured on J2-3T3 feeder cells with their respective genotypes indicated to the left of the panels. Black arrows indicate the positions of keratinocyte colonies. (F–J) Show immunofluorescence obtained with an antibody to keratin 5 (red), a marker of basal epithelial cells; cells were counterstained with DAPI (blue). (K) Indicates the proliferation rates of cultured keratinocytes of the following genotypes: wild type (WT, blue), ArfGFP/GFP (Arf, red), Ink4a−/− (Ink4a, yellow), p63−/− (p63, green), p53−/−;p63−/− (brown), ArfGFP/GFP;p63−/− (Arf;p63, purple), and Ink4a−/−;p63−/− (Ink4a;p63, orange). (L) Computes the cumulative population doublings of keratinocyte cultures passaged every 5 days for seven passages. The average of results obtained with three independent keratinocyte lines each assayed in triplicate is shown. The genotypes are indicated as in (K).
Although p63−/− keratinocytes at passage-2 did not proliferate further, cells of all the other genotypes continued to accumulate over an 8-day time course (Figure 3K). The most rapidly expanding keratinocyte populations were those derived from the Ink4a−/− and ArfGFP/GFP embryos, but the ArfGFP/GFP;p63−/−, and Ink4a−/−;p63−/− keratinocytes divided as rapidly as wild-type keratinocytes. In addition to measuring their proliferative rates, we passaged the cultured keratinocytes every 5 days, and assessed their cumulative population doublings for seven passages (Figure 3L). The p63−/− keratinocytes senesced at passage-1, whereas cells of all other genotypes continued to proliferate on further passage. The wild-type keratinocytes senesced at passage-3, but the Ink4a−/−, ArfGFP/GFP, Ink4a−/−;p63−/−, and ArfGFP/GFP; p63−/− keratinocytes reached 12–14 accumulated population doublings by passage-7 (Figure 3L). Notably, in both assays, the p53−/−;p63−/− keratinocytes behaved similarly to p63−/− keratinocytes (Figure 3K and L) providing further evidence that loss of p53 does not rescue the ability of p63−/− keratinocytes to proliferate. Taken together, these results show that loss of either Arf or Ink4a can rescue the proliferative defect of p63-deficient keratinocytes and that this rescue is not phenocopied by p53 inactivation.
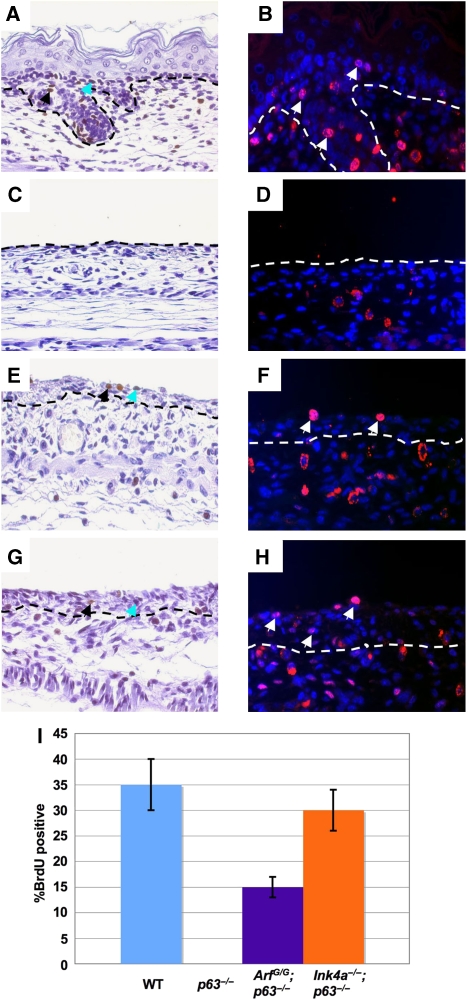
The ability of rescued epidermal cells to proliferate in vivo was determined by the incorporation of the thymidine analogue, bromodeoxyuridine (BrdU), into the DNA of the developing epidermis of wild type, p63−/−, ArfGFP/GFP;p63−/−, and Ink4a−/−;p63−/− embryos at day E18.5 (Figure 4A–H). BrdU-positive cells could be detected in multiple layers of the epidermis in ArfGFP/GFP;p63−/− and Ink4a−/−;p63−/− embryos (Figure 4E–H). Consistent with the findings made with cultured keratinocytes, inactivation of Ink4a or Arf significantly rescued the proliferation of p63-null cells, with elimination of Ink4a having the greater effect and restoring the proliferative capacity of the cells to wild-type levels (Figure 4I).
Figure 4.
In vivo proliferation of epidermal cells in embryos at E18.5. (A, C, E, G) Show cross sections of skin labeled with BrdU (brown) and counterstained with haematoxylin (purple). Black arrows indicate positive cells and blue arrows indicate cells that stain weakly for BrdU. (B, D, F, H) Show immunofluorescence staining of skin sections with BrdU (red) and counterstained with DAPI (blue). Genotypes of the embryos are as follows: wild type (A, B), p63−/− (C, D), ArfGFP/GFP;p63−/− (E, F), and Ink4a−/−;p63−/− (G, H). White arrows indicate positive cells. Both were used to calculate the mitotic index shown in the bar graph in (I). The mitotic index is shown on the x-axis as the percentage of BrdU-positive cells on the surface of the epidermis.
p63 downregulates p19Arf and p16Ink4a to rescue the proliferation and differentiation defects of cells lacking p63
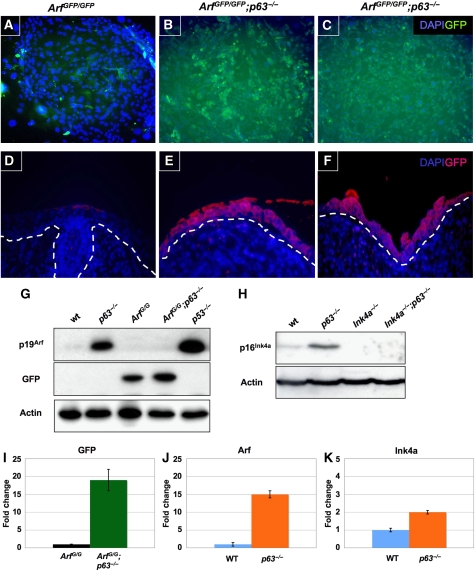
The failure of p63−/− mice to generate proliferative keratinocytes together with the rescue of this process in Arf-null and Ink4a-null backgrounds suggests that p63 negatively regulates p19Arf and p16Ink4a expression. Presumably, in the absence of p63, the untimely expression of p16Ink4a and p19Arf inhibits the outgrowth of keratinocytes, leading to the profound defects in epidermal development. Indeed, upregulation of p16Ink4a in cultured primary keratinocytes was observed earlier after conditional p63 ablation, but in this setting, shRNA-mediated knockdown of Ink4a expression did not bypass cellular senescence; p19Arf expression was not studied (Keyes et al, 2005). We made use of the GFP reporter built into the ArfGFP/GFP knock-in mouse. Arf expression is not detectable during normal mouse epithelial development but is induced when cells are explanted into culture (Zindy et al, 2003). Direct measurements of GFP fluorescence indicated that ArfGFP/GFP;p63−/− keratinocytes expressed significantly higher GFP levels than those detected in age-matched ArfGFP/GFP keratinocyte cultures (Figure 5A–C). Most pertinent, GFP levels were significantly elevated in the epithelium of ArfGFP/GFP;p63−/− embryos when compared with those in ArfGFP/GFP embryos at E18.5 (Figure 5D–F). Immunoblotting of lysates from cultured keratinocytes documented a two-fold increase in GFP protein expression in ArfGFP/GFP;p63−/− cells versus that in their ArfGFP/GFP counterparts (Figure 5G, lanes 3 and 4). Strikingly, the difference in the level of p19Arf expressed in wild type versus p63−/− cells was much greater (Figure 5G, lanes 1 and 2) indicating that p63 suppressed p19Arf expression. The p19Arf protein levels were also elevated in MEFs lacking p53 (Figure 5G, lane 5) as reported earlier (Kamijo et al, 1998; Stott et al, 1998). Likewise, similar increases in the levels of p16Ink4a were detected in the absence of p63 also indicating that p63 downregulates p16Ink4a expression (Figure 5H).
Figure 5.
Expression of p19Arf, p16Ink4a, and GFP in keratinocytes and stratified epithelium. (A–C) Illustrate GFP expression in keratinocytes of the indicated genotypes (top). Cells were counterstained with DAPI (blue). Representative colonies of cultured ArfGFP/GFP;p63−/− keratinocytes (B, C) were vividly fluorescent when compared (matched exposures) with cultures at the same passage derived from ArfGFP/GFP;p63+/+ mice (A). (D–F) Show cross sections of stratified epithelium from E18.5 embryos stained with antibodies to GFP (red). DAPI (blue) was used as a counterstain. The respective genotypes correspond to those indicated above (A–C). Matched exposures are shown. (G, H) Illustrate immunoblotting analysis of selected proteins (indicated at the left) identified in detergent lysates of age-matched passage-2 keratinocytes (B) of the indicated genotypes (top). Passage-2 p53−/− MEFs were used as a positive control (G). Actin was used as a loading control. (I–K) Show results from qRT–PCR of the indicated mRNAs (top) extracted from E18.5 dermis/epidermis of the indicated genotypes (bottom). GAPDH was used as an internal control.
Quantitative real-time (qRT) PCR was performed on RNA extracted from embryonic skin containing the dermis and attached epidermal cells of embryos of the indicated genotypes (Figure 5I–K). Notably, the analysis of RNA expressed in dermal/epidermal tissues taken directly from embryos bypasses any inductive effects of ‘culture shock' seen with explanted cells propagated in vitro. Indeed, we found that levels of GFP RNA were 18-fold higher in ArfGFP/GFP;p63−/− versus ArfGFP/GFP embryos (Figure 5I). Similarly, Arf levels were found to be 15-fold higher in p63−/− compared with wild-type embryos (Figure 5J). Likewise, Ink4a RNA levels were increased in the absence of p63, although to a lesser extent (Figure 5K).
p63 directly represses Ink4a and Arf expression
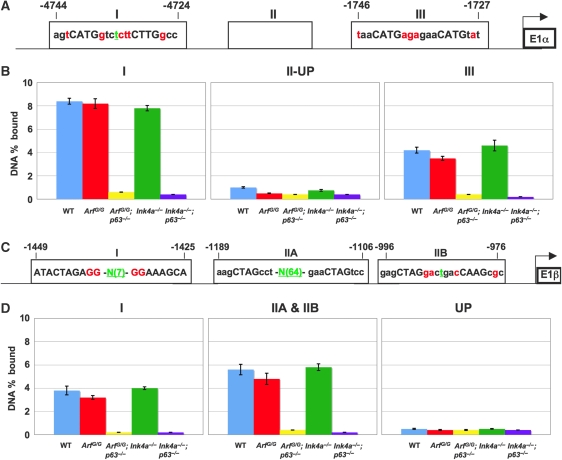
Both the Ink4a and Arf promoters were found to have elements that match the p53 consensus binding site (Figure 6A and C). Chromatin immunoprecipitation (ChIP) analysis using an antibody for p63 and primers to amplify the discovered sites by qRT–PCR identified p63-binding sites upstream of the Ink4a and Arf promoters (Figure 6). Two sites were identified in both promoters (Figure 6B and D). The sites bound by p63 on the Ink4a promoter were located between nucleotides -4835 and -4621 (Figure 6A and B[I]) and -1850 and -1539 (Figure 6A and B[III]). Other sites analysed within the Ink4a promoter from nucleotides -3996 and -3845 (Figure 6A and B [II-UP]), between -3632 and -3362 (Supplementary Figure 1A[II-DOWN]), and -1519 and -1157 (Supplementary Figure 1B[IV]) did not show p63 binding. The p63-binding sites identified on the Arf promoter reside between -1533 and -1331 (Figure 6C and D[I]) and between -1276 and -943 (Figure 6C and D[IIA and IIB]). Other amplicons analysed upstream, -1811 and -1622 (Figure 6C and D[UP]), or in intervening sequences, -1417 and -1303 (Supplementary Figure 1C[INT]), were negative for p63 binding. As a positive control, ChIP analysis was performed using an antibody for p63 and primers to amplify known transcriptional targets of p63, p21, and Perp, by qRT–PCR (Supplementary Figure 2).
Figure 6.
p63 binds to response elements located on the Ink4a and Arf promoters. (A) Illustrates two p53/p63 consensus binding sites in the Ink4a promoter upstream of exon 1α. (B) Shows the percentage of total input DNA bound (DNA percentage bound) to p63. A graph for each region amplified (1-nucleotides -4835 to -4621, II-UP-nucleotides -3996 to -3845, and III-nucleotides -1850 to -1539) is shown. (C) Shows three p53/p63 consensus binding sites in the Arf promoter upstream of exon 1β. (D) Shows the percentage of total input DNA bound (DNA percentage bound) to p63. A graph for each region amplified (1-nucleotides -1533 to -1331, IIA and IIB-nucleotides -1276 to -943, and UP denotes a site upstream of site (I) from nucleotides -1811 to -1622). Genotypes of keratinocytes analysed are indicated below each bar on the graph. Nucleotides shown in red are those that do not match the consensus. Nucleotides in green correspond to those found in the spacer region. If the spacer was greater than 5, N(X) is shown where X=number of nucleotides in the spacer region.
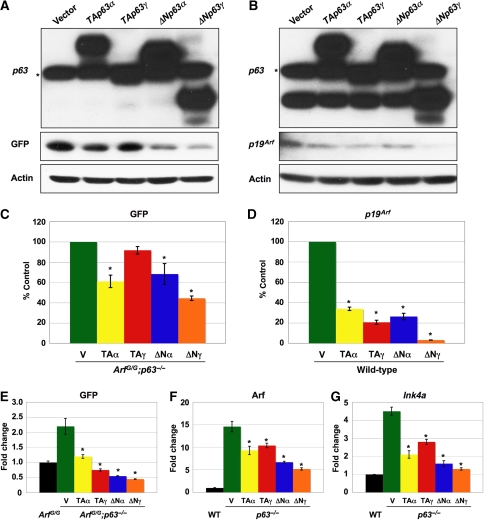
Transfection of the p63 isoforms in ArfGFP/GFP;p63−/− MEFs showed a marked repression of GFP protein and RNA levels (Figure 7A, C, and E) with ΔNp63α and -γ having the greatest effect. Similarly, introduction of the p63 isoforms into wild type and p63−/− MEFs repressed expression of Arf protein and mRNA (Figure 7B, D, and F). p63 can also repress Ink4a. p63−/− MEFs express Ink4a mRNA and protein at high levels. Transfection of the p63 isoforms resulted in a reduction in Ink4a mRNA (Figure 7G). Although all isoforms of p63 repressed Ink4a, the ΔNp63 isoforms repressed to the greatest extent (Figure 7G).
Figure 7.
p63 represses Arf and Ink4a. (A) Shows GFP expression in passage-2 ArfGFP/GFP;p63−/− MEFs transfected with the indicated expression vectors (top). (B) Illustrates p19Arf expression in passage-2 wild-type MEFs transfected with the indicated expression vectors (top). Both western blots in (A, B) were probed with a p63 antibody (4A4) specific for all isoforms of p63. The asterisk denotes a non-specific band. Actin was used as a loading control. (C, D) Show quantification of western blots, including the ones shown in (A, B), performed in triplicate. Actin was used as a loading control. (E–G) Show results from qRT–PCR of the indicated mRNAs (top) extracted from the sepcified MEFs transfected with the indicated expression vectors (bottom). GAPDH was used as an internal control. The asterisk denotes statistical significance (P<0.01).
These data, taken together, indicate that p63 directly represses Ink4a and Arf gene expression and that the ectopic expression of these genes when p63 is absent results in a failure to form stratified epithelium.
Discussion
The p63 protein regulates an extensive transcriptional network of genes required for the formation and integrity of epithelium in the skin and other organs, such as the breast, prostate, thymus, and urothelium (Mills et al, 1999; Yang et al, 1999). Complete inactivation of p63 globally compromises epithelial development and leads as well to craniofacial and limb deformities. However, it is particularly difficult to discern whether ablation of p63 affects epithelial stem cell self-renewal, lineage commitment, proliferation, keratinocyte differentiation, cell adhesion, or as seems likely, several of these functions.
The p63 gene encodes various isoforms that contain (TA) or lack (ΔN) a transactivation domain. The TAp63α, TAp63β, and TAp63γ isoforms are the first to be expressed during embryogenesis and are required for the initialization of epithelial stratification (Koster et al 2004). In contrast, the ΔNp63α isoform is the predominant species expressed in the epidermis, and it is detected in the bulge region of hair follicles and intermittently along the basal layer of the interfollicular epidermis, two regions in which epidermal stem cells are known to reside (Clayton et al, 2007; Senoo et al, 2007; Fuchs 2008). Once developing epidermal cells become suprabasal, ΔNp63α expression is extinguished, pointing to its possible involvement in gating stem cell proliferation (Fuchs, 2008). When ΔNp63α synthesis is knocked down with shRNAs, cultured epidermal progenitors form smaller colonies with reduced rates of proliferation (Senoo et al, 2007). However, opinions differ as to the relative contributions of the various p63 isoforms in skin development (Candi et al, 2007) and as to whether the failure of the primitive ectoderm to differentiate in p63-null mice reflects a defect in stem cell self-renewal (Yang et al, 1999; Senoo et al, 2007) or an early block in epidermal differentiation (Mills et al, 1999; Keyes et al, 2005; Koster et al, 2007).
Here, we now provide evidence that germ line disruption of either Ink4a or Arf significantly alleviates the phenotypic consequences of p63 ablation during embryonic development. Double mutant mice lacking p63 and either Ink4a or Arf produced K5-, K14-, K10-, K15-, and filaggrin-expressing epidermal cell sheets that encircled the embryos. Rescue was more impressive in the Ink4a−/−;p63−/− mice, which also exhibited rudimentary hair follicle formation not detected in their Arf−/−;p63−/− counterparts. However, the skin of these mice was fragile and was easily detached from the embryo as shown by discontinous expression of E-cadherin. Given that p63 also regulates genes involved in cell adhesion (Ihrie et al, 2005; Candi et al, 2007), our findings suggest that loss of Ink4a or Arf only counters the initial proliferative defect of keratinocytes deficient in p63 but not the additional consequences of p63 inactivation that are manifested as the double null cells differentiate.
Primary keratinocytes explanted from p63-null embryos cannot be propagated in culture, but those derived from Ink4a−/−;p63−/− or ArfGFP/GFP;p63−/− embryos exhibited a proliferative capacity approaching those of their wild-type counterparts. Conversely, and consistent with the notion that p63 negatively regulates Ink4a and Arf gene expression to facilitate skin development, expression of Ink4a and Arf mRNA, their encoded proteins, and of the product of the ArfGFP allele were upregulated in cultured keratinocytes lacking p63. Strikingly, GFP was expressed in situ in the epidermis of genotypically Arf-null ArfGFP/GFP;p63−/− embryos, arguing that induction of the Arf promoter was physiologically relevant and not simply because of explantation of primary keratinocytes into culture. Negative regulation of the Ink4a–Arf locus by p63 is reminiscent of the effects of Bmi-1, which has an essential role in the maintenance of stem cell function in HSCs and NSCs (Jacobs et al, 1999; Molofsky et al, 2003, 2005; Park et al, 2003; Bruggeman et al, 2005; Bracken et al, 2007) but has no such function during skin development.
p53 is also a potent negative feedback regulator of Arf gene expression (Kamijo et al, 1998; Stott et al, 1998), and yet despite a decade of investigation, there is still no evidence that p53 binds directly to Arf promoter sequences. We found that p63 can bind to potential response elements upstream of the transcriptional start sites of both Arf and Ink4a. These sequences conform to p53/p63 consensus binding sites that have been predicted to negatively regulate transcription (Johnson et al, 2001; Yang et al, 2006), raising the possibility that p63 acts as a direct repressor of the Ink4a–Arf locus.
p63 has been shown to have p53-dependent and -independent activities that promote cell survival and stem cell maintenance (Truong et al, 2006). Intriguingly, although Arf disruption in the mouse germ line partially rescues the p63-null epidermal phenotype, germ line inactivation of p53 exerts no such effects (Keyes et al, 2005; this paper). This implies that Arf ablation during early embryonic development affects a crucial p63-dependent but p53-independent function. Many p53-independent functions have been attributed to Arf, although much of the extant data remain controversial (Sherr, 2006). However, there is some evidence that Arf and p53 loss-of-function can contribute independently to the formation of carcinogen-induced squamous cell carcinomas (Kelly-Spratt et al, 2004). In contrast, cellular senescence induced by the acute inactivation of p63 in cultured primary keratinocytes was rescued when p53 expression was ablated by use of shRNA (Keyes et al, 2005). Thus, in the absence of p63, Arf inactivation may well exert both p53-independent and p53-dependent effects that are, respectively, manifested during early stages of skin development in utero or, differentially, in mature keratinocytes in which an Ink4a–Arf-dependent senescence programme is abruptly induced by somatic p63 disruption.
p63+/− mice have a shortened lifespan and display many features of accelerated aging and declining fitness (Flores et al, 2005; Keyes et al, 2005). Whether the aging phenotype depends on accelerated expression of the Ink4–Arf gene cluster is unknown, although it is now well appreciated that p16Ink4a and p19Arf upregulation can induce age-dependent declines in the regenerative potential of various tissues (Janzen et al, 2006; Krishnamurthy et al, 2006; Molofsky et al, 2006; Signer et al, 2008). Unfortunately, though inhibition of Ink4b–Arf–Ink4a functions may counter the impact of advancing age on tissue stem cell function, cancer development, including the formation of malignant skin tumours (Krimpenfort et al 2007), is a consequence.
Materials and methods
Mouse husbandry
p63+/− mice on (C57BL/6 × 129SvJae) background enriched for C57BL/6 (95%) (Yang et al, 1999; Flores et al, 2005) were intercrossed with Ink4a−/− (Sharpless et al, 2001), ArfGFP/GFP (Zindy et al, 2003) or p53−/− (Jacks et al, 1994) mice to obtain compound mutant mice and embryos of the following genotypes: Ink4a−/−; p63−/−, ArfGFP/GFP;p63−/−, and p53−/−;p63−/−. Wild type and p63−/− mice were also generated from each cross and analysed. All animal studies were approved by the Institutional Animal Care and Use Committee.
Histology and immunostaining of E18.5 embryos
Pregnant females at day 18.5 of gestation were injected three times at 1-h intervals with 100 μg of BrdU per gram of total body weight. Embryos were extracted 1 h later and fixed in 10% formalin at room temperature overnight, sectioned, and stained with H&E for microscopic analysis. For IF, paraffin-embedded sections were rehydrated in xylene and decreasing concentrations of ethanol. Sections were subjected to antigen unmasking in citrate buffer unmasking solution (Vector Laboratory) followed by a 1-h incubation with blocking solution, and overnight incubation at 4°C with primary antibodies to K5 (1:1000) (Abcam), K14 (1:500) (LifeSpan BioSciences), collagen IV (1:80) (Abcam), E-cadherin (1:200) (Abcam), K10 (1:1000) (Covance), K15 (1:500) (Covance), and filaggrin (1:1000) (Abcam). For staining with collagen IV antibody, unmasking was performed with Protease XXV (Fisher, AP-9006-002) at 37°C for 10 min. Visualization was performed using an anti-rabbit secondary antibody conjugated to Texas-red (1:5000) (Jackson ImmunoResearch Laboratories), an anti-guinea pig secondary antibody conjugated to FITC (1:1000, Jackson ImmunoResearch), or an anti-chicken secondary antibody conjugated to Alexa 488 (1:1000) (Molecular Probes) followed by counterstaining with DAPI (Vector Laboratory). Incorporation of BrdU was quantified using the BrdU detection kit II (Roche). For BrdU analysis by IF, an anti-rabbit secondary antibody conjugated to Texas Red was used with the BrdU detection kit II (Roche). GFP immunofluorescence was performed on frozen sections prepared from E18.5 embryos. Briefly, the epidermis was peeled from day 18.5 embryos and frozen in OCT compound (Tissue Tek). Frozen sections were fixed in 2% formaldehyde/0.2% glutaraldehyde solution for 2 min before incubation with anti-GFP (1:1000) (Invitrogen) at 4°C overnight followed by incubation with anti-rabbit secondary antibody conjugated to Texas-red (1:2000) (Jackson ImmunoResearch Laboratories) and counterstaining with DAPI (Vector Laboratory).
Skeleton preparation and staining
Embryos were collected at E18.5. Skin and internal organs were discarded, and the remaining embryo was fixed overnight in 95% ethanol at room temperature. The following day, the cartilage was stained in 0.05% alcian blue (Sigma) in 95% ethanol in acetic acid for 6 h and then destained in 1% KOH overnight. Bone staining was performed in 0.015% alizarin red (Sigma) in 1% KOH. The embryonic skeleton was washed in 20% glycerol/1% KOH for 5 days and changed twice per day until the solution was clear.
MEF and keratinocyte isolation and culture
Wild type, p53−/−, p63−/−, ArfGFP/GFP, and ArfGFP/GFP;p63−/− MEFs were isolated from embryos at day 13.5 and cultured in DMEM with 10% FBS as described earlier (Flores et al, 2002). Wild type, p63−/−, p53−/−;p63−/−, ArfGFP/GFP, ArfGFP/GFP;p63−/−, Ink4a−/−, and Ink4a−/−; p63−/− keratinocytes were isolated from E18.5 embryos. Briefly, the skin was peeled off and incubated in dispase II (Roche) for 18 h at 4°C to separate the epidermis from the dermis. Epidermal sheets were cut into small pieces and incubated in 0.25% trypsin–EDTA (Gibco-BRL) for 20 min at 37°C. Cells were plated on J2-3T3 feeder cells and cultured in F media (Sigma) supplemented with 0.4 μg/ml hydrocortisone, 24 ng/ml adenine, 8.4 ng/ml cholera toxin, 5 μg/ml insulin, 13 ng/ml 3,3,5-triiodo-L-thyronine, and 10 ng/ml EGF as described earlier (Barrandon and Green, 1987; Flores et al, 2000).
Immunostaining of keratinocytes
Keratinocytes seeded at a density of 4 × 104 cells/6 cm diameter dish were fixed after 8 days in culture in 10% formalin for 30 min. Immunostaining was performed by blocking and permeabilization of cells using 8% bovine serum albumin in 0.3% Triton X-100 for 30 min at room temperature. Keratinocytes were incubated with anti-K5 (1:1000) (Abcam) or anti-GFP (1:200) (Invitrogen) overnight at 4oC in a humidified chamber. Visualization of K5 was performed using an anti-rabbit secondary antibody conjugated to Texas-red (1:500) (Jackson ImmunoResearch laboratories) and GFP using an anti-rabbit secondary antibody conjugated to FITC (1:500) followed by counterstaining with DAPI (Vector Laboratory).
Keratinocyte proliferation assays
Passage-2 keratinocytes of 1 × 104 were plated in triplicate on J2-3T3 feeder cells. Keratinocytes were counted every day for 8 days. Keratinocytes were also subjected to seven serial passages at 5-day intervals. After isolation, 1 × 104 keratinocytes were plated in triplicate, passaged, and the cumulative number of population doublings was calculated.
Immunoblotting
MEFs and keratinocytes were lysed in buffer (100 mM Tris–HCl pH 8.0, 100 mM NaCl, 1% NP40, protease inhibitor cocktail tablet (Roche), and 0.2 M phenylmethylsulfonyl fluoride) and 100 μg of lysate were resolved by electrophoresis on a 12.5% SDS–PAGE gel, transferred to a PVDF membrane and probed with antibodies against p19Arf (5C3-1) (Bertwistle et al, 2004), GFP (1:1000) (Invitrogen), p16Ink4a (1:200) (Santa Cruz), p63-4A4 (1:500) (Santa Cruz), or Actin (1:5000) (Sigma) followed by goat anti-rat immunoglobulin G (IgG) or goat anti-rabbit IgG secondary antibodies conjugated to HRP (1:5000) (Jackson ImmunoResearch)
RNA extraction and real–time PCR
RNA was prepared from the dermis/epidermis peeled from wild type, p63−/−, ArfGFP/GFP, and ArfGFP/GFP;p63−/− 18.5 day embryos using an RNeasy plus kit (Qiagen Ltd.). cDNA was synthesized from 800 ng of total RNA using the SuperScript® III First-Strand Synthesis kit (Invitrogen) according to the manufacturer's protocol. The RNA levels of Ink4a, Arf, and GFP encoded by the cellular Arf promoter were determined by qRT–PCR and performed in triplicate using an ABI 7900 machine. The following primers were used for performing real-time PCR: Ink4a sense 5′-CGTACCCCGATTCAGGTGAT-3′, Ink4a antisense 5′-TTGAGCAGAAGAGCTGCTACGT-3′ (Bruggeman et al, 2005), GFP sense 5′-GTCCGCCCTGAGCAAAGA-3′, GFP antisense 5′-TCCAGCAGGACCATGTGATC, and p19ARF sense 5′-GCCGCACCGGAATCCT-3′, p19ARF antisense 5′-TGGAGCAGAAGAGCTGCTACGT-3′ (Bruggeman et al, 2005). Primers for GAPDH were used as an internal control.
Chromatin immunoprecipitation
Wild type and ArfGFP/GFP;p63−/− keratinocytes were grown to near confluence on J2-3T3m feeder cells in F media as described above. Twenty-four hours before collecting chromatin, the feeder cells were removed with 0.02% EDTA. Cellular proteins were crosslinked to DNA using 1% formaldehyde and chromatin was prepared as described earlier (Flores et al, 2002). p63–DNA complexes were diluted 10-fold in ChIP dilution buffer and incubated overnight at 4°C with 2 μg of pan-p63 antibody (4A4, Abcam) or 2 μg IgG (no antibody). Resulting chromatin was resuspended in 300 μl of double-distilled H2O. Each ChIP was performed in triplicate using keratinocytes from three embryos from each genotype. qRT–PCR was performed by using primers specific for the indicated regions of the Ink4a and Arf promoters (primer sequences below), p21 (Flores et al, 2002), and Perp promoters (Ihrie et al, 2005) using a 96 well plate ABI Step One Plus real-time PCR machine and 1 × SYBR green PCR master mix (Applied Biosystems). The percentage DNA bound was calculated using the following formula:
Percentage total bound=(2(ΔCT) × 2.5)p63antibody −(2(ΔCT) × 2.5)no antibody
where ΔCT=CTinput chromatin−CTsample and CT is cycle threshold.
Primers for qRT–PCR are as follows:
A. Ink4a promoter
(1) Site I
-4835F-5′-ACCTCTCCAGGGTGGTCACAGGC-3′
-4621R-5′-AAAGGCCGAGTCAGATGATCTGGCC-3′
(2) Site II-upstream (II-UP)
-3996F-5′-ACACTGCACTGGAAGAGGACATCA-3′
-3845R-5′-GTTAAAAATGTGTAGACATGGGCC-3′
(3) Site II-downstream (II-DOWN)
-3632F-5′-CCACAGTTTGAACAGCGTGAAGCA-3′
-3362R-5′-AGGAAACGGATGTGATCCCAGGAA-3′
(4) Site III
-1850F-5′-AACTAGTAGAAGGGAGATTACTTATTG-3′
-1539R-5′-ATAAACTATGTAAATTTTCTGAGGC-3′
(5) Site IV
-1519F-5′-TGGTAGCAGCTTCTAACCCAGCA-3′
-1157R-5′-TTTGTATGTGTGTGGTGAGAATGTC-3′
B. Arf promoter
(1) Upstream regions (UP)
-1811F-5′-AGACATCAGGCAACAGGGAATGGA-3′
-1622R-5′-TTCGGGCTACCCTACCTGGAATTT-3′
(2) Site I
-1533F-5′-ATACTCAGGGAGCCCTCAAAACTC-3′
-1331R-5′-TAGACATAGGAAGATGGCAAGGCC-3′
(3) Intervening sequences (INT)
-1417F-5′-ACCTGCCCTGCGGCTCTCAGTTGC-3′
-1303R-5′-TCTTATAGAGCGGATTCCAAATGC-3′
(4) Sites IIA and IIB
-1276F-5′-ATTGCTACTTTACTGCAGCCAGAC-3′
-943R-5′-TTGATCGGAGCGCCGGCCGGGGAC-3′
Transfection of MEFs
MEFs were plated on a six-well dish at a density of 4 × 105 cells/well. Twenty-four hours later, 2 μg of pcDNA3-p63 expression vectors (TAp63α, TAp63γ, ΔNp63α, or ΔNp63γ) were individually transfected using FuGENE HD Transfection Reagent (Roche) following the manufacturer's protocol. Cells were collected 24 h after transfection for analysis by western blotting and qRT–PCR.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Review Process File
Acknowledgments
ERF acknowledges Dr Ken Tsai for advice and comments on the pathology of skin samples and critical reading of the paper. ERF also acknowledges Dr Paul F Lambert and Denis Lee for the gift of J2-3T3 feeder cells. This work was supported by grants from the American Cancer Society (RSG-07-082-01-MGO), March of Dimes (Basil O′Connor Scholar), and Susan G Komen Foundation (BCTR600208) to ERF, and in part by NCI-Cancer Center Core Grant (CA-16672). ERF is a scholar of the Rita Allen Foundation and the V Foundation for Cancer Research. CJS, an investigator of the Howard Hughes Medical Institute, is also supported in part by ALSAC of St. Jude Children's Research Hospital.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akala OO, Park I-K, Qian D, Pihalja M, Becker MW, Clarke MF (2008) Long-term hematopoietic reconstitution by Trp53−/−, p16Inka−/−, p19Arf−/− multipotent progenitors. Nature 453: 228–232 [DOI] [PubMed] [Google Scholar]
- Barrandon Y, Green H (1987) Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA 84: 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertwistle D, Zindy F, Sherr CJ, Roussel MF (2004) Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybrid Hybridomics 23: 293–300 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M (2005) Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev 19: 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Mueller M, Krammer PH, Melino G (2007) TAp63 and ΔNp63 in cancer and epidermal development. Cell Cycle 6: 274–285 [DOI] [PubMed] [Google Scholar]
- Carroll DK, Brugge JS, Attardi LD (2007) p63, cell adhesion and survival. Cell Cycle 6: 255–261 [DOI] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW (2006) p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 8: 551–561 [DOI] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH (2007) A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189 [DOI] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF (2000) The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J Virol 74: 6622–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7: 363–373 [DOI] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T (2002) p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416: 560–564 [DOI] [PubMed] [Google Scholar]
- Fuchs E (2008) Skin stem cells: rising to the surface. J Cell Biol 180: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD (2005) Perp is a p63-regulated gene essential for epithelial integrity. Cell 120: 843–856 [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA (1994) Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168 [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT (2006) Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443: 421–426 [DOI] [PubMed] [Google Scholar]
- Johnson RA, Ince TA, Scotto KW (2001) Transcriptional repression by p53 through direct binding to a novel DNA element. J Biol Chem 276: 27716–27720 [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ (1998) Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA 95: 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ (2004) p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent mechanisms. PLoS Biol 2: E242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA (2005) P63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev 19: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR (2007) p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA 104: 3255–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR (2004) p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 18: 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR (2004) Genetic pathways required for epidermal morphogenesis. Eur J Cell Biol 83: 625–629 [DOI] [PubMed] [Google Scholar]
- Krimpenfort P, Ijpenberg A, Song JY, van der Valk M, Nawjin M, Zevenhoven J, Berns A (2007) p15Ink4b is a critical tumour suppressor inb the absence of p16Ink4a. Nature 448: 943–946 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey NR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE (2006) p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443: 404–405 [DOI] [PubMed] [Google Scholar]
- Liu Y, Lyle S, Yang Z, Cotsarelis G (2003) Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J Invest Dermatol 121: 963–968 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713 [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Miyazacki K, Itoi M, Katoh Y, Guo Y, Kanno R, Katoh-Fukui Y, Honda H, Amagai T, van Lohuizen M, Kawamoto H, Kanno M (2008) Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi1-mediated Cdkn2a repression. Immunity 28: 231–245 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R (2005) Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev 19: 1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ (2003) Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Kishnamurthy J, Sharpless NE, Morrison SJ (2006) Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during aging. Nature 443: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ (2008) Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-K, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T (1995) A subset of p53-deficient embryos exhibit exencephaly. Nat Genet 10: 175–180 [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F (2007) p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA (2001) Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413: 86–91 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (2006) Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 6: 663–673 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F (2002) The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K (2008) Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev 22: 3115–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G (1998) The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J 17: 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, Mantovani R (2006) New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J 25: 5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano MA, Mantovani R (2007) Hitting the numbers: the emerging network of p63 targets. Cell Cycle 6: 233–239 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K (2006) Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24: 593–602 [DOI] [PubMed] [Google Scholar]
- Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ (2003) Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci USA 100: 15930–15935 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Review Process File