Abstract
Ubiquitously encountered in a wide variety of cellular processes, the Grb2-Sos1 interaction is mediated through the combinatorial binding of nSH3 and cSH3 domains of Grb2 to various sites containing PXψPXR motifs within Sos1. Here, using isothermal titration calorimetry, we demonstrate that while the nSH3 domain binds with affinities in the physiological range to all four sites containing PXψPXR motifs, designated S1, S2, S3 and S4, the cSH3 domain can only do so at S1 site. Further scrutiny of these sites yields rationale for the recognition of various PXψPXR motifs by the SH3 domains in a discriminate manner. Unlike the PXψPXR motifs at S2, S3 and S4 sites, the PXψPXR motif at S1 site is flanked at its C-terminus with two additional arginine residues that are absolutely required for high-affinity binding of cSH3 domain. In striking contrast, these two additional arginine residues augment the binding of nSH3 domain to S1 site but their role is not critical for the recognition of S2, S3 and S4 sites. Site-directed mutagenesis suggests that the two additional arginine residues flanking the PXψPXR motif at S1 site contribute to free energy of binding via the formation of salt bridges with specific acidic residues in SH3 domains. Molecular modeling is employed to project these novel findings into the 3D structures of SH3 domains in complex with a peptide containing the PXψPXR motif and flanking arginine residues at S1 site. Taken together, this study furthers our understanding of the assembly of a key signaling complex central to cellular machinery.
Keywords: Grb2 adaptor, Sos1 nucleotide exchange factor, SH3 specificity and promiscuity, SH3-ligand thermodynamics
Grb2-Sos1 interaction, mediated by the canonical binding of N-terminal SH3 (nSH3) and C-terminal SH3 (cSH3) domains of Grb2 to proline-rich motifs within Sos1, plays a central role in relaying external signals from receptor tyrosine kinases (RTKs) at the cell surface to downstream effectors and regulators such as Ras within the cytosol (1–4). Comprised of a central SH2 domain flanked between nSH3 and cSH3 domains (Figure 1a), Grb2 recognizes activated RTKs by virtue of its SH2 domain to bind to tyrosine-phosphorylated (pY) sequences in the context of pYXN motif located within the cytoplasmic tails of a diverse array of receptors, including EGF and PDGF receptors (5, 6). Upon binding to RTKs, the SH3 domains live up to Grb2’s reputation and grab a wide variety of proteins, containing proline-rich sequences, in an attempt to recruit them to the inner membrane surface — the site of initiation of a plethora of signaling cascades (3, 7–14). Among them, the guanine nucleotide exchange factor Sos1 is by far the best characterized downstream partner of Grb2 (3, 7). Upon recruitment to the inner membrane surface, Sos1 catalyzes the GDP-GTP exchange within the membrane-bound GTPase Ras and thereby switches on a key signaling circuit that involves the activation of MAP kinase cascade central to cellular proliferation, survival and differentiation (15, 16).
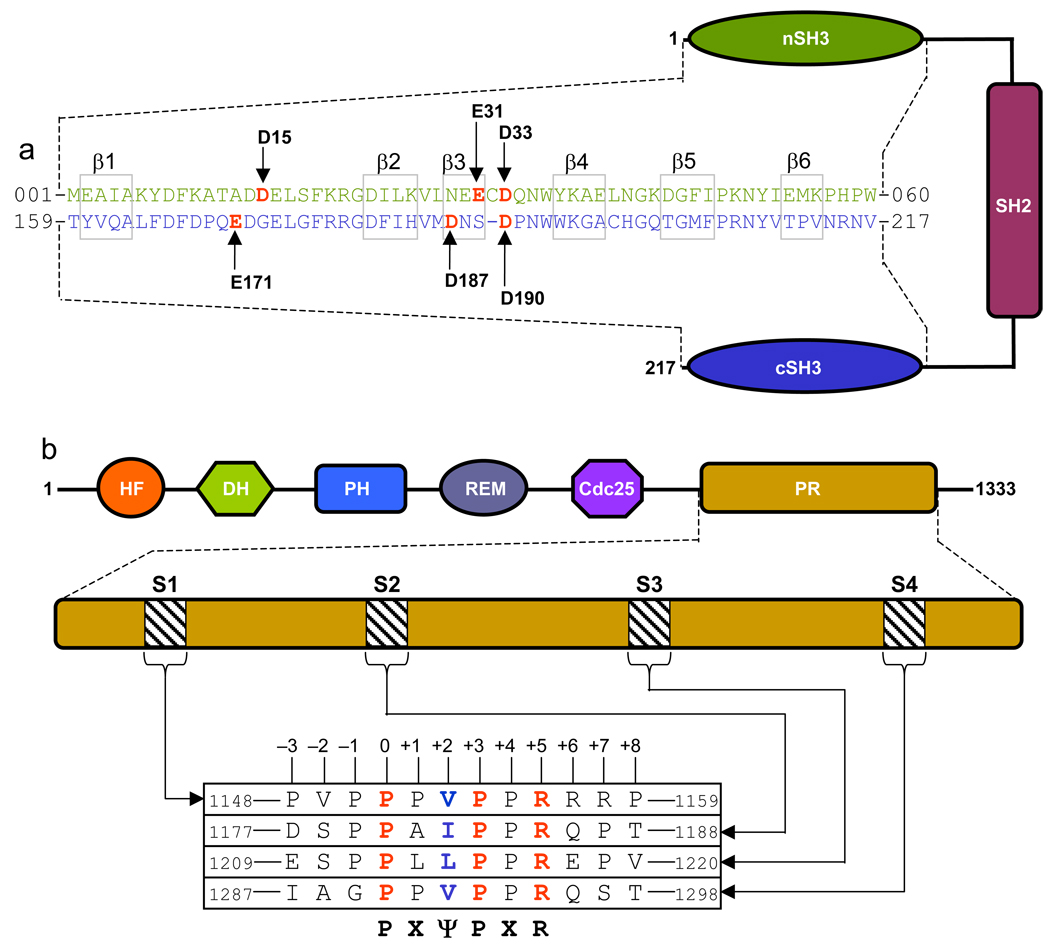
Figure 1.
Domain organization and sequence analysis of Grb2 and Sos1. (a) Grb2 is comprised of a central SH2 (Src homology 2) domain flanked between SH3 (Src homology 3) domains. The sequence alignment of nSH3 and cSH3 domains highlights various structurally equivalent residues within the β-barrel fold. The residues constituting the β1–β6 strands are boxed. Vertical arrows indicate acidic residues whose roles in recognizing Sos1 are being investigated in this study. (b) The proline-rich (PR) domain of Sos1 lies at the extreme C-terminal end. The PR domain contains four distinct sites (designated S1, S2, S3 and S4) characterized by the PXψPXR consensus motif. The complete sequences of these sites are shown. The position of various residues relative to the first proline within the PXψPX motif, which is designated zero, is also indicated. Other domains within Sos1 shown are HF (histone fold), DH (Dbl homology), PH (pleckstrin homology), REM (Ras exchange motif) and Cdc25.
Sos1 contains four distinct sites within its proline-rich (PR) domain for binding to the SH3 domains of Grb2 (Figure 1b). These sites, designated S1, S2, S3 and S4, conform to the PXψPXR consensus motif, where X is any residue and ψ is valine, leucine or isoleucine. In accordance with the nomenclature suggested by Schreiber and co-workers (17), the residues within the PXψPXR motif are assigned 0 for the N-terminal proline through to +5 for the C-terminal arginine. Although site-directed mutagenesis studies suggest that PXψPXR is the minimal motif (with the prolines at 0 and +3 positions, ψ at +2 position and arginine at +5 position) required for high-affinity binding to nSH3 domain (18), the extent to which residues flanking this motif may be non-redundant for binding to the cSH3 domain is not understood. On the basis of structural studies of the nSH3 domain of Grb2 in complex with peptides derived from S1 site in Sos1 (18–21), the nSH3 domain folds into a characteristic β-barrel architecture resulting in the formation of a hydrophobic cleft on one face of the domain for accommodating the incoming peptide. While the β-barrel is comprised of a pair of nearly-orthogonal β-sheets, with each β- sheet containing three anti-parallel β-strands, the peptide adapts a relatively open left-handed polyproline type II (PPII) helical conformation upon binding. Our previous studies suggest that the cSH3 domain of Grb2 is likely to bind to S1 peptide in a manner akin to that observed for the nSH3 domain (22).
Although biophysical and structural analysis of the binding of SH3 domains to peptides derived from S1 site has been extensively carried out (18, 19, 22, 23), little is understood about Grb2-Sos1 interaction at S2, S3 and S4 sites. In an effort to fully address Grb2-Sos1 interaction in biophysical terms, we have employed here isothermal titration calorimetry (ITC) to study the binding of SH3 domains of Grb2 to peptides derived from S1–S4 sites within Sos1. Our data demonstrate that while the nSH3 domain binds with affinities in the physiological range to all four sites containing PXψPXR motifs, the cSH3 domain can only do so at S1 site. Further scrutiny of these sites yields rationale for the recognition of various PXψPXR motifs by the SH3 domains in a discriminate manner. Unlike the PXψPXR motifs at S2, S3 and S4 sites, the PXψPXR motif at S1 site is flanked at its C-terminus with two additional arginine residues that are absolutely required for high-affinity binding of cSH3 domain. In striking contrast, these two additional arginine residues augment the binding of nSH3 domain to S1 site but their role is not critical for the recognition of S2, S3 and S4 sites. Site-directed mutagenesis suggests that the two additional arginine residues flanking the PXψPXR motif at S1 site contribute to free energy of binding via the formation of salt bridges with specific acidic residues in SH3 domains. Molecular modeling is employed to project these novel findings into the 3D structures of SH3 domains in complex with a peptide containing the PXψPXR motif and flanking arginine residues at S1 site. Taken together, this study furthers our understanding of the assembly of a key signaling complex central to cellular machinery.
MATERIALS and METHODS
Sample preparation
Wildtype and mutant SH3 domains of human Grb2 were expressed, purified and characterized as described earlier (22). HPLC-grade 12-residue peptides spanning S1, S2, S3 and S4 sites within the human Sos1 were commercially obtained from GenScript Corporation. The sequences of these peptides are shown in Figure 1b. The peptide concentrations were measured gravimetrically. Circular dichroism (CD) analysis of SH3 domains and Sos1 peptides revealed that the introduction of various alanine substitutions at specific positions had no observable effect on their secondary structural conformations.
ITC measurements
Isothermal titration calorimetry (ITC) experiments were performed on Microcal VP-ITC instrument and data were acquired and processed using fully automized features in Microcal ORIGIN software. All measurements were repeated 2–3 times. Briefly, SH3 domain samples were prepared in 50mM Tris, 200mM NaCl, 1mM EDTA and 5mM β-mercaptoethanol at pH 8.0. The experiments were initiated by injecting 25 × 10µl aliquots of 2–8mM of each peptide from the syringe into the calorimetric cell containing 1.8ml of 50–200µM of an SH3 domain solution at 25°C. The binding affinity (Kd) and the enthalpy change (ΔH) were extracted from the data by the fit of a one-site model, derived from the binding of a ligand to a macromolecule (24), using Microcal Origin. All other control experiments and data processing were performed as described earlier (22).
Molecular modeling
3D structures of SH3 domains of Grb2 in complex with S1 peptide were modeled using the MODELLER software based on homology modeling (25). In each case, the NMR structure of nSH3 domain of Grb2 in complex with a peptide corresponding to S1 site in Sos1 (with a PDB code of 4GBQ) was used as a template. Briefly, MODELLER employs molecular dynamics and simulated annealing protocols to optimize the modeled structure through satisfaction of spatial restraints derived from amino acid sequence alignment with a corresponding template in Cartesian space. For amino acid sequence identity between 25–50% between the template and target, MODELLER can generate 3D structures with accuracy comparable to NMR and X-ray structures for small proteins such as SH3 domains of around 50 amino acids. It is of worthy note that the nSH3 domain shares 35% sequence identity with the cSH3 domain, while the peptide ligand is identical between the template and modeled structures. Thus, the modeled structures of SH3 domains of Grb2 in complex with S1 peptide should be expected to adapt 3D folds similar to the template structure except for the sidechain conformations of specific amino acids due to the introduction of specific hydrogen bonding restraints between specific pairs of basic residues in the S1 peptide and acidic residues in the SH3 domains. Such hydrogen bonding restraints being introduced herein are necessary to bring the sidechain atoms of respective residues within optimal hydrogen bonding distance in agreement with our thermodynamic data reported here and in the previous study (22). The atomic distances set for hydrogen bonding restrains between a specific pair of oxygen and nitrogen atoms were 2.8±0.5Å. Thus, MODELLER will force the sidechain oxygen and nitrogen atoms of specific hydrogen bonding partners to lie within approximately 2.8Å of each other through the rotation of backbone N-Cα and Cα-C’ bonds with little effect on the overall global fold of S1 peptide and SH3 domains. To generate the 3D structural model of nSH3 domain in complex with S1 peptide, hydrogen bonding restraints were added between the OD1 and OD2 atoms of D15 in the nSH3 domain and NH1 and NH2 atoms of R+5 in S1 peptide, and between the OD1 and OD2 atoms of D33 in the nSH3 domain and NH1 and NH2 atoms of R+7 in the S1 peptide. To generate the 3D structural model of cSH3 domain in complex with S1 peptide, hydrogen bonding restraints were added between the OE1 and OE2 atoms of E171 in the cSH3 domain and NH1 and NH2 of R+5 in the S1 peptide, between the OD1 and OD2 atoms of D187 in the cSH3 domain and NH1 and NH2 atoms of R+6 in the S1 peptide, and between the OD1 and OD2 atoms of D190 in the cSH3 domain and NH1 and NH2 atoms of R+7 in the S1 peptide. To generate the 3D structural model of the cSH3_G173D mutant domain (that behaves like nSH3-mimetic) in complex with S1 peptide, hydrogen bonding restraints were added between the OE1 and OE2 atoms of E171 in the cSH3_G173D domain and NH1 and NH2 of R+5 in the S1 peptide, between the OD1 and OD2 atoms of G173D in the cSH3_G173D domain and NH1 and NH2 atoms of R+5 in the S1 peptide, between the OD1 and OD2 atoms of D187 in the cSH3_G173D domain and NH1 and NH2 atoms of R+6 in the S1 peptide, and between the OD1 and OD2 atoms of D190 in the cSH3_G173D domain and NH1 and NH2 atoms of R+7 in the S1 peptide. In each case, a total of 100 structural models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further energy minimization in MODELLER prior to analysis. The modeled structures were rendered using RIBBONS (26). All calculations were performed on the lowest energy structural model.
Motif search
The PXψPXR and PXψPXRRR motifs within the human proteome were searched using the ScanProsite software available at Expasy online server. X was set to any residue while ψ was set to valine, leucine or isoleucine. The protein database searched contained a total of 402,482 entries.
RESULTS and DISCUSSION
nSH3 and cSH3 domains pose distinct requirements for binding to PXψPXR motifs within Sos1
To further characterize the Grb2-Sos1 interaction in biophysical terms, we measured the binding of nSH3 and cSH3 domains to peptides derived from S1–S4 sites containing the PXψPXR motifs using ITC. Figure 2 and Figure 3 show representative data obtained upon conducting such measurements, while corresponding thermodynamic parameters are reported in Table 1. It is clearly evident from our data that the binding of both SH3 domains at S1–S4 sites within Sos1 is exclusively under enthalpic control and, with the exception of binding of cSH3 domain to S1 site as noted previously (22), binding at all other sites is also accompanied by entropic penalty as is often observed in enthalpically-driven biological processes. This observation is consistent with the notion that enthalpy drives SH3-ligand interactions in general, while entropic changes provide unfavorable contributions to the free energy (19, 27–32). That enthalpy drives the SH3-peptide interaction implies that both hydrophobic forces and electrostatic interactions play a key role in the assembly of this signaling complex. However, the dominance of entropic penalty accompanying the SH3-peptide interaction is most likely attributable to the greater loss of degrees of motions available to these molecules when free in solution but becoming more compact upon binding. Close scrutiny of data presented in Table 1 indicates that both SH3 domains have a preference for binding to S1 site. However, while the nSH3 domain binds to S1–S4 sites with very similar affinities that extend over nearly three folds, the cSH3 domain not only binds to S1 site in an exclusive manner but also with an affinity that is weaker that that observed for the binding of nSH3 domain to any of these sites. The binding of cSH3 domain to S2, S3 and S4 sites occurs with affinities in the millimolar range, implying that the interaction of cSH3 domain with these sites is not likely to be physiologically relevant. It should be noted here that the affinities observed for the binding of nSH3 domain to S1–S4 sites and cSH3 domain to S1 site within Sos1 are consistent with the binding of SH3 domains to their cognate ligands with affinities typically in the 10–100µM range (17, 33–42). However, in some cases, SH3 domains have also been shown to bind to their ligands with affinities in the sub-µM range (9, 43–49). Given the ubiquitous nature of SH3 domains within the mammalian proteome, it is believed that the rather weak SH3-ligand interactions are central to the temporal and spatial regulation of signaling pathways that constitute the bedrock of cellular communication.
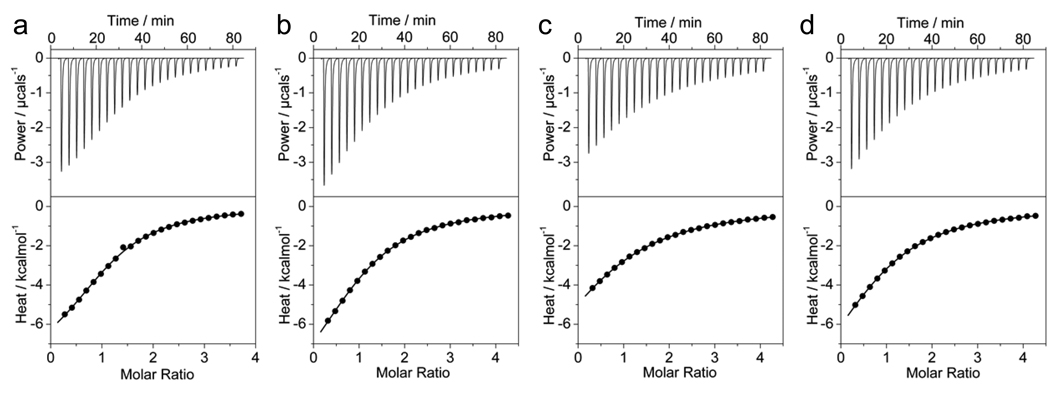
Figure 2.
ITC analysis for the binding of nSH3 domain of Grb2 to Sos1-derived peptides S1 (a), S2 (b), S3 (c) and S4 (d). The solid lines show the fit of data to a one-site model based on the binding of a ligand to a macromolecule as incorporated in the Microcal Origin software.
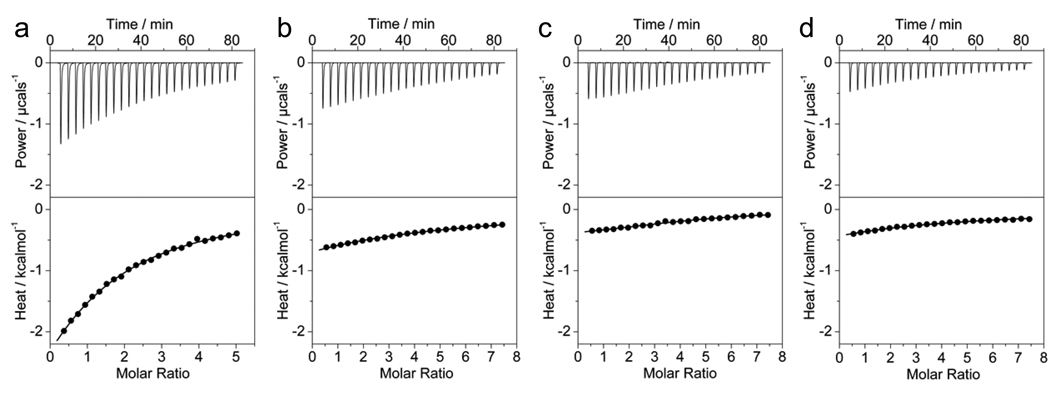
Figure 3.
ITC analysis for the binding of cSH3 domain of Grb2 to Sos1-derived peptides S1 (a), S2 (b), S3 (c) and S4 (d). The solid lines show the fit of data to a one-site model based on the binding of a ligand to a macromolecule as incorporated in the Microcal Origin software.
Table 1.
Experimentally determined thermodynamic parameters for the binding of wildtype nSH3 and cSH3 domains of Grb2 to various Sos1 peptides obtained from ITC measurements at 25°C and pH 8.0
| nSH3 | cSH3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptide | Sequence | Kd/µM | ΔH/kcalmol−1 | TΔS/kcalmol−1 | ΔG/kcalmol−1 | Kd/µM | ΔH/kcalmol−1 | TΔS/kcalmol−1 | ΔG/kcalmol−1 |
| S1 | PVPPPVPPRRRP | 39±1 | −9.41±0.02 | −3.39±0.03 | −6.02±0.01 | 125±13 | −4.45±0.15 | +0.88±0.09 | −5.29±0.06 |
| S2 | DSPPAIPPRQPT | 56±5 | −11.63±0.94 | −5.83±0.88 | −5.80±0.05 | 1396±87 | −9.94±0.84 | −6.04±0.88 | −3.87±0.04 |
| S3 | ESPPLLPPREPV | 117±2 | −11.38±0.24 | −6.01±0.25 | −5.36±0.01 | 1718±33 | −16.63±0.16 | −12.85±0.17 | −3.78±0.01 |
| S4 | IAGPPVPPRQST | 82±1 | −11.45±0.09 | −5.87±0.10 | −5.58±0.01 | 1318±44 | −11.52±0.41 | −7.59±0.43 | −3.92±0.02 |
| S1_AAA | PVPPPVPPAAAP | 2557±335 | −89.13±7.02 | −85.58±7.10 | −3.54±0.08 | 3089±121 | −35.66±0.21 | −32.23±0.19 | −3.42±0.02 |
| S1_AA | PVPPPVPPRAAP | 84±1 | −11.42±0.35 | −5.85±0.36 | −5.57±0.01 | 1574±27 | −20.75±1.70 | −16.92±1.73 | −3.82±0.03 |
| S1_R+5A | PVPPPVPPARRP | 490±20 | −9.42±0.61 | −4.90±0.59 | −4.52±0.02 | 962±26 | −2.92±0.12 | +1.20±0.13 | −4.12±0.02 |
| S1_R+6A | PVPPPVPPRARP | 53±1 | −9.10±0.25 | −3.26±0.27 | −5.84±0.02 | 331±2 | −3.96±0.41 | +0.80±0.42 | −4.75±0.01 |
| S1_R+7A | PVPPPVPPRRAP | 72±1 | −9.50±0.05 | −3.84±0.05 | −5.66±0.01 | 318±23 | −3.53±0.56 | +1.25±0.52 | −4.78±0.04 |
The residues within the S1 peptide subjected to alanine substitution are underlined for clarity. The values for the binding affinity (Kd) and the enthalpy change (ΔH) were obtained from the fit of a function, based on the binding of a ligand to a macromolecule (24), to the ITC isotherms. Free energy of binding (ΔG) was calculated from the relationship ΔG=RTlnKd, where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (K). Entropic contribution (TΔS) to binding was calculated from the relationship TΔS=ΔH−ΔG. The binding stoichiometries to the fits agreed to within±10%. Errors were calculated from 2–3 independent measurements. All errors are given to one standard deviation. All parameters are reported to no more than four significant figures.
The differential behavior of SH3 domains toward their putative sites within Sos1 must correlate with amino acids residing within or flanking the PXψPXR motifs. In view of the fact that one or more basic residues flanking consensus proline-rich motifs provide an additional level of modulating their binding affinity and specificity toward SH3 domains, we reasoned that the differential behavior of nSH3 and cSH3 domains toward S1–S4 sites might be attributable to the presence of arginine residues at positions +6 (R+6) and +7 (R+7) within the S1 site but absent in other sites (Figure 1b). Supporting this credence is the observation that the affinity of S1 peptide lacking all three arginine residues at positions +5, +6 and +7 (S1_AAA) is significantly compromised toward both SH3 domains (Table 1). To directly test that the high affinity and specific binding of cSH3 domain to S1 site is largely afforded by the presence of R+6 and R+7, we measured the binding of S1 peptide containing alanine substitutions at these two arginine positions (S1_AA) to SH3 domains. Our results reveal that while the binding of S1_AA peptide to nSH3 domain is only slightly weaker than the S1 peptide, it binds to the cSH3 domain with an affinity that is over an order of magnitude weaker relative to S1 peptide (Table 1). This salient observation unequivocally demonstrates that R+6 and R+7 flanking the C-terminal of the PXψPXR motif at S1 site are absolutely required for the binding of cSH3 domain within the physiological context. In a leap of further curiosity, we also analyzed the extent to which R+5, R+6 and R+7 within the S1 site were individually critical for binding to SH3 domains (Table 1). The substitution of R+5 to alanine within S1 peptide (S1_R+5A) reduced the binding affinities to both the nSH3 and cSH3 domains by an order of magnitude, implying that R+5 is critical for the binding of both SH3 domains to S1 site. In the case of substitution of R+6 to alanine within S1 peptide (S1_R+6A), the binding to nSH3 domain was virtually indistinguishable from that observed for the S1 peptide but underwent a close to three-fold reduction in affinity upon binding to the cSH3 domain relative to S1 peptide.
On the basis of these observations, the most straightforward conclusion is that while R+6 is important for the binding of cSH3 domain to S1 site, it is redundant in the case of nSH3 domain. Finally, in the case of the S1 peptide containing an alanine substitution at R+7 (S1_R+7A), the binding to both SH3 domains suffers between two-to-three fold reduction, implying that R+7 plays a non-redundant role in the binding of both the nSH3 and cSH3 domains to S1 site. Taken together, our data demonstrate that while all three R+5, R+6 and R+7 arginine residues are critical for the binding of cSH3 domain to S1 site, the nSH3 domain only strictly requires R+5 and R+7.
D33-R+7 salt bridge enhances the binding of nSH3 domain to S1 site
Our data presented above suggest strongly that both R+5 and R+7 arginine residues within S1 site are required for optimal binding of the nSH3 domain. The simplest mechanism by which these basic residues in the S1 peptide are likely to contribute to the free energy of binding is by virtue of their ability to engage in the formation of ion pairs or salt bridges with specific acidic residues in the nSH3 domain. It should be noted that such charged residues do not exist in solitude but in a symbiotic relationship with counterions when free in solution. Upon the formation of ion pairs with oppositely charged residues, usually through intermolecular association, the release of counterions into solution contributes to the free energy of binding through entropic gain. We have previously shown that R+5 ion pairs with D15 in the nSH3 domain (22). But which acidic residue in the nSH3 domain does R+7 ion pair with?
Analysis of available 3D atomic coordinates of nSH3 domain in complex with a Sos1-derived peptide flanking the S1 site reveals that the two most likely suspects for this role could be either E31 or D33 (Figure 1a) (18–21). Both of these acidic residues are located close to the exit of the hydrophobic groove in the nSH3 domain that accommodates the peptide and lie within stretching distance of R+7. To determine which of these two potential acidic residues is actually responsible for neutralizing the positive charge on R+7, we measured the binding of mutant nSH3 domains containing either an alanine substitution at E31 (nSH3_E31A) or at D33 (nSH3_D33A) to S1 peptide using ITC (Table 2). Our data reveal that while nSH3_E31A binds to S1 peptide with an affinity that is very similar to that observed for the binding of nSH3 domain, the binding of nSH3_D33A to S1 peptide occurs with an affinity that is over four-fold weaker relative to nSH3 domain. This finding suggests that it is D33 and not E31 that engages in the formation of a salt bridge with R+7. To further demonstrate that this is so, we also measured the binding of S1_R+6A and S1_R+7A peptides to nSH3_E31A and nSH3_D33A mutant domains (Table 2). The fact that the S1_R+6A peptide binds to nSH3_E31A and nSH3_D33A mutant domains with affinities that are similar to those observed for the binding of S1 peptide implies that R+6 is not involved in the formation of an ion pair as noted above. In contrast, the S1_R+7A peptide binds to nSH3_E31A with an affinity that is nearly two-fold weaker than that observed for the binding of S1 peptide, implying that this reduction in affinity is most likely due to the disruption of an ion pair involving R+7 with an acidic residue in the nSH3_E31A domain other than E31. This mysterious acidic residue involved in the formation of an ion pair with R+7 is thus likely to be D33 due to the fact that the nSH3_D33A mutant domain binds with very similar affinities to both the S1 and S1_R+7A peptides.
Table 2.
Experimentally determined thermodynamic parameters for the binding of mutant cSH3 domains of Grb2 to various Sos1 peptides obtained from ITC measurements at 25°C and pH 8.0
| nSH3_E31A | nSH3_D33A | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptide | Sequence | Kd/µM | ΔH/kcalmol−1 | TΔS/kcalmol−1 | ΔG/kcalmol−1 | Kd/µM | ΔH/kcalmol−1 | TΔS/kcalmol−1 | ΔG/kcalmol−1 |
| S1 | PVPPPVPPRRRP | 56±0.1 | −7.49±0.13 | −1.69±0.12 | −5.80±0.01 | 157±0.1 | −8.15±0.07 | −2.96±0.07 | −5.19±0.01 |
| S1_R+6A | PVPPPVPPRARP | 67±0.3 | −8.10±0.14 | −2.40±0.14 | −5.70±0.01 | 137±7.6 | −8.54±0.18 | −3.27±0.21 | −5.28±0.03 |
| S1_R+7A | PVPPPVPPRRAP | 91±1.2 | −7.99±0.12 | −2.47±0.11 | −5.52±0.01 | 169±4.5 | −8.62±0.19 | −3.47±0.20 | −5.15±0.02 |
The residues within the S1 peptide subjected to alanine substitution are underlined for clarity. The values for the binding affinity (Kd) and the enthalpy change (ΔH) were obtained from the fit of a function, based on the binding of a ligand to a macromolecule (24), to the ITC isotherms. Free energy of binding (ΔG) was calculated from the relationship ΔG=RTlnKd, where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (K). Entropic contribution (TΔS) to binding was calculated from the relationship TΔS=ΔH−ΔG. The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from 2–3 independent measurements. All errors are given to one standard deviation. All parameters are reported to no more than four significant figures.
In light of these considerations and data reported previously (22), the binding of nSH3 domain to S1 site in Sos1 is driven by the formation of D15-R+5 and D33-R+7 salt bridges. However, the D33-R+7 salt bridge is not critical allowing the nSH3 domain to also bind to S2, S3 and S4 sites in Sos1 that are devoid of R+7.
D187-R+6 and D190-R+7 salt bridges cooperate to drive the binding of cSH3 domain to S1 site
While the nSH3 domain only requires the ion pairing of R+5 and R+7 in binding to S1 site, R+6 also appears to be necessary for the binding of cSH3 domain (Table 1). In a previous study, we demonstrated that R+5 forms a salt bridge with E171 in the cSH3 domain (22). Here we set out to identify the potential partners of R+6 and R+7 in the cSH3 domain. The most likely candidates for this role are D187 and D190 within the cSH3 domain in agreement with structure-based sequence alignment with nSH3 domain (Figure 1a). To test this hypothesis, we introduced alanine substitutions within the cSH3 domain at D187 (cSH3_D187A) and D190 (cSH3_D190A) and measured the binding of these mutant domains to S1 peptide (Table 3). The fact that both cSH3_D187A and cSH3_D190A mutant domains bind to S1 peptide with about two-fold reduction in binding affinity relative to the wildtype cSH3 domain suggests strongly that both D187 and D190 are involved in forming ion pairs with R+6 and R+7.
Table 3.
Experimentally determined thermodynamic parameters for the binding of mutant cSH3 domains of Grb2 to various Sos1 peptides obtained from ITC measurements at 25°C and pH 8.0
| cSH3_D187A | cSH3_D190A | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptide | Sequence | Kd/µM | ΔH/kcalmol−1 | TΔS/kcalmol−1 | ΔG/kcalmol−1 | Kd/µM | ΔH/kcalmol−1 | TΔS/kcalmol−1 | ΔG/kcalmol−1 |
| S1 | PVPPPVPPRRRP | 230±4 | −4.55±0.11 | +0.42±0.12 | −4.97±0.01 | 272±11 | −4.24±0.42 | +0.63±0.45 | −4.87±0.02 |
| S1_R+6A | PVPPPVPPRARP | 544±102 | −3.35±0.03 | +1.12±0.08 | −4.46±0.11 | 910±57 | −14.80±0.27 | −10.65±0.23 | −4.15±0.04 |
| S1_R+7A | PVPPPVPPRRAP | 2388±73 | −9.94±0.11 | −6.36±0.09 | −3.58±0.02 | 482±47 | −5.12±0.41 | −0.59±0.47 | −4.53±0.06 |
The residues within the S1 peptide subjected to alanine substitution are underlined for clarity.The values for the binding affinity (Kd) and the enthalpy change (ΔH) were obtained from the fit of a function, based on the binding of a ligand to a macromolecule (24), to the ITC isotherms. Free energy of binding (ΔG) was calculated from the relationship ΔG=RTlnKd, where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (K). Entropic contribution (TΔS) to binding was calculated from the relationship TΔS=ΔH−ΔG. The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from 2–3 independent measurements. All errors are given to one standard deviation. All parameters are reported to no more than four significant figures.
To identify the specific residues involved in the formation of these ion pairs, we also measured the binding of cSH3_D187A and cSH3_D190A mutant domains to S1_R+6A and S1_R+7A peptides (Table 3). The fact that the cSH3_D187A binds to the S1_R+7A peptide with an affinity that is over four-fold weaker than that observed for its binding to the S1_R+6A peptide implies that it is R+6 and not R+7 that must ion pair with D187. This observation is further corroborated upon the binding of cSH3_D190A to S1_R+6A peptide with an affinity that is nearly two-fold weaker than that observed for its binding to S1_R+7A, implying that it is R+7 and not R+6 that forms an ion pair with D190. Taking these observations together in light of the previous data (22), the binding of cSH3 domain to S1 site in Sos1 is driven by the formation of E171-R+5, D187-R+6 and D190-R+7 salt bridges. Given that all of these three salt bridges are critical for the binding of nSH3 domain to S1 site implies that it cannot bind to S2, S3 and S4 sites in Sos1 devoid of R+6 and R+7.
Arginine residues within S1 site contribute differentially to the free energy of binding of nSH3 and cSH3 domains
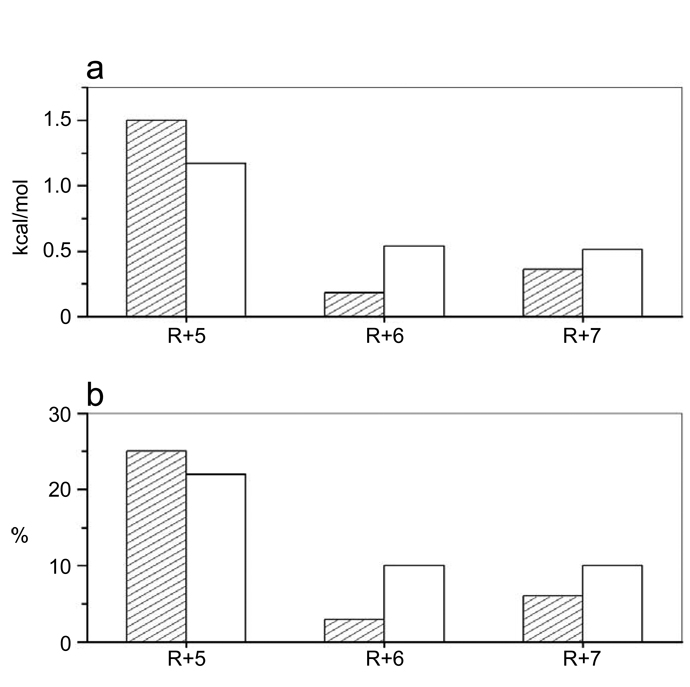
The foregoing argument suggests strongly that the arginine residues R+5, R+6 and R+7 within the S1 site play a key role in driving the Grb2-Sos1 interaction. As illustrated in Figure 4, it can be further seen that these arginine residues contribute between 30–40% of the total free energy available to drive the binding of nSH3 and cSH3 domains to S1 site in a distinct manner. Thus, while R+5 is the major contributor to the overall free energy of binding of nSH3 domain to S1 site, R+7 and R+6 play lesser roles, with the contribution of the latter falling within the experimental error of the measurements being reported here. In contrast, while R+5 also heavily contributes to the overall free energy of binding of cSH3 domain to S1 site, R+6 and R+7 clearly play significant roles and, together, the energetic contributions of R+6 and R+7 almost match those of R+5. The specificity of nSH3 and cSH3 domains toward Sos1 thus seems to be largely due to the distinct contributions of arginine residues R+5, R+6 and R+7 to the free energy of binding. In other words, electrostatic interactions in lieu of hydrophobic contacts appear to define the distinguishing features of the binding of nSH3 and cSH3 domains to Sos1. This is further corroborated by the fact that the S1_AAA peptide, in which all three arginine residues are substituted by alanine, binds to both nSH3 and cSH3 domains with very similar affinities, albeit in a non-physiologically-relevant manner (Table 1).
Figure 4.
Comparison of energetic contributions of arginine residues R+5, R+6 and R+7 within the S1 site to the free energy of binding of nSH3 (shaded columns) and cSH3 (unshaded columns) domains. (a) Energetic contributions relative to the total free energy (ΔGTotal) of binding of nSH3 and cSH3 domains to S1 peptide expressed in absolute terms (kcal/mol). The energetic contribution of R+5 (ΔGR+5) was calculated from the relationship ΔGR+5=ΔGR+5A−ΔGTotal, where ΔGR+5A is the free energy of binding of S1_R+5A peptide to the nSH3 or the cSH3 domain. The energetic contribution of R+6 (ΔGR+6) was calculated from the relationship ΔGR+6=ΔGR+6A−ΔGTotal, where ΔGR+6A is the free energy of binding of S1_R+6A peptide to the nSH3 or the cSH3 domain. The energetic contribution of R+7 (ΔGR+7) was calculated from the relationship ΔGR+7=ΔGR+7A−ΔGTotal, where ΔGR+7A is the free energy of binding of S1_R+7A peptide to the nSH3 or the cSH3 domain. (b) Energetic contributions relative to the total free energy (ΔGTotal) of binding of nSH3 and cSH3 domains to S1 peptide expressed as a percentage (%). The energetic contribution of R+5 (%R+5) was calculated from the relationship %R+5=[(ΔGR+5A/ΔGTotal)×100], where ΔGR+5A is the free energy of binding of S1_R+5A peptide to the nSH3 or the cSH3 domain. The energetic contribution of R+6 (%R+6) was calculated from the relationship %R+6=[(ΔGR+6A/ΔGTotal)×100], where ΔGR+6A is the free energy of binding of S1_R+6A peptide to the nSH3 or the cSH3 domain. The energetic contribution of R+7 (%R+7) was calculated from the relationship %R+7=[(ΔGR+7A/ΔGTotal)×100], where ΔGR+7A is the free energy of binding of S1_R+7A peptide to the nSH3 or the cSH3 domain. All energetic contributions were calculated using data provided in Table 1.
D15 underscores the high-affinity binding of nSH3 domain to PXψPXR motifs within Sos1
Our data presented above provide the rationale underlying the binding of nSH3 domain to all four S1–S4 sites with Sos1, while the cSH3 domain can only bind to S1 site. But what features within the nSH3 domain enable it to only strictly require R+5, while the cSH3 domain has an obligate requirement of all three R+5, R+6 and R+7 arginine residues within the putative binding sites in Sos1? Given that the binding affinity of S1_R+5A peptide to nSH3 domain is reduced by more than an order of magnitude relative to S1 peptide (Table 1), we reasoned that the energetic contribution resulting from the formation of D15-R+5 salt bridge may be largely responsible for driving the binding of nSH3 domain to all four S1–S4 sites in contrast to cSH3 domain that can only bind to S1 site. This seems logical in light of the fact that the cSH3 domain contains a glycine (G173) instead of an acidic residue, let alone an aspartate, at the structurally equivalent position occupied by D15 in the nSH3 domain (Figure 1a). The cSH3 domain rather relies on a neighboring glutamate (E171) to engage in the formation of a salt bridge with R+5. Because D15 and E171 are structurally and chemically non-equivalent, it is very likely that such differences also translate into D15-R+5 and E171-R+5 salt bridges being energetically non-equivalent and thus may underlie the distinct behavior of nSH3 and cSH3 domains toward S1–S4 sites. As discussed above and illustrated in Figure 4, this is indeed the case.
To further test the extent to which the ability of nSH3 domain to bind to all four S1–S4 sites is attributable to D15, we introduced alanine substitution at G173 in the cSH3 domain (cSH3_G173D) and measured the binding of cSH3_G173D mutant domain to S1–S4 peptides containing PXψPXR motifs. Comparison of affinities and various associated thermodynamic parameters for the binding of nSH3 and cSH3_G173D domains to S1–S4 peptides is provided in Figure 5. It is clearly apparent from our data that the G173D substitution renders the cSH3 domain to behave very much like the nSH3 domain in its ability to recognize all four S1–S4 sites with binding affinities in the physiologically relevant range. This suggests strongly that D15 is largely responsible for the ability of nSH3 domain to recognize all four S1–S4 sites, while the placement of a glycine residue at this structurally equivalent position within the cSH3 domain deprives it of recognizing S2, S3 and S4 sites.
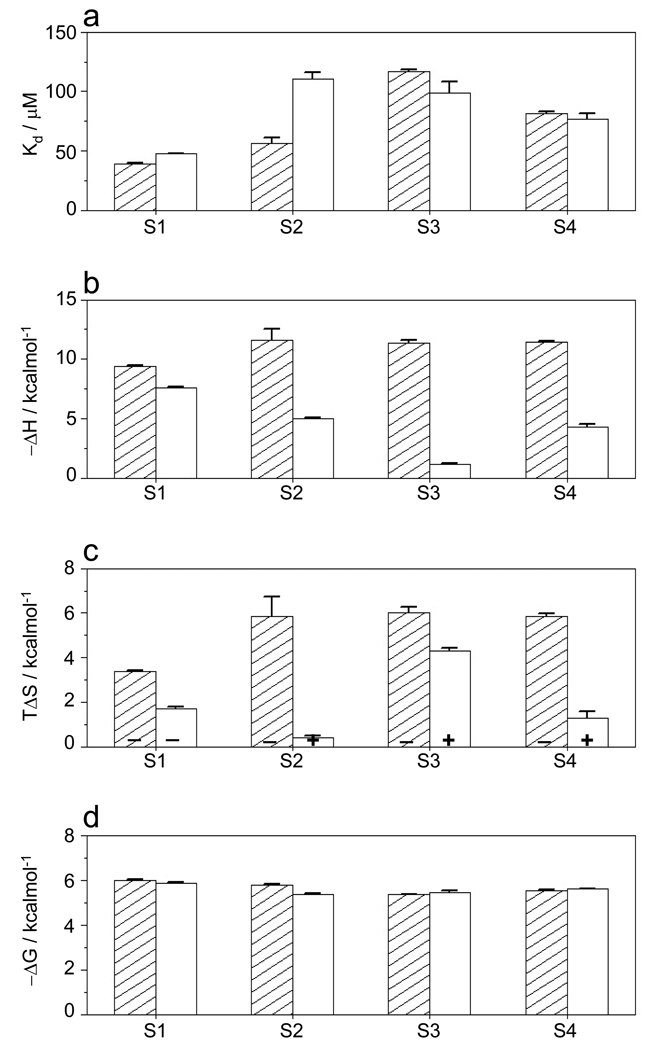
Figure 5.
Comparison of energetics of binding of Sos1-derived peptides S1–S4 to wildtype nSH3 domain (shaded columns) and cSH3_G173D mutant domain (unshaded columns). (a) Binding affinity (Kd); (b) Enthalpic contribution to binding (ΔH); (c) Entropic contribution to binding (TΔS) with the + and − signs indicating entropic gain and entropic penalty, respectively; and (d) Overall free energy of binding (ΔG). All parameters were directly determined from ITC analysis. Error bars were calculated from 2–3 independent measurements. All error bars are given to one standard deviation.
It is of worthy note that although the nSH3-mimetic cSH3_G173D may appear to exhibit similar binding energetics (Figures 5a and 5d), the underlying thermodynamic forces are quite distinct (Figures 5b and 5c). Thus, while the binding of nSH3 domain to all four S1–S4 sites is under enthalpic control accompanied by entropic penalty, this only holds true in the case of cSH3_G173D binding to S1 site, whereas binding of cSH3_G173D to S2, S3 and S4 sites is accompanied by favorable entropic contributions. This is indicative of the fact that although D15 may be largely responsible for the ability of nSH3 domain to bind to all four S1–S4 sites, other residues may also play a role in defining its binding specificity.
3D atomic models offer structural insights into the distinct mechanisms employed by SH3 domains in binding to Sos1
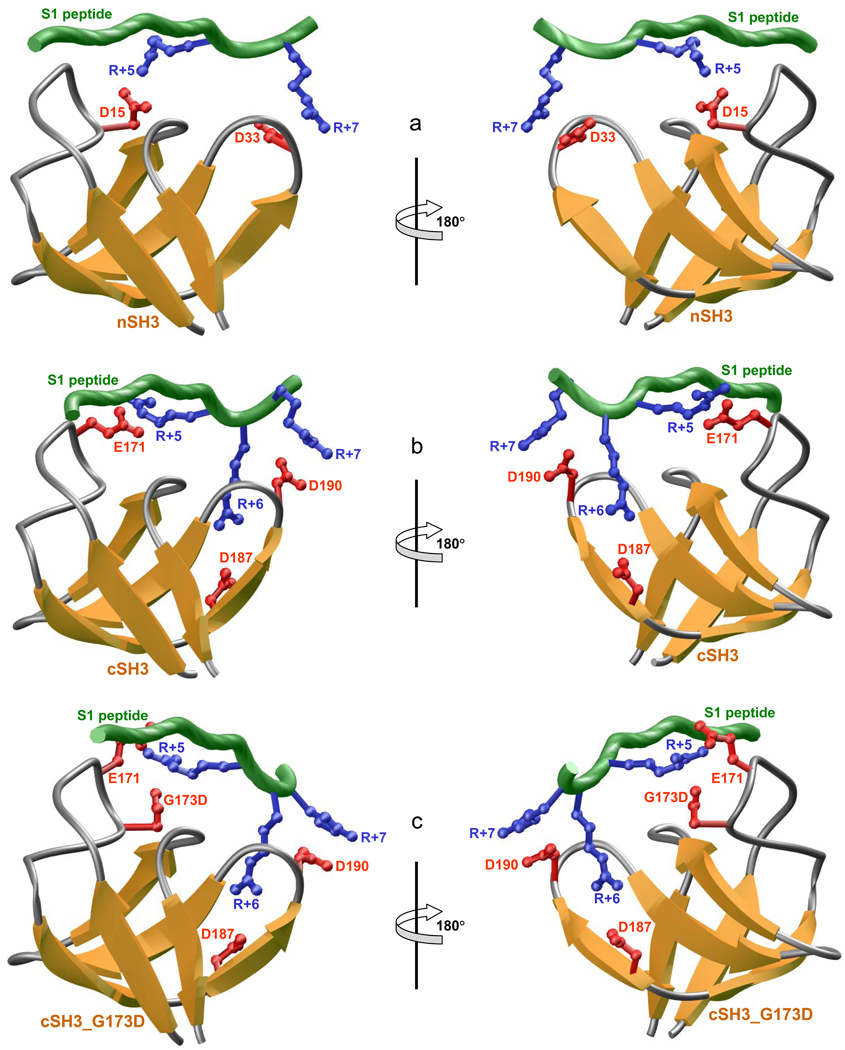
In an attempt to rationalize the distinct mechanisms employed by SH3 domains of Grb2 in recognizing Sos1, we modeled 3D structures of nSH3, cSH3 and cSH3_G173D domains in complex with S1 peptide (Figure 6). In these models, the SH3 domains adapt the characteristic β-barrel fold and the peptide is accommodated into a hydrophobic groove with a relatively open left-handed polyproline type II (PPII) helical conformation. Although the nature of intermolecular hydrophobic forces stabilizing nSH3-peptide and cSH3-peptide complexes is virtually identical, as discussed earlier (22), the major differences surface in the nature of intermolecular electrostatic forces. Thus, while the nSH3 domain employs respectively D15 and D33 in the formation of ion pairs with R+5 and R+7 in the peptide (Figure 6a), the cSH3 domain relies on E171 and D190 in accomplishing what may seem to be the same feat but it is far from that (Figure 6b). The reason being that while D190 is structurally and chemically equivalent to D33 with the effect that the D33-R+7 and D190-R+7 salt bridges are more or less energetically equivalent, D15 and E171 are structurally non-equivalent and chemically distinct, with G173 in the cSH3 domain being structurally equivalent to D15. Such differences within the two SH3 domains result in the D15-R+5 and E171-R+5 salt bridges being energetically non-equivalent and therefore being the major source of the differential behavior of these two domains toward various potential binding sites within Sos1. As if these residues were insufficient to distinguish their biological roles, the cSH3 domain entertains one additional trick to further differentiate its behavior from the nSH3 domain. Such coup de grace is delivered in the form of D187 involved in the formation of a third salt bridge with R+6 in the peptide (Figure 6b).
Figure 6.
3D structural models of S1 peptide in complex with nSH3 (a), cSH3 (b), and cSH3_G173D (c) domains of Grb2. The β-strands in SH3 domains are shown in yellow with loops depicted in gray and the sidechains of acidic residues involved in salt bridging with the peptide in red. The backbone of Sos1 peptide is colored green with the sidechains of arginine residues involved in salt bridging with SH3 domains in blue. Two alternative orientations, related by an 180°-rotation about the vertical axis, are shown for clarity and close scrutiny.
Taking these considerations into account, it would suffice to add that the nSH3 and cSH3 domains employ two-prong and three-prong mechanisms to engage in electrostatic interactions with S1 site in Sos1, respectively. Clearly, the lack of R+6 and R+7 at S2, S3 and S4 sites in Sos1 would imply that not only nSH3 would bind to these sites in an exclusive manner but that it would do so through the engagement of a single D15-R+5 salt bridge, albeit with no effect on the nature of intermolecular hydrophobic forces. It is also of worthy note that while the nSH3 domain and its mimetic cSH3_G173D mutant domain bind to S1–S4 peptides with very similar affinities, the latter displays distinct contributions from the underlying enthalpic and entropic components (Figure 5). Although structural thermodynamics is still in its infancy, it is interesting to note that such differences in the underlying thermodynamic components may be due to the fact that while nSH3 domain engages in only two intermolecular salt bridges (Figure 6a), the cSH3_G173D mutant domain employs three (Figure 6c). Additionally, the R+5 residue in S1 peptide likely bifurcates the E171 and G173D ion pairs within the cSH3_G173D mutant domain, while R+5 merely engages in the formation of an ion pair with D15 — the residue that is structurally equivalent to G173D — due to the substitution of an alanine (A13) at the structurally equivalent position occupied by E171 in the cSH3_G173D mutant domain. In short, our 3D atomic models offer a closer glimpse into the structural basis of the exclusivity of S1 site for binding to cSH3 domain, while the nSH3 domain can bind to all S1–S4 sites indiscriminately.
SH3 domains appear to combine fidelity and promiscuity under the flagship of Grb2 in cellular signaling
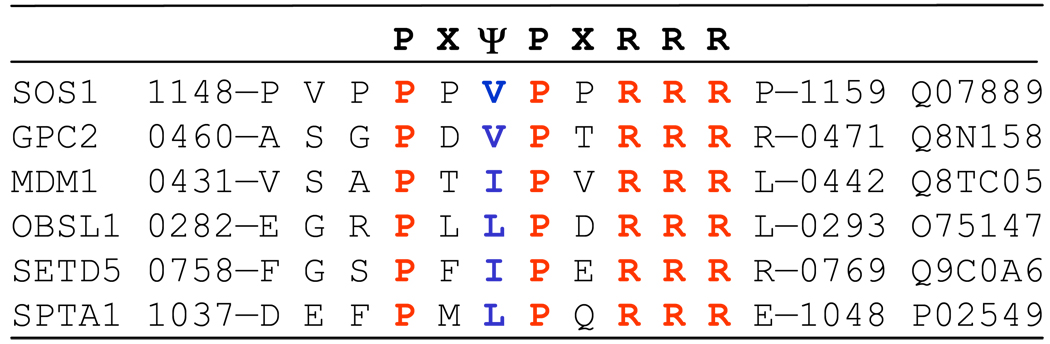
The data presented herein suggest strongly that the cSH3 domain strictly requires the PXψPXRRR motif for binding to Sos1, while the nSH3 domain will do so only upon the presentation of PXψPXR motif under physiological context. In light of these observations, it is tempting to add that the SH3 domains may have evolved to bring the best of both worlds to Grb2. Thus, while nSH3 domain may be ideally suited for signaling promiscuity due to requirement of only one arginine residue within its proline-rich ligands, the cSH3 domain most certainly imparts signaling fidelity upon Grb2. To further confirm this notion, we searched the human proteome for the occurrence of PXψPXR and PXψPXRRR motifs. Our findings reveal that while there are 753 PXψPXR motifs within the human proteome across nearly as many proteins, only six of them contain the PXψPXRRR motif (Figure 7). In addition to Sos1, these include GPC2, MDM1, OBSL1, SETD5 and SPTA1 (50–54). With the exception of Sos1, none of these proteins contain the PXψPXR motif, implying that the cSH3 domain of Grb2 may bind to them in an exclusive manner and thereby leading to build-up of weak interactions. Such a design may be an important aspect of transient signaling allowing Grb2 to fine-tune various overlapping cascades via its interaction with the aforementioned partners.
Figure 7.
Amino acid sequence alignment showing the cccurrence of PXψPXRRR motif in human proteome. Absolutely conserved residues within the PXψPXRRR motif are shown in red, the ψ residue is depicted in blue and all other residues are colored black. The proteins containing this motif are listed in the far left column, while the far right column provides the corresponding Expasy codes. The numerals hyphenated to amino acid sequence at each end denote the residue number within the protein sequence.
Our analysis presented above clearly suggests that while the interaction of Grb2 with other cellular partners through its nSH3 domain may be highly promiscuous, the cSH3 domain provides a striking contrast by virtue of its ability to bind to only a handful of partners adding much needed fidelity to signaling via Grb2. It should be borne in mind that while the proteins containing the PXψPXRRR motif should be expected to bind to the cSH3 domain, the lack of such motif in cellular proteins may not necessarily rule out their interaction with Grb2. This is due to the fact that SH3 domains have also adapted an alternative mechanism independent of the consensus proline-rich sequence PXXP for recognizing some of their partners. In particular, the cSH3 domain of Grb2 has been shown to bind to key signaling modulators Gab1 and SLP76 via the recognition of a non-consensus PXXXRXXKP motif (9, 47). Structural analysis reveals that the PXXXRXXKP motif binds to the cSH3 domain in a manner akin to the binding of PXψPXRRR motif, with the both motifs sharing the same binding groove and orientation (48, 49). However, there are some discernable differences. While the PXψPXRRR motif adapts a relatively open left-handed polyproline type II (PPII) helical conformation, the PXXXRXXKP motif adapts a 310-helical conformation upon binding to the cSH3 domain. The respective conformations are required to orient the critical residues within each motif for optimal interactions within the binding groove of the cSH3 domain. Overall, the PXXXRXXKP motif engages in additional contacts within the binding groove of cSH3 domain relative to the PXψPXRRR motif and, by virtue of these distinguishing interactions, the cSH3 domain binds to Gab1 and SLP76 with higher affinity than Sos1. Such differential binding of cSH3 domain to Gab1 and SLP76 versus Sos1 may be an important determinant of monitoring the ratios of Grb2-Sos1 pool versus Grb2-Gab1 and Grb2-SLP76 pools with consequences for activation of Ras versus other signaling pathways.
CONCLUSIONS
Although the central role of Grb2-Sos1 signaling complex in mitogenic signaling was first demonstrated over a decade ago (3, 4, 55, 56), the precise mechanism by which it is assembled remains largely obscure. The prevailing view for the past decade has been that Grb2 recognizes Sos1 by virtue of its SH3 domains to bind to various PXψPXR motifs within Sos1. Our present study suggests that although there are four such putative sites within Sos1 (S1–S4), they may not all be used indiscriminately to recognize the SH3 domains of Grb2. Thus, while the nSH3 domain binds to all four sites with very similar affinities in the physiological range, the cSH3 domain can only do so at S1 site. The reason for such high specificity for the cSH3 domain is not clear but it may be an important feature of Grb2 in signaling cascades that demand high fidelity. Nonetheless, the mechanism by which the cSH3 domain has acquired such a high level of specificity appears to be remarkably simple. The substitution of D15 within the RT loop of nSH3 domain to a glycine at the structurally equivalent position in the cSH3 domain seems to be all that is required to specifically direct the cSH3 domain to S1 site and prevent it from binding to other potential sites containing the PXψPXR motifs. Because of this substitution, the cSH3 domain employs a distinct mechanism in recognizing the S1 site compared with the nSH3 domain. Thus, while the nSH3 domain employs only two intermolecular salt bridges for binding to S1 site, the cSH3 domain relies on three. Electrostatic interactions are a general feature of SH3-ligand specificity due to the presence of one or more charged residues flanking the PXXP motif as well as those lining the binding groove within the SH3 domains (17, 34, 40, 42). In fact, substitution of these charged residues with alanine almost always abrogates SH3-ligand interactions. Consistent with these observations, our present data epitomize the role of multiple salt bridges in the assembly of Grb2-Sos1 complex. In particular, our data suggest that the salt bridges contribute between 30–40% of the total free energy available to drive the binding of nSH3 and cSH3 domains to S1 site. The specificity of nSH3 and cSH3 domains toward Sos1 thus largely appears to be due to the distinct contributions of arginine residues R+5, R+6 and R+7 to the free energy of binding. In other words, electrostatic interactions in lieu of hydrophobic contacts appear to define the distinguishing features of the binding of nSH3 and cSH3 domains to Sos1. That this is so implies that intracellular salt concentrations may be an important determinant of Grb2-Sos1 assembly. We have indeed previously shown that salt tightly modulates the binding of nSH3 and cSH3 domains of Grb2 to Sos1 (22).
It has been previously suggested that Grb2 binds to Sos1 with a 1:1 stoichiometry (57). In light of this observation coupled with our data presented herein, we propose a model of how Grb2 and Sos1 might assemble at the inner membrane surface upon growth factor receptor stimulation. The exclusivity of binding of cSH3 domain to S1 site would allow the nSH3 domain to bind to one of the other available S2–S4 sites with little or no preference. In this manner, the binding of cSH3 domain to S1 site could restrict the orientation of Grb2 relative to Sos1, while the binding of nSH3 domain to one of the three available S2–S4 sites may result in the formation of various structurally and conformationally distinct Grb2-Sos1 complexes. Such orientational constraint imposed upon Grb2-Sos1 assembly by the cSH3 domain coupled with conformational heterogeneity resulting from the promiscuity of nSH3 domain may be an important determinant of regulating the activation of downstream molecules such as Ras. For example, the preference of cSH3 domain toward S1 site may orient Grb2 in such a manner that it does not interfere with the GDP-GTP nucleotide exchange function of Sos1 required for the activation of membrane-bound Ras (58, 59). Given that Sos1 competes with a diverse array of other downstream effectors for binding to Grb2, including the adaptor proteins Gab1 and SLP76 (8, 9), the endocytic GTPase dynamin1 (10, 11), the ubiquitin ligase Cbl (9, 12, 13) and the cell cycle inhibitor p27kip1 (14), the ability of cSH3 domain to dictate the binding of Grb2 to Sos1 in an oriented manner may also play an important role in determining the fraction of Grb2-Sos1 pool. It should be noted that the role of molecular orientation and conformational heterogeneity of protein-protein complexes is largely under-appreciated in the temporal and spatial regulation of signaling complexes. Our present study thus sets a precedent for the design and guidance of further experiments to unravel the regulatory role of molecular orientation and conformational heterogeneity in the assembly of signaling complexes.
ABBREVIATIONS
- CD
Circular dichroism
- EGF
Epidermal Growth Factor
- Grb2
Growth factor Receptor Binder 2
- ITC
Isothermal Titration Calorimetry
- MAP
Mitogen-Activated Protein
- NMR
Nuclear Magnetic Resonance
- PDGF
Platelet-Derived Growth Factor
- RTK
Receptor Tyrosine Kinase
- SH2
Src Homology 2
- SH3
Src Homology 3
- Sos1
Son of Sevenless 1
Footnotes
This work was supported by funds from the National Institutes of Health (Grant# R01-GM083897), the American Heart Association (Grant# 0655087B) and the UM/Sylvester Braman Family Breast Cancer Institute to AF.
REFERENCES
- 1.Chardin P, Cussac D, Maignan S, Ducruix A. The Grb2 adaptor. FEBS Lett. 1995;369:47–51. doi: 10.1016/0014-5793(95)00578-w. [DOI] [PubMed] [Google Scholar]
- 2.Nimnual A, Bar-Sagi D. The two hats of SOS. Sci STKE. 2002;2002:PE36. doi: 10.1126/stke.2002.145.pe36. [DOI] [PubMed] [Google Scholar]
- 3.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 4.Gale NW, Kaplan S, Lowenstein EJ, Schlessinger J, Bar-Sagi D. Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras. Nature. 1993;363:88–92. doi: 10.1038/363088a0. [DOI] [PubMed] [Google Scholar]
- 5.Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thoma S, Brugge J, Pelicci PG, Schlessinger J, Pawson T. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the ras pathway by tyrosine kinases. Nature. 1992;360:689–692. doi: 10.1038/360689a0. [DOI] [PubMed] [Google Scholar]
- 6.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 7.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 8.Schaeper U, Gehring NH, Fuchs KP, Sachs M, Kempkes B, Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J Cell Biol. 2000;149:1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewitzky M, Kardinal C, Gehring NH, Schmidt EK, Konkol B, Eulitz M, Birchmeier W, Schaeper U, Feller SM. The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P-x-x-P core motif. Oncogene. 2001;20:1052–1062. doi: 10.1038/sj.onc.1204202. [DOI] [PubMed] [Google Scholar]
- 10.Seedorf K, Kostka G, Lammers R, Bashkin P, Daly R, Burgess WH, van der Bliek AM, Schlessinger J, Ullrich A. Dynamin binds to SH3 domains of phospholipase C gamma and GRB-2. J Biol Chem. 1994;269:16009–16014. [PubMed] [Google Scholar]
- 11.Vidal M, Montiel JL, Cussac D, Cornille F, Duchesne M, Parker F, Tocque B, Roques BP, Garbay C. Differential interactions of the growth factor receptor-bound protein 2 N-SH3 domain with son of sevenless and dynamin. Potential role in the Ras-dependent signaling pathway. J Biol Chem. 1998;273:5343–5348. doi: 10.1074/jbc.273.9.5343. [DOI] [PubMed] [Google Scholar]
- 12.Odai H, Sasaki K, Iwamatsu A, Hanazono Y, Tanaka T, Mitani K, Yazaki Y, Hirai H. The proto-oncogene product c-Cbl becomes tyrosine phosphorylated by stimulation with GM-CSF or Epo and constitutively binds to the SH3 domain of Grb2/Ash in human hematopoietic cells. J Biol Chem. 1995;270:10800–10805. doi: 10.1074/jbc.270.18.10800. [DOI] [PubMed] [Google Scholar]
- 13.Park RK, Kyono WT, Liu Y, Durden DL. CBL-GRB2 interaction in myeloid immunoreceptor tyrosine activation motif signaling. J Immunol. 1998;160:5018–5027. [PubMed] [Google Scholar]
- 14.Moeller SJ, Head ED, Sheaff RJ. p27Kip1 inhibition of GRB2-SOS formation can regulate Ras activation. Mol. Cell. Biol. 2003;23:3735–3752. doi: 10.1128/MCB.23.11.3735-3752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 16.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 18.Wittekind M, Mapelli C, Lee V, Goldfarb V, Friedrichs MS, Meyers CA, Mueller L. Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts. J. Mol. Biol. 1997;267:933–952. doi: 10.1006/jmbi.1996.0886. [DOI] [PubMed] [Google Scholar]
- 19.Wittekind M, Mapelli C, Farmer BT, 2nd, Suen KL, Goldfarb V, Tsao J, Lavoie T, Barbacid M, Meyers CA, Mueller L. Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy. Biochemistry. 1994;33:13531–13539. doi: 10.1021/bi00250a004. [DOI] [PubMed] [Google Scholar]
- 20.Goudreau N, Cornille F, Duchesne M, Parker F, Tocque B, Garbay C, Roques BP. NMR structure of the N-terminal SH3 domain of GRB2 and its complex with a proline-rich peptide from Sos. Nat Struct Biol. 1994;1:898–907. doi: 10.1038/nsb1294-898. [DOI] [PubMed] [Google Scholar]
- 21.Terasawa H, Kohda D, Hatanaka H, Tsuchiya S, Ogura K, Nagata K, Ishii S, Mandiyan V, Ullrich A, Schlessinger J, et al. Structure of the N-terminal SH3 domain of GRB2 complexed with a peptide from the guanine nucleotide releasing factor Sos. Nat Struct Biol. 1994;1:891–897. doi: 10.1038/nsb1294-891. [DOI] [PubMed] [Google Scholar]
- 22.McDonald CB, Seldeen KL, Deegan BJ, Farooq A. Structural Basis of the Differential Binding of the SH3 Domains of Grb2 Adaptor to the Guanine Nucleotide Exchange Factor Sos1. Arch Biochem Biophys. 2008;479:52–62. doi: 10.1016/j.abb.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Kohda D, Terasawa H, Ichikawa S, Ogura K, Hatanaka H, Mandiyan V, Ullrich A, Schlessinger J, Inagaki F. Solution structure and ligand-binding site of the carboxy-terminal SH3 domain of GRB2. Structure. 1994;2:1029–1040. doi: 10.1016/s0969-2126(94)00106-5. [DOI] [PubMed] [Google Scholar]
- 24.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 26.Carson M. Ribbons 2.0. J. Appl. Crystallogr. 1991;24:958–961. [Google Scholar]
- 27.Arold S, O'Brien R, Franken P, Strub MP, Hoh F, Dumas C, Ladbury JE. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37:14683–14691. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Pawley NH, Nicholson LK. The role of backbone motions in ligand binding to the c-Src SH3 domain. J Mol Biol. 2001;313:873–887. doi: 10.1006/jmbi.2001.5083. [DOI] [PubMed] [Google Scholar]
- 29.Renzoni DA, Pugh DJ, Siligardi G, Das P, Morton CJ, Rossi C, Waterfield MD, Campbell ID, Ladbury JE. Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase. Biochemistry. 1996;35:15646–15653. doi: 10.1021/bi9620969. [DOI] [PubMed] [Google Scholar]
- 30.Ferreon JC, Hilser VJ. The effect of the polyproline II (PPII) conformation on the denatured state entropy. Protein Sci. 2003;12:447–457. doi: 10.1110/ps.0237803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreon JC, Hilser VJ. Ligand-induced changes in dynamics in the RT loop of the C-terminal SH3 domain of Sem-5 indicate cooperative conformational coupling. Protein Sci. 2003;12:982–996. doi: 10.1110/ps.0238003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreon JC, Hilser VJ. Thermodynamics of binding to SH3 domains: the energetic impact of polyproline II (PII) helix formation. Biochemistry. 2004;43:7787–7797. doi: 10.1021/bi049752m. [DOI] [PubMed] [Google Scholar]
- 33.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- 34.Cesareni G, Panni S, Nardelli G, Castagnoli L. Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett. 2002;513:38–44. doi: 10.1016/s0014-5793(01)03307-5. [DOI] [PubMed] [Google Scholar]
- 35.Panni S, Dente L, Cesareni G. In vitro evolution of recognition specificity mediated by SH3 domains reveals target recognition rules. J Biol Chem. 2002;277:21666–21674. doi: 10.1074/jbc.M109788200. [DOI] [PubMed] [Google Scholar]
- 36.Brannetti B, Via A, Cestra G, Cesareni G, Helmer-Citterich M. SH3-SPOT: an algorithm to predict preferred ligands to different members of the SH3 gene family. J Mol Biol. 2000;298:313–328. doi: 10.1006/jmbi.2000.3670. [DOI] [PubMed] [Google Scholar]
- 37.Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat Struct Biol. 2001;8:998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer BJ. SH3 domains: complexity in moderation. J. Cell. Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 39.Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Sci STKE. 2003;2003:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 40.Sparks AB, Rider JE, Hoffman NG, Fowlkes DM, Quillam LA, Kay BK. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci U S A. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 42.Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 43.Kami K, Takeya R, Sumimoto H, Kohda D. Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. Embo J. 2002;21:4268–4276. doi: 10.1093/emboj/cdf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CH, Leung B, Lemmon MA, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. Embo J. 1995;14:5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal Structure of the Conserved Core of HIV-1 Nef Complexed with a Src Family SH3 Domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 46.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 47.Berry DM, Nash P, Liu SK, Pawson T, McGlade CJ. A high-affinity Arg-X-X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr Biol. 2002;12:1336–1341. doi: 10.1016/s0960-9822(02)01038-2. [DOI] [PubMed] [Google Scholar]
- 48.Harkiolaki M, Lewitzky M, Gilbert RJ, Jones EY, Bourette RP, Mouchiroud G, Sondermann H, Moarefi I, Feller SM. Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76. Embo J. 2003;22:2571–2582. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Berry D, Nash P, Pawson T, McGlade CJ, Li SS. Structural basis for specific binding of the Gads SH3 domain to an RxxK motif-containing SLP-76 peptide: a novel mode of peptide recognition. Mol Cell. 2003;11:471–481. doi: 10.1016/s1097-2765(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 50.Geisler SB, Robinson D, Hauringa M, Raeker MO, Borisov AB, Westfall MV, Russell MW. Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin. Genomics. 2007;89:521–531. doi: 10.1016/j.ygeno.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang B, Mandal MN, Chavali VR, Hawes NL, Khan NW, Hurd RE, Smith RS, Davisson ML, Kopplin L, Klein BE, Klein R, Iyengar SK, Heckenlively JR, Ayyagari R. Age-related retinal degeneration (arrd2) in a novel mouse model due to a nonsense mutation in the Mdm1 gene. Hum Mol Genet. 2008;17:3929–3941. doi: 10.1093/hmg/ddn295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodfish P, Warne D, Nyberg K, Spurr NK. Dinucleotide repeat polymorphism at the human erythroid alpha-spectrin (SPTA1) mRNA gene detected using PCR. Hum Mol Genet. 1992;1:287. doi: 10.1093/hmg/1.4.287-a. [DOI] [PubMed] [Google Scholar]
- 53.Hentati A, Hu P, Asgharzadeh S, Siddique T. Dinucleotide repeat polymorphism at the human erythroid alpha spectrin (SPTA1) locus. Hum Mol Genet. 1992;1:218. doi: 10.1093/hmg/1.3.218-a. [DOI] [PubMed] [Google Scholar]
- 54.Kurosawa N, Chen GY, Kadomatsu K, Ikematsu S, Sakuma S, Muramatsu T. Glypican-2 binds to midkine: the role of glypican-2 in neuronal cell adhesion and neurite outgrowth. Glycoconj J. 2001;18:499–507. doi: 10.1023/a:1016042303253. [DOI] [PubMed] [Google Scholar]
- 55.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 56.Schlaepfer DD, Hanks SK, Hunter T, van der Geer P. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 57.Chook YM, Gish GD, Kay CM, Pai EF, Pawson T. The Grb2-mSos1 complex binds phosphopeptides with higher affinity than Grb2. J Biol Chem. 1996;271:30472–30478. doi: 10.1074/jbc.271.48.30472. [DOI] [PubMed] [Google Scholar]
- 58.Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 59.Sondermann H, Soisson SM, Boykevisch S, Yang SS, Bar-Sagi D, Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]