Abstract
The tumor suppressor p53 is a master transcriptional regulator that affects a diverse range of cellular events. Surprisingly, even with >100 validated p53 response element (RE) sequences available, the effect of p53 binding on transcriptional behavior is seldom predictable and no functional rules have been described. Here, we report a systematic study on the role of specific nucleotides within the p53RE by using p21, a well-known target for p53 activation and contrasting it with Lasp1, a gene recently identified to be repressed by p53. Functional assays revealed a specific dinucleotide core combination within the CWWG motif of the p53RE to be the key factor that determines whether p53 transcriptionally activates or represses a target gene. The triplet RRR and YYY sequences flanking the core CWWG motif were also shown to play an important role in modulating the transcriptional behavior of p53. With the establishment of a set of predictive rules, we were able to reassess 162 published p53REs and showed that the attributed function for 20/162 p53REs studied were in fact erroneous. A significant proportion of p53REs (39/162) were found to be repressive, which is substantially higher than what is currently thought. Hence this clearer definition of the transcriptional behavior of p53 interaction with its RE will provide better insight toward the understanding of its fundamental role in cellular networks.
Keywords: cancer, gene regulation, transcription, activation, repression
The p53 protein is a tumor suppressor that regulates transcription of a broad range of target genes involved in apoptosis, growth control, senescence, and angiogenesis (1, 2). It recognizes and binds to DNA containing p53 response elements (REs) that comprise 2 decamer motif RRRCWWGYYY separated by a spacer of 0–13 bp, where W is A or T, R is C or G, and Y is C or T (3, 4). High-affinity binding of p53 to the RE frequently requires the conserved CWWG motif but nucleotide polymorphisms within the rest of the RE can affect transcriptional behavior of p53 (5–7).
To orchestrate the genomic response to cellular stress signals, p53 binds to a large number of RE variants and subsequently either activates or represses promoter activity of its target genes (8, 9). Although the activating function of p53 has been well studied and easily linked to the classical p53RE motifs (9–11), the repressing function has not been well characterized because of the difficulty in identifying associated p53RE motifs (12–15). A study using overexpression of p53 in human ovarian cancer cells reported that ≈80% of 1,501 responsive genes were repressed by p53 (16). However, such observations lack validation of identifiable p53REs and the reported numbers are likely to be the result of secondary or indirect effects. Hence there is still a lack of understanding surrounding the extent of p53 repressed genes.
Although a number of studies have attempted to predict patterns of p53RE sequences (17–19) or SNPs (20, 21) to link with function, accurate determination has been unsuccessful. From a theoretical perspective, it has been estimated that the complexity of a 20-bp p53RE sequence could generate 410 variants based on the 10-bp half-site and that there can be several thousand copies of p53REs in the human genome (22). Using ChIP and microarray analysis, Wei et al. (23) identified 542 p53 binding loci with relative high confidence. However, there still exists a gap in identifying and validating bona fide p53REs, and the ability to differentiate between activating and repressing p53REs remains a challenge. We reasoned that this failure lies in the lack of a robust approach to discern subtle differences in the sequence composition of the p53REs and specific nucleotides can exert significant effect on p53 transcriptional behavior. In this study, we report the systematic functional analysis of nucleotides within the p53RE and the identification of a dinucleotide core motif that plays a key role in determining the transcriptional behavior of p53.
Results
Identification of a Bona Fide Repressive p53RE.
To compare and contrast the functional properties of p53RE sequences, the p21 gene was selected as a model for p53 activation, and for the repression counterpart a recently identified Lasp1 gene was used (24). Because previous studies on p53-repressed genes could not always identify a p53RE, we took additional steps to verify that Lasp1 p53RE is indeed repressive. To test whether p53 functionally represses Lasp1 gene expression, pCMV-p53 or pCMV-p53-R175H expression constructs were transfected into HCT116 (p53−/−) cells. Fig. 1A illustrates that the overexpression of p53 induced opposite effects on p21 and Lasp1 transcription in HCT116 (p53−/−) cells. Wild-type p53 but not p53-R175H mutant repressed Lasp1 protein expression, whereas p21 transcription was activated by wild-type p53 but not p53-R175H mutant. The use of p53-specific siRNA in HCT116 (p53+/+) cells also elicited the same divergent pattern of p53 transcriptional behavior, where Lasp1 mRNA was up-regulated to 154%, whereas p21 mRNA was repressed by p53 siRNA by 63% relative to that of control (P < 0.05; Fig. 1B).
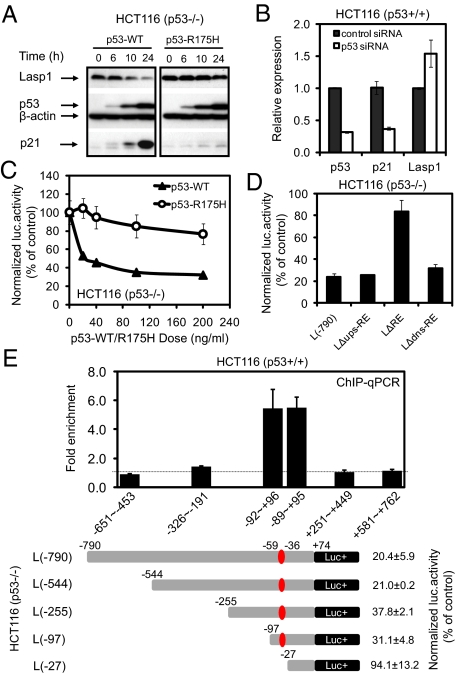
Fig. 1.
p53 represses Lasp1 transcription. (A) Western blot analysis of Lasp1, p21, and p53 protein expression from HCT116 (p53−/−) cells with overexpressed wild-type or mutant p53. (B) Real-time qRT-PCR analysis of Lasp1, p21, and p53 mRNA expression from HCT116 (p53+/+) cells transfected with p53-specific siRNA. (C) Wild-type p53 but not p53-R175H mutant down-regulated Lasp1 promoter activity in a dose-dependent manner. (D) Loss of p53 response by deletion of the putative p53RE (LΔRE) but not the 24 bp immediately upstream (Lups-ΔRE) or downstream (Ldns-ΔRE) sequences. (E) ChIP-qPCR analysis showed p53 occupancy with the region containing the identified Lasp1 p53RE (Upper), which corresponds to the p53 response region further confirmed by luciferase assay using serial deletion constructs (Lower).
To confirm the presence of a p53RE, luciferase assay using a pGL3-luciferase reporter construct L(−790) containing the (−790 ≈ +74 bp) fragment of Lasp1 gene was performed, and the results showed that wild-type p53 but not p53-R175H mutant inhibited the Lasp1 promoter activity in a dose-dependent manner (Fig. 1C). Luciferase assay using internal deletion construct LΔRE confirmed that removing the putative p53RE site (−59 ≈ −36 bp, AGGCCAGTTCcccaGCTCCAGCCG) significantly abrogated p53-mediated transcriptional repression (P < 0.01). As controls, 2 additional deletion constructs targeting 24 bp either upstream (LΔups-RE) or downstream (LΔdns-RE) of the RE remained repressed by p53 (Fig. 1D). To verify whether the identified Lasp1 p53RE is associated with p53 interaction in vivo, we examined the region (−651 to +762 bp) of the Lasp1 gene by using ChIP-quantitative PCR (qPCR) in HCT116 cells. The results confirmed p53 binding with the highest level of occupancy coinciding with the region containing the Lasp1 p53RE (−59 to −36bp) and further supported by serial deletion analysis (Fig. 1E). Taken together, the above data show that the (AGGCCAGTTCcccaGCTCCAGCCG) sequence functions to mediate repression by p53.
A Regulatory Dinucleotide Core Is Located Within the p53RE.
To understand the basis behind the differences between p53-mediated activation and repression, we compared the repressing p53RE of Lasp1 with the well-known activating p53RE of p21. In addition to the nucleotide sequence differences, the Lasp1 p53RE has a 4-bp spacer separating the 2 decamer motifs. To examine whether this spacer has any impact on p53RE function, successive nucleotide deletion of the 4-bp spacer was performed. The results showed that deleting part or all of the spacer did not abrogate p53-dependent repression (Fig. 2A).
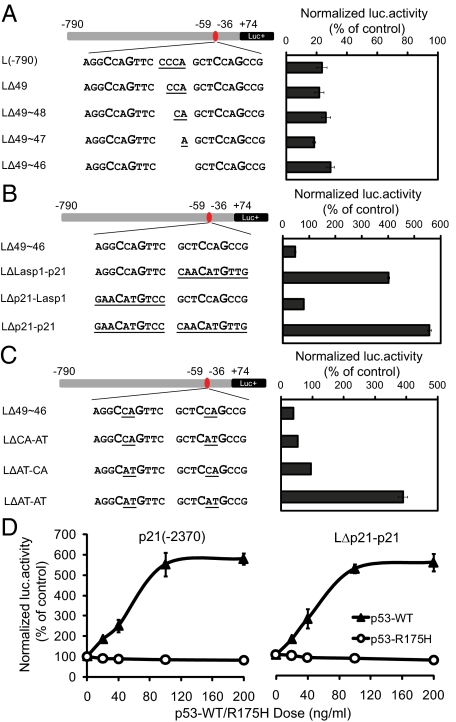
Fig. 2.
Dinucleotide core within CWWG motif determines p53 transcriptional behavior. (A) Sequential deletion of the 4-bp spacer in the repressing RE did not affect p53 repression. (B) Replacement of either or both decamers in the repressing Lasp1 RE with that of activating p21 RE resulted in conversion of p53 function from repression to activation. (C) Change of dinucleotide core within the CWWG motif in the repressing Lasp1 RE from CA to AT resulted in conversion of p53 function from repression to activation. (D) Both p21 promoter construct p21(−2370) containing the p21 p53RE and the chimeric LΔp21-p21 have a comparable level of activation in response to different doses of p53.
Next, we proceeded to test the properties of the 2 half-sites of p53RE by performing domain swaps. Three chimera constructs (LΔLasp1-p21, LΔp21-Lasp1, and LΔp21-p21) were generated, in which one or both half-sites of the Lasp1 p53RE were replaced with that of p21 p53RE. Surprisingly, replacement of the second decamer half-site resulted in conversion from repression to activation and the effect was increased further when both half-sites were replaced (Fig. 2B). Because changing the half-sites alone resulted in such functional conversion, we asked the question of whether part or all of the decamer is required for this function. Interestingly, the sequence difference between these 2 p53REs is essentially a dinucleotide core within the conserved CWWG motif, where it is CA in Lasp1 and AT in p21. Fig. 2C shows that when the CA dinucleotide core alone was substituted by AT in both half-sites (LΔAT-AT), it resulted in a dramatic conversion from repression to activation. This effect is mediated principally by the dinucleotide core as a p21 promoter region (2.4 kb) containing p21 p53RE when compared with the (−790 bp) Lasp1 promoter containing the p21 p53RE (LΔp21-p21) insert showed comparable levels of activation in response to p53 (Fig. 2D). These results suggest that the promoter sequence outside of the p53RE has relatively minor or no effect on the function of the RE itself.
To further probe the properties of the dinucleotide core, all combinatorial permutations were generated and functionally tested against both repressing and activating p53RE half-site backgrounds (Fig. 3 A and B). The results revealed that p53RE is strongly activating if its dinucleotide core is either AT, AA, or TT, whereas CG, GG, TG, CC, GC, or CA are repressive. The remaining dinucleotides, TA, AC, AG, CT, GT, TC, and GA, can be activating or repressing depending on the p53RE half-site background where it is located.
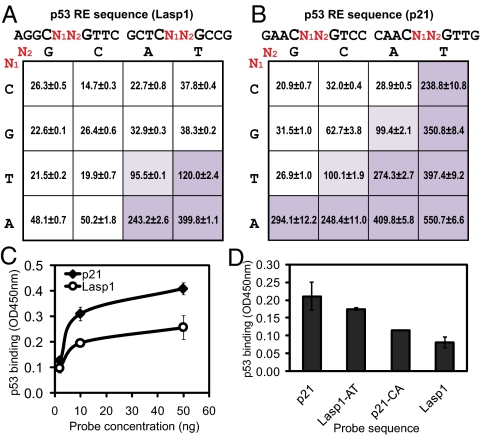
Fig. 3.
Effect of combinatorial dinucleotide core on transcription. (A) Matrix showing the normalized luciferase activity of 16 luciferase constructs bearing all dinucleotide combinations in the core CWWG motif using Lasp1 RE background based on L(−790). (B) Matrix showing the normalized luciferase activity of 16 luciferase constructs bearing all dinucleotide combinations in the core CWWG motif using p21 RE background based on L(−790). (C) DNA-protein binding ELISA showing dose-dependent p53 binding to oligonucleotides of repressing (Lasp1) or activating (p21) RE. p53 protein was standardized to 25 ng per well reaction, while the DNA probe concentrations range from 2 to 50 ng per reaction. (D) DNA-protein binding ELISA showing p53 binding to test the effect of dinucleotide core change in Lasp1 and p21 RE, where 50 ng of each probe and 25 ng of p53 protein were applied in each reaction.
To investigate whether there is a physical basis to account for the differential function of p53 binding, oligonucleotides of p21 and Lasp1 p53RE were coated onto ELISA microtiter plates and tested for their ability to bind p53 protein (25). Fig. 3C shows that p53 had a distinctly higher binding affinity to p21 p53RE oligonucleotides compared with Lasp1 p53RE. Moreover, replacement of the AT dinucleotide core of p21 RE with CA (p21-CA) resulted in a significant decrease in p53 binding, whereas converse replacement of CA with AT in Lasp1 RE (Lasp1-AT) resulted in an increased level of p53 binding (Fig. 3D). These results demonstrate that the dinucleotide core is important in stabilizing the binding of p53 to its RE.
Triplet Flanking Sequences Modulate p53 Transcriptional Behavior.
A series of chimera half-site constructs were generated to functionally evaluate the role of the triplet RRR and YYY flanking sequences. Lasp1 p53RE was first replaced by either duplicated half-site 1 or duplicated half-site 2. Although these replacements did not change the overall p53 repressive function, slight loss of repression was observed when Lasp1 p53RE half-site 1 was duplicated, whereas the duplicated half-site 2 gave rise to an even stronger repression effect (Fig. 4A). Because the triplet flanking sequences in the half-site 2 of Lasp1 p53RE is RYYCCAGYYR, which differs from the YRRCATGYYR pattern in p21 p53RE half-site 2, we generated 3 additional constructs based on the Lasp1 p53RE background to replace only the flanking triplet of Lasp1 p53RE half-site 2 with that of p21 p53RE. Such replacement, however, did not result in any change in p53 repression (Fig. 4B).
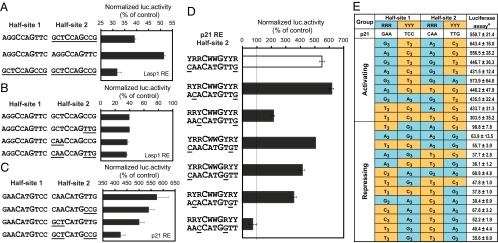
Fig. 4.
Triplet sequences flanking the core CWWG motif modify p53 transcriptional behavior. (A) Duplicate Lasp1 RE half-site 1 reduced p53-mediated repression, whereas duplicate half-site 2 enhanced repression. (B) Replacement of the triplet flanking sequences in Lasp1 RE half-site 2 with that from p21 RE did not affect the p53 repression. (C) Replacement of the triplet flanking sequences in p21 RE half-site 2 with that from Lasp1 RE diminished p53 activation. (D) Positional substitution of nucleotides within the triplet flanking sequences in the p21 RE half-site 2 can abrogate p53 activation. (E) The normalized luciferase activity of 20 luciferase constructs bearing different nucleotide combinations of the triplet flanking sequences using p21 RE background. G3 = GGG, A3 = AAA, C3 = CCC, T3 = TTT. *, Normalized luciferase activity (% of control).
Next, either one or both of the triplet flanking sequences in the half-site 2 of p21 p53RE was replaced by that of Lasp1 p53RE. Although the CAACATGTTG sequence was changed to CAACATGCCG, they both conform to the same YRRCATGYYR pattern, and thus there was only a slight loss of activation observed. In contrast, the change of triplet sequence pattern from YRRCATGYYR (CAACATGTTG) to RYYCATGYYR (GCTCATGTTG or GCTCATGCCG) resulted in a significant reduction in p53-mediated activation (Fig. 4C). Although the p21 RE is well studied, its half-site 1 possesses the canonical RRRCWWGYYY motif whereas half-site 2 is YRRCWWGYYR. Using this natural variation in half-site 2, the positional effects of R and Y nucleotides were evaluated by creating different triplet combinations (Fig. 4D). The results show that the activating property of the dinucleotide core AT can be abrogated by changing the nucleotide adjacent to the CWWG motif (YRRCWWGYYR to RRYCWWGRYY). Hence the data indicate that the positional weight of individual nucleotides within the triplet flanking sequence is stronger when closer to the CWWG motif and decreases when further away from it.
Arising from this observation, a comprehensive set of synthetic triplet flanking motifs was constructed to assess the combinatorial possibilities on p53RE function (Fig. 4E). As expected, the RRR-YYY:RRR-YYY combination, which conforms to the classical canonical motif, is most favorable for activation. Alterations to this motif and in particular the YYY-RRR:YYY-RRR combination will disrupt activation even when the dinucleotide motif is of the activating type. Taken together, these results reveal a set of general principles or rules governing the roles of nucleotides within the p53RE: (i) the dinucleotide core within CWWG motif is the major factor determining activation or repression function, (ii) the triplet flanking sequences can exert a modulating effect, and (iii) the nucleotide adjacent to the CWWG motif has the strongest positional effect. In repressive p53REs, the flanking triplet can only modulate the degree of repression but not reverse it; whereas in activating p53REs selected triplet combinations can convert activation to repression.
Reassignment of 20 Discrepant p53REs.
To test the robustness of these rules, a list of published p53REs were compiled and grouped into activating or repressing types. When the above rules were applied to published data, 142/162 (87.7%) were found to be concordant (Table S1), whereas 20 REs (16 activating, 2 repressing, 2 unknown) (26–28) were discrepant. To experimentally verify these discrepancies, all 20 p53REs were synthesized and individually cloned into pGL3-Lasp1–790 luciferase reporter plasmid. The validation assays showed that the assigned roles of these published p53REs turned out to be erroneous (Table S2). For instance, IER3/IEX-1 gene has a reported p53RE that functions as a repressor (29), however, the sequence CCACATGCCT:CGACATGTGC contains the classical activating dinucleotide core motif of AT in both decamers. Functional assay performed on this p53RE confirmed that the sequence is indeed activating rather than repressing. Another example is the second CTSD/IRDD p53 binding motif (AAGCTGGGCC:GGGCTGACCC), where it was reported to be activating (30), but the functional assay proved it to be of a repressing type. This result agrees with the repressing dinucleotide core combination (Fig. 3 A and B), which is TG in both decamers. The role of triplet flanking nucleotides in modulating the p53RE function can be seen in the p53RE of BCL2L14/BCL-G (31), where both dinucleotide cores are of the activating type, but the RE was functionally repressing. This observation can be explained by the positional effect of the nucleotides adjacent to the CWWG motif as shown in Fig. 4 D and E where instead of the canonical RRRCWWGYYY:RRRCWWGYYY, the BCL2L14/BCL-G p53RE (AGCCAAGGCT:GGTCTTGAAC) is RRYCWWGRYY:RRYCWWGRRY. Similarly, a second example can be seen in the p53RE of PLK2/SNK gene, GGTCATGATT:TAACTTGCCT, i.e., RRYCWWGRYY:YRRCWWGYYY (32), in which the dinucleotide core is also activating type, but the p53 function was modulated by the nucleotides adjacent to CWWG motif in half-site 1 to become repressive.
Activating and Repressing p53REs Have Characteristic Nucleotide Distribution Patterns.
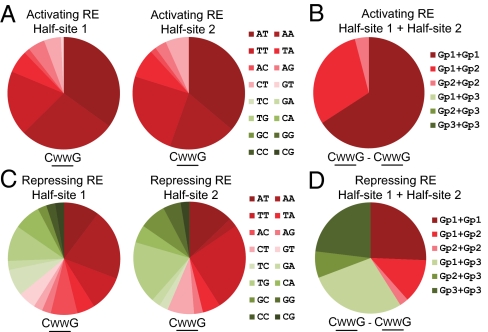
To capture the general principles and rules of p53RE behavior, we analyzed the distribution frequency of the dinucleotide core among the 123 activating p53REs and 39 repressing p53REs. To do this, dinucleotide cores were grouped into 3 categories. Group 1 dinucleotides include AT, AA, and TT, which had consensus activating properties in both repressing (Fig. 3A) and activating (Fig. 3B) p53RE backgrounds. Similarly, group 3 dinucleotides (TC/GA/TG/CA/GC/GG/CC/CG) showed consensus repressing functions in both backgrounds, whereas group 2 (TA/AC/AG/CT/GT) are those that are relatively neutral in activity. It was found that the 123 activating p53REs have predominantly AT/AA/TT in each half-site (Fig. 5A), and when taken together as a complete RE, only group 1 and/or group 2 dinucleotide combinations were observed (Fig. 5B). The 39 repressing p53REs, however, have very different dinucleotide core composition with a high proportion of TG/TC/GC/CA/GG/CG motifs that are rarely found in activating p53REs (Fig. 5C). As a complete RE, >60% of the repressing p53REs contain at least 1 group 3 dinucleotide core combination (Fig. 5D).
Fig. 5.
Distribution analysis of dinucleotide core within CWWG motif in activating and repressing p53REs. (A and B) Distribution analysis of dinucleotide core combinations from 123 validated activating REs at half-site 1 and half-site 2 (A) and complete RE (B). Group 1 (Gp1): AT/AA/TT; group 2 (Gp2): TA/AC/AG/CT/GT; group 3 (Gp3): TC/GA/TG/CA/GC/GG/CC/CG. (C and D) Distribution analysis of dinucleotide core combinations from 39 validated repressing REs at half-site 1 and half-site 2 (C) and complete RE (D).
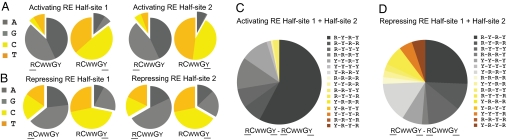
Turning our attention to the nucleotides adjacent to the CWWG motif, Fig. 6A shows that in 123 activating p53REs the frequencies of these nucleotides agree well with the canonical sequence. In contrast, the repressing p53REs have a much higher frequency of noncanonical nucleotides in the position immediately adjacent to the CWWG motif (Fig. 6B). Similarly, at the complete RE level, these nucleotides in activating p53REs conform well to the canonical RCWWGY-RCWWGY pattern (Fig. 6C), whereas among the 39 repressing p53REs, this pattern is seldom intact (Fig. 6D). Therefore, the repressing p53REs were found to differ from the activating p53REs in 2 aspects. First, the dinucleotide core is likely to be of the repressing type, and second, the flanking nucleotides adjacent to the CWWG motif are frequently found to depart from the canonical p53RE motif.
Fig. 6.
Distribution analysis of nucleotides adjacent to CWWG motif in activating and repressing p53REs. (A) Distribution analysis of the nucleotides immediately adjacent the core CWWG motif from 123 validated activating REs. (B) Distribution analysis of the nucleotides immediately adjacent the core CWWG motif from 39 validated repressing REs. (C) Distribution analysis of combinations of the 4 nt derived from A. (D) Distribution analysis of combinations of the 4 nt derived from B.
Discussion
It is widely accepted that p53 binds to REs located in the promoter of target genes to activate or repress transcription of these genes. However, it is not clear which genes will be activated or repressed even when the canonical sequence motif of the p53RE is known. By contrasting the relative efficiencies of an activating p21 p53RE against that of a repressing Lasp1 p53RE to drive luciferase gene transcription, we showed that the primary factor determining the outcome of p53 binding lies in the dinucleotide core of the CWWG motif. Although it has been observed that the fourth position (C) and seventh position (G) of the p53RE decamer are essential to the integrity of the p53RE, the importance of the internal dinucleotide core (WW) has not been described. The crucial role of the dinucleotide core was shown in 2 ways. First, the substitution of the dinucleotide core of a repressing RE with that from an activating RE was sufficient to convert it to an activating mode and the converse exchange resulted in the respective conversion. Second, a more refined analysis in which all 16 combinations of the dinucleotide core were tested in both repressing and activating p53RE half-site backgrounds (Fig. 3 A and B, respectively) displayed a clear pattern of dinucleotide combinations that were favorable to eliciting activating or repressing activity in either background. ELISA studies showed the dinucleotide core from the p21 activating p53RE (AT) was able to bind p53 protein with higher affinity compared with the dinucleotide core from the Lasp1 repressing p53RE (CA). A possible mechanism to explain the role of specific nucleotide changes in the p53RE causing activation or repression is the result of reduced dwell time or promoter occupancy of p53 tetramer when engaging specific RE sequences. This proposition is supported by studies using fluorescence anisotropy that showed that most REs with high binding affinity to p53 have CATG core motif (22, 33). Atomic force microscopy studies also show that different dinucleotide cores affect DNA flexibility and tetrameric binding of p53 (34). It is possible that cofactor recruitment is altered by the conformational outcome and contributes to further differential transactivation or repression effects.
The role of triplet RRR and YYY sequences flanking the CWWG motif also needed clearer definition. The first indication that the triplet flanking sequences have a modulating effect is seen in Fig. 4A, where duplication of Lasp1 RE half-site 1 led to loss of repression, whereas duplication of half-site 2 resulted in stronger repression. Nevertheless both chimeric REs remained repressive in function. Replacing the triplet flanking sequences in half-site 2 of Lasp1 RE with that of p21 RE resulted in little change to the repression (Fig. 4B), suggesting that the triplet flanking sequences from an activating RE does not confer activation to a repressive RE. A similar dampening effect can be seen when the RRR and YYY sequences in the half-site 2 of p21 RE were replaced with those from the repressive Lasp1 RE (Fig. 4C). Because the triplet flanking sequences in half-site 1 for both Lasp1 and p21 RE are similar, only half-site 2 was investigated with replacement studies.
Using this positional weightage of specific nucleotides within the p53RE to re-examine the functional attributes of 162 published p53RE, we showed that 142/162 were in agreement with the predicted function (Table S1). The predictions for 20 p53REs differed from the published data with 16 activating, 2 repressing, and 2 unknown. To determine the reliability of our prediction approach, sequences of these 20 p53REs were synthesized and functionally assayed (Table S2). The outcome of this validation exercise for the 20 p53REs was in perfect agreement with the predicted profile. To our surprise, all 16 activating p53REs turned out to be repressing in nature, whereas 2 previously reported repressing p53REs turned out to be activating. Two p53REs with unknown function were predicted and validated to be repressing. It was observed that a number of these p53REs did not conform to the canonical motif such that the triplet flanking sequence is RRYCWWGRYY or RRYCWWGRRY, in which case the nucleotide closest to the CWWG motif was able to modulate and convert the activating function to repressing type. Hence, activating p53REs need to conform to a stricter sequence requirement and deviations from it can lead to abrogation of this function. These results suggest that repressing p53REs may have been difficult to ascertain with a good degree of confidence in the past and their numbers were underestimated.
Distinct patterns of nucleotide frequency distribution emerged when the dinucleotide core combinations were analyzed in activating and repressing p53RE groups. Among activating p53REs the dinucleotide core combination AT/AA/TT predominates, whereas in the repressing p53RE group combinations of TG/TC/GC/CA/GG/CG were most frequent. Another critical element is the nucleotide closest to the CWWG motif, and Fig. 6A shows that the frequency of this nucleotide conforms well to the canonical sequence in activating REs. This is not so in repressing REs (Fig. 6B) where Y(C/T) nucleotides can occur in R(G/A) positions and vice versa. When viewed at the complete RE level, the occurrence of these nucleotides in activating REs fit well with the canonical RCWWGY-RCWWGY pattern, whereas in repressing REs, this pattern is disrupted (Fig. 6 C and D). This explains that even though ≈25% of the repressing p53REs possess activating dinucleotide cores (Fig. 5D), variations at the triplet flanking sequence are able to modulate overall p53RE behavior.
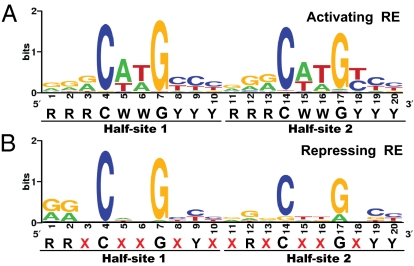
In conclusion, our study provides strong evidence that p53RE should no longer be described as a single canonical entity but redefined as 2 distinct activating and repressing types, each with its own characteristic features. Although the functional outcome of p53 binding to its RE is still subject to influences such as p53 mutations (35), transcriptional cofactors (9, 36), and variable spacer lengths (37), the primary determinant for activating p53REs is the canonical RRRCWWGYYY-RRRCWWGYYY sequence, whereas repressing p53REs are RRXCXXGXYX-XRXCXXGXYY (Fig. 7 A and B). The surprising proportion of validated repressing p53REs (39/162, 24.1%) shown here suggests that this aspect of p53 function needs to be better understood in the context of gene regulatory networks. With the clearer definition of what constitutes a repressing p53RE, more p53 repressed genes are likely to be identified. The findings from this study allow the functional role of a p53RE to be determined with a high degree of confidence.
Fig. 7.
Redefined activating and repressing p53RE. p53 RE motifs from 123 validated activating p53REs (A) and 39 validated repressing p53REs (B) drawn with WebLogo software (http://weblogo.berkeley.edu/logo.cgi) (38, 39).
Materials and Methods
Cell Lines.
The human colon cancer cell line HCT116, its derived p53−/− cells, the wild-type p53 (pCMV-p53), and mutant p53 (pCMV-p53-R175H) expression vectors were kindly provided by Bert Vogelstein (The Johns Hopkins University, Baltimore).
Luciferase Reporter Assay.
DNA fragments containing 5′ flanking region (−790 to +74 bp) of Lasp1 promoter were first amplified by PCR and subsequently subcloned into pGL3-Basic vector. Different luciferase constructs used in this study were then generated based on L(−790) template. Different promoter luciferase reporter constructs (2 μg/mL) were cotransfected with pCMV-p53, pCMV-p53-R175H expression vectors, or pcDNA3.1 control plasmid (unless specified, 200 ng/mL of the expression vectors was used), together with 20 ng/mL of Renilla vector PRL-null into HCT116 (p53−/−) cells plated in 96-well plates. The luciferase reporter activity was measured by using the Dual-Luciferase Reporter Assay System (Promega). For all luciferase assays, the normalized luciferase activity of each construct (Firefly/Renilla ratio) in the pCMV-p53 or pCMV-R175H cotransfected cells was then compared with that of the pcDNA3.1 control cotransfected cells.
ChIP-qPCR.
ChIP assays were carried out with HCT116 (p53+/+) cells treated with 5-fluorouracil (5-FU) (375 μM) for 24 h as described (23).
ELISA.
To analyze binding of p53 protein to DNA sequences from different p53REs, a DNA-protein binding ELISA was performed as described (25) with modifications. Briefly, 25 ng of purified wild-type p53 protein (Aviva Systems Biology) was incubated with anti-p53 antibody DO-1 and then mixed with different dilutions of annealed double-stranded biotin labeled DNA probes (2, 10, or 50 ng) before plated into streptavidin-coated 96-well plates. After incubation with goat-anti-mouse HRP-conjugated IgG, measurement of color resulting from the enzymatic reaction (TMB substrate) was carried out in a Tecan plate reader at 450/570-nm wavelength.
For additional information see SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Bert Vogelstein for providing p53+/+ and p53−/− HCT116 cells and pCMV-p53, pCMV-p53-R175H vectors, Jianming Jiang for help with ChIP assay, and Su Qin Peh for technical assistance. This work was supported by the Agency for Science, Technology and Research (A*STAR), Singapore. Z.X. is supported by a President Graduate Fellowship, National University of Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903284106/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lu X. Live or let die: The cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 4.Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol. 2002;22:8612–8625. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan JJ, et al. Noncanonical DNA motifs as transactivation targets by wild-type and mutant p53. PLoS Genet. 2008;4:e1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osada M, et al. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumor suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 10.Muller M, et al. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sax JK, et al. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- 12.Gopalkrishnan RV, Lam EW, Kedinger C. The p53 tumor suppressor inhibits transcription of the TATA-less mouse DP1 promoter. J Biol Chem. 1998;273:10972–10978. doi: 10.1074/jbc.273.18.10972. [DOI] [PubMed] [Google Scholar]
- 13.Krause K, et al. The tumor suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Lin M, Liu J. Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene. 2004;23:9336–9347. doi: 10.1038/sj.onc.1208114. [DOI] [PubMed] [Google Scholar]
- 15.May P, May E. Twenty years of p53 research: Structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 16.Mirza A, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- 17.Spurgers KB, et al. Identification of cell cycle regulatory genes as principal targets of p53-mediated transcriptional repression. J Biol Chem. 2006;281:25134–25142. doi: 10.1074/jbc.M513901200. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, et al. Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J Biol Chem. 2001;276:43604–43610. doi: 10.1074/jbc.M106570200. [DOI] [PubMed] [Google Scholar]
- 19.Zhao R, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 20.Menendez D, et al. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci USA. 2006;103:1406–1411. doi: 10.1073/pnas.0508103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomso DJ, et al. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc Natl Acad Sci USA. 2005;102:6431–6436. doi: 10.1073/pnas.0501721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veprintsev DB, Fersht AR. Algorithm for prediction of tumor suppressor p53 affinity for binding sites in DNA. Nucleic Acids Res. 2008;36:1589–1598. doi: 10.1093/nar/gkm1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Feng P, Xiao Z, Ren EC. LIM and SH3 protein 1 (Lasp1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J Hepatol. 2009;50:528–537. doi: 10.1016/j.jhep.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 25.Jagelska E, Brazda V, Pospisilova S, Vojtesek B, Palecek E. New ELISA technique for analysis of p53 protein/DNA binding properties. J Immunol Methods. 2002;267:227–235. doi: 10.1016/s0022-1759(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 26.Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: Cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 29.Im HJ, Pittelkow MR, Kumar R. Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. J Biol Chem. 2002;277:14612–14621. doi: 10.1074/jbc.M109414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu GS, Saftig P, Peters C, el-Deiry WS. Potential role for cathepsin D in p53-dependent tumor suppression and chemosensitivity. Oncogene. 1998;16:2177–2183. doi: 10.1038/sj.onc.1201755. [DOI] [PubMed] [Google Scholar]
- 31.Miled C, Pontoglio M, Garbay S, Yaniv M, Weitzman JB. A genomic map of p53 binding sites identifies novel p53 targets involved in an apoptotic network. Cancer Res. 2005;65:5096–5104. doi: 10.1158/0008-5472.CAN-04-4232. [DOI] [PubMed] [Google Scholar]
- 32.Burns TF, Fei P, Scata KA, Dicker DT, el-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23:5556–5571. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol. 2005;348:589–596. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Balagurumoorthy P, Lindsay SM, Harrington RE. Atomic force microscopy reveals kinks in the p53 response element DNA. Biophys Chem. 2002;101–102:611–623. doi: 10.1016/s0301-4622(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 35.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: Cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 36.Latchman DS. Transcription factors: Bound to activate or repress. Trends Biochem Sci. 2001;26:211–213. doi: 10.1016/s0968-0004(01)01812-6. [DOI] [PubMed] [Google Scholar]
- 37.Tokino T, et al. p53 tagged sites from human genomic DNA. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.