Abstract
The impact of antibiotics on the host's protective microbiota and the resulting increased susceptibility to mucosal infection are poorly understood. In this study, antibiotic regimens commonly applied to murine enteritis models are used to examine the impact of antibiotics on the intestinal microbiota, the time course of recovery of the biota, and the resulting susceptibility to enteric Salmonella infection. Molecular analysis of the microbiota showed that antibiotic treatment has an impact on the colonization of the murine gut that is site and antibiotic dependent. While combinations of antibiotics were able to eliminate culturable bacteria, none of the antibiotic treatments were effective at sterilizing the intestinal tract. Recovery of total bacterial numbers occurs within 1 week after antibiotic withdrawal, but alterations in specific bacterial groups persist for several weeks. Increased Salmonella translocation associated with antibiotic pretreatment corrects rapidly in association with the recovery of the most dominant bacterial group, which parallels the recovery of total bacterial numbers. However, susceptibility to intestinal colonization and mucosal inflammation persists when mice are infected several weeks after withdrawal of antibiotics, correlating with subtle alterations in the intestinal microbiome involving alterations of specific bacterial groups. These results show that the colonizing microbiotas are integral to mucosal host protection, that specific features of the microbiome impact different aspects of enteric Salmonella pathogenesis, and that antibiotics can have prolonged deleterious effects on intestinal colonization resistance.
The mammalian host is colonized by trillions of microbes that live in a predominantly symbiotic relationship with their host (18, 43). The majority of these microbes inhabit the gastrointestinal (GI) tract (12, 17), and a large percentage of these microbes cannot be cultured by currently available methods, necessitating the use of molecular approaches for the identification and quantification of these organisms (40). The recent application of 16S rRNA gene sequences for the study of complex microbial ecosystems has greatly advanced the understanding of intestinal microbial ecology (2, 40). Current analyses of the intestinal microbiota suggest that the gut is colonized by more than 1,000 different bacterial species (12). The intestinal microbiotas are involved in mucosal and immunological growth and development, nutrition, and mucosal protection (14, 29, 48) and have been implicated in pathophysiology as well.
The importance of an intact biota for mucosal protection from bacterial infection has been demonstrated with animal models and the human host. Germfree animals have stunted mucosal and immune development and are highly susceptible to enteric infection (14). Recently, associations between the ability of an enteric pathogen to disrupt the microbial ecology of the gut and the ability of the pathogen to cause enteritis have been shown (4, 25, 39). In humans, treatment with broad-spectrum oral antibiotics may result in the development of Clostridium difficile infections, a common colonizer of the human gut whose growth is held in check by the normal biota but which overgrows the biota upon antibiotic use (6, 21). Many mouse models of enteritis employ the use of antibiotics to eliminate and/or perturb the indigenous biota to allow consistent enteric infection by a variety of pathogens including Salmonella enterica (5, 8, 30, 36), Vibrio cholerae (26), Escherichia coli (46, 47), and Enterococcus faecalis (45) and have demonstrated the importance of colonization resistance by an intact microbiota.
The effects of antibiotics on the intestinal microbiota have often focused on analyses of culturable bacterial species (27, 46, 47). More-recent studies using antibiotics to sterilize the gut have used culture techniques to suggest the loss of all colonizing bacteria (31). Because a large percentage of the microbiota cannot be cultured, there is a limited understanding of the impact of antibiotics on intestinal microbial ecology and the relationship between perturbation of the microbiota and susceptibility to enteric infection. We hypothesized that the ability of Salmonella to colonize the murine intestinal tract and the severity of enteritis and systemic spread would be correlated with the extent of the disruption of the protective microbiota.
In this study, we treated mice with three different regimens of antibiotics that are commonly used to disrupt the microbiota in mouse models of enteric infection and inflammation and evaluated their effect on intestinal microbial colonization by several dominant bacterial groups (35). Using the same regimens, mice were challenged with oral Salmonella infection to investigate the role of antibiotic-induced biota disruption in host susceptibility to infection. We found that antibiotics varied in their abilities to reduce total bacterial numbers in the gut, but none of the regimens tested completely eliminated the microbiota. There was also variation in the impact of antibiotic on specific dominant bacterial species in the microbiota. Unexpectedly, all the antibiotic regimens used to perturb the microbial ecosystem enhanced Salmonella colonization of the gut, mucosal inflammation, and invasion irrespective of the antibiotic regimen or its relative ability to eliminate colonizing bacteria. These findings were confirmed by antibiotic recovery experiments. Analysis of the recovery of the intestinal biome after withdrawal of antibiotics demonstrated a rapid recovery of total bacterial numbers but persistent changes in the biome composition over several weeks. The enhanced ability of Salmonella to translocate the intestinal tract diminished rapidly after antibiotic withdrawal. However, even after the biome had recovered in many aspects, including total numbers, the ratio of aerobes/anaerobes, and the abundance of several dominant bacterial groups, the mice retained their susceptibility to Salmonella colonization and enteritis. These results suggest that total numbers of bacteria comprising the microbiota contribute to limit pathogen invasion but that complete colonization resistance depends on the correct complex balance of bacterial diversity and quantity and that the use of antibiotics can have lasting deleterious effects on the capacity of the intestinal microbiome to resist infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serotype Typhimurium strain ATCC 14028 cells (ATCC) were cultured aerobically at 37°C in Luria-Bertani (LB) broth as described previously (34). Briefly, Salmonella bacteria were grown in a 10-ml standing culture for 3.5 h at 37°C to select for the most motile bacteria. The top 5 ml of the culture was transferred into 45 ml of prewarmed LB broth and grown with shaking for 1.5 h at 37°C. The quantity of Salmonella cells was determined using a Petroff-Hauser chamber (Hauser Scientific, PA), and the appropriate dose was prepared in sterile 0.2 M phosphate buffer (pH 8).
Animals.
Female FvB mice were obtained from Taconic Laboratories at 4 weeks of age. Animals were housed under specific-pathogen-free conditions in the Medical College of Wisconsin Biomedical Resource Center vivarium. The husbandry and diet for all study animals were controlled and unchanged through the course of this study to reduce the variable environmental impact on the intestinal microbiome. The animal care and use committee at the Medical College of Wisconsin approved all animal-related experiments and procedures.
Antibiotic treatment experiments.
The following antibiotics were used: streptomycin sulfate (Fisher), bacitracin (Fluka), vancomycin hydrochloride hydrate (Sigma-Aldrich), ampicillin trihydrate (Sigma-Aldrich), neomycin sulfate (Fisher), and metronidazole (Sigma-Aldrich). Four different antibiotic regimens were used to treat mice: control (no antibiotics), streptomycin (0.5 g/250 ml drinking water), streptomycin-bacitracin (0.5 g of each/250 ml drinking water), and vancomycin (0.125 g)-neomycin (0.25 g)-metronidazole (0.25 g)-ampicillin (0.25 g, combined in 250 ml water). These are drugs and combinations that have been commonly used to disrupt intestinal microbial ecology (5, 8, 31, 47). Streptomycin, used most commonly in animal infection models, is not effectively absorbed from the GI tract; streptomycin is most effective against gram-negative bacteria but also has some activity against gram-positive bacteria and Mycobacterium tuberculosis. Bacitracin is also poorly absorbed from the gut and is effective primarily against gram-positive bacteria. The four-drug regimen of ampicillin, vancomycin, neomycin, and metronidazole (AVNM) demonstrates broad-spectrum coverage. Ampicillin is moderately well absorbed from the gut but is rapidly eliminated systemically and shows activity against gram-positive and -negative aerobes and anaerobes. Metronidazole is well absorbed from the gut but is rapidly cleared from systemic circulation and shows activity against gram-positive and -negative anaerobes. Neither vancomycin nor neomycin is well absorbed by the GI tract. Vancomycin has activity against gram-positive bacteria, while neomycin has broad-spectrum activity but is particularly effective against gram-negative bacteria. At 5 weeks of age, groups of mice (five mice per group) were treated with antibiotics, as specified, in their drinking water for 7 days. Fecal pellets were obtained from the mice for aerobic and anaerobic culture. Animals were sacrificed, and intestinal tissue was taken for histology, aerobic and anaerobic culture, fluorescence in situ hybridization (FISH), total bacterial genomic isolation, and quantitative PCR (qPCR).
Antibiotic recovery experiments.
Groups of mice (five mice per group) were treated with streptomycin-bacitracin in their drinking water for 7 days as described above and then placed on regular water without antibiotics. Age-matched groups of control mice received no antibiotics in their drinking water. Groups of control animals and treated animals were sacrificed at 1, 3, 5, 7, 14, and 21 days after the cessation of antibiotic treatment. The intestinal tracts were removed, divided, and analyzed as described above. The experiment was repeated, and results were reproducible.
Bacterial culture of feces and large-intestinal contents.
Fresh stool pellets or large-intestinal tissue samples were collected from mice from each treatment group. The specimens were then homogenized in 2.0 ml of 1× phosphate-buffered saline (PBS). The homogenized samples were then plated onto LB agar for aerobic bacterial growth and onto Schaedler agar for anaerobic bacterial growth as previously described (16, 28). The LB plates were incubated at 35°C for 24 h, and photographs of the resulting growth were taken. The Schaedler plates were preincubated in a GasPak anaerobic chamber (BD Diagnostic Systems) for 18 h, and after plating, they were placed into a GasPak anaerobic chamber and incubated at 35°C for 48 h. Photographs were taken after incubation.
Salmonella infection experiments.
As in the antibiotic treatment experiments described above, groups of mice (five mice per group) were treated with antibiotics, as specified, for 7 days in their drinking water. Control mice received water without antibiotic supplementation. On day 7, the antibiotics were withdrawn from the treatment group, and all mice received untreated water. Mice were deprived of food overnight. Each group of animals was inoculated with 108 CFU of Salmonella by intragastric gavage. Mice were sacrificed 3 days postinoculation to allow a measurable translocation of Salmonella to the liver and spleen but prior to the animals becoming moribund. Intestinal tissue was isolated and processed as described above for the antibiotic experiment. Spleen and liver were removed and analyzed for the presence and abundance of translocated Salmonella cells. Salmonella burden was determined by homogenization of spleen and liver in sterile PBS and plating in dilution onto Salmonella-Shigella agar. The experiment was repeated, and results were reproducible.
Salmonella infection after antibiotic recovery.
Groups of FvB mice (five mice per group) were given untreated drinking water or drinking water containing streptomycin-bacitracin for 1 week as described above. Antibiotic-treated animals and control animals were inoculated with 108 Salmonella enterica serovar Typhimurium cells by oral gavage 1, 3, 5, 7, 14, and 21 days after antibiotic withdrawal, as described above. Three days postinfection, animals were sacrificed and analyzed as described above. The experiment was repeated, and results were reproducible.
FISH.
FISH was performed on mouse terminal ileum and cecum as described previously (4). Briefly, intestinal tissue from each mouse was fixed in Carnoy's fixative, 3-μm sections were mounted onto slides, and FISH was performed using a combination of a 6-carboxyfluorescein (FAM)-labeled oligonucleotide probe for segmented filamentous bacteria (SFB) (SFB1008 [FAM-GCGAGCTTCCCTCATTACAAGG]) (37) and a Texas Red-labeled universal bacterial probe (Bact338 [Texas Red-GCTGCCTCCGTAGGAGT]) (4) (Operon Technologies, Huntsville, AL). Slides were viewed by fluorescence microscopy using a Nikon E400 upright microscope. Images were captured using a Photometrics CoolSnap ES charge-coupled-device camera and analyzed using Metaview software (Universal Imaging Corporation, Molecular Devices). Representative sections from mice belonging to each study group were photographed.
Histology.
Three-micrometer sections of zinc formalin- or Carnoy's fixative-fixed distal small intestine (DSI), cecum, and large intestine (LI) were mounted onto slides and stained with hematoxylin and eosin. Slides were examined by an anatomic pathologist (N.H.S.) using a Nikon E400 upright microscope. Images were captured using a Spot camera and analyzed using Spot software, version 3.5.4 (Diagnostic Instruments, Inc.). Cecal pathology was scored in a blinded fashion, grading the extent of edema (0, no edema; 1, less than 50% of mucosa involved; 2, more than 50% of mucosa involved; 3, total mucosal involvement), inflammation (0, no acute inflammation; 1, focal acute inflammation in lamina propria; 2, extensive submucosal neutrophilic infiltrate; 3, transmural neutrophilic infiltrate), and hyperplasia (0, no epithelial hyperplasia; 1, twofold increase in thickness; 2, threefold increase in thickness; 3, fourfold or greater increase in thickness). Cecal sections from each mouse (five mice per group) were scored for each criterion and combined for a total enteritis score.
Bacterial genomic DNA extraction.
The DSI, cecum, and LI isolated from the experimental animals were weighed and then homogenized using a Polytron PT 10-35 homogenizer (Kinematica Switzerland) in 2 ml sterile PBS. Bacterial genomic DNA was extracted from the DSI, cecum, and LI using the Qiagen stool kit according to the kit directions, using the optional high-temperature step.
Quantitative real-time PCR amplification of 16S rRNA gene sequences.
The abundances of specific intestinal bacterial groups were measured by qPCR using the MyiQ single-color real-time PCR detection system (Bio-Rad, Hercules, CA) using group-specific 16S rRNA gene primers (Operon Technologies, Huntsville, AL) (Table 1), as previously described (4). Briefly, the real-time PCR, done using IQ SYBR green supermix (Bio-Rad), started with an initial step at 95°C for 3 min, followed by 40 cycles of 10 s at 95°C and 45 s at 63°C. Data were acquired in the final step at 63°C. Using the same genomic DNA from each sample, real-time PCRs were completed using group-specific primers to determine the amount of bacteria in each of the following major groups: the Eubacterium rectale-Clostridium coccoides group, Lactobacillus sp., Bacteroides sp., mouse intestinal Bacteroides (MIB), SFB, Enterobacteriaceae, Salmonella enterica, and total bacteria (eubacteria) (Table 1). Bacterial numbers were determined using standard curves constructed with reference bacteria specific for each bacterial group analyzed (Table 1). qPCR measures 16S gene copies per sample and not actual bacterial numbers or CFU. Nevertheless, these values are directly related and correlate well (4).
TABLE 1.
16S rRNA gene group-specific and kingdom-specific primers for qPCR
| Group | Reference strain | Primer | Sequence (5′-3′) | Temp (°C) at last step | Reference |
|---|---|---|---|---|---|
| Eubacteria (All bacteria) | Ruminococcus productus (ATCC 27340D) | UniF340 | ACTCCTACGGGAGGCAGCAGT | 63 | 1 |
| UniR514 | ATTACCGCGGCTGCTGGC | 63 | |||
| Eubacterium rectale-Clostridium coccoides | R. productus (ATCC 27340D) | UniF338 | ACTCCTACGGGAGGCAGC | 60 | 13 |
| C.cocR491 | GCTTCTTTAGTCAGGTACCGTCAT | 60 | |||
| Lactobacillius/Lactococcus | Lactobacillus acidophilus (ATCC 4357D) | LabF362 | AGCAGTAGGGAATCTTCCA | 56 | 33 |
| LabR677 | CACCGCTACACATGGAG | 56 | |||
| Bacteroides | Bacteroides fragilis (ATCC 25285D) | BactF285 | GGTTCTGAGAGGAGGTCCC | 61 | 11 |
| UniR338 | GCTGCCTCCCGTAGGAGT | 61 | |||
| MIB | MIB plasmid CT11-6 | Uni516F | CCAGCAGCCGCGGTAATA | 58 | 35 |
| MIBR677 | CGCATTCCGCATACTTCTC | 58 | |||
| SFB | SFB plasmid CTL5-6 | SFB736F | GACGCTGAGGCATGAGAGCAT | 58 | 37 |
| SFB844R | GACGGCACGGATTGTTATTCA | 58 | |||
| Salmonella enterica serovar Typhimurium | S. Typhimurium (ATCC 700720-D) | Sal454 | TGTTGTGGTTAATAACCGCA | 56 | 24 |
| Uni785R | GACTACCAGGGTATCTAATCC | 56 | 3 | ||
| Enterobacteriaceae | E. coli (ATCC 10798D) | 515F | GTGCCAGCMGCCGCGGTAA | 67 | 23 |
| 826R | GCCTCAAGGGCACAACCTCCAAG | 67 |
Bacterial RNA isolation and reverse transcription (RT)-qPCR.
Fresh intestinal samples were isolated from mice and snap-frozen in liquid nitrogen. Bacterial RNA was isolated using the RNeasy Plus RNA isolation kit (Qiagen). First-strand synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA was then analyzed using the qPCR procedures described above.
Statistical analysis of data.
Analyses of changes in total bacterial number and total Salmonella colonization were performed using a two-way analysis of variance, with a P value of <0.05 being considered significant. Analyses of Salmonella invasion experiments were performed using a Wilcoxon rank-sum test followed by a Tukey's multiple-comparison test, with a P value of <0.05 being considered significant. Analysis of histological scoring of enteritis was performed using a Student's t test, with a P value of <0.05 being considered significant. Analysis of changes of specific bacterial groups in response to antibiotic treatment was done as follows. For each organ, a global mixed-effects model was fitted to the log counts; the response was allowed to vary by bacterial class and treatment group, and the within-animal correlation was modeled by an unstructured correlation matrix. A Bonferroni correction was used within each organ to control for multiple testing. The pairwise comparisons of treatments within each bacterial class were protected by overall F tests.
RESULTS
Antibiotic treatment alters but does not eliminate the intestinal microbiota.
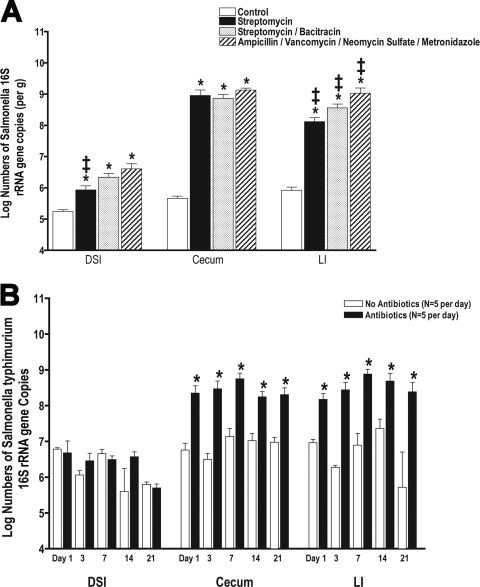
Antibiotic regimens (streptomycin, streptomycin-bacitracin, and AVNM) were selected because of their activities against bacterial groups common to the intestinal microbiota and their frequent use in animal models of intestinal infection and inflammation. Aerobic and anaerobic culturings of feces after antibiotic treatment showed parallel results, with significant numbers of culturable aerobes and anaerobes remaining in the feces of mice treated with streptomycin. However, the combination of streptomycin with bacitracin or the AVNM regimen resulted in the complete or near-complete elimination of culturable bacteria (not shown). Gross examination of the GI tract revealed characteristic findings, with notable swelling of the cecum ascribed to an accumulation of undigested mucus secondary to the reduction in levels of colonizing bacteria (not shown) (5). The impact of antibiotic treatment on DSI bacteria was minimal, and only the AVNM regimen resulted in significant losses represented by less than 1 log of total bacteria. The effect of antibiotics on the cecum and LI was more profound and varied depending on the specific antibiotic regimen (Fig. 1A), with total bacterial losses ranging from 1 to 3 logs. Quantitative studies using qPCR demonstrated that streptomycin had the most modest effects on the total biota (1-log loss), while the combination of streptomycin-bacitracin or AVNM had a similarly greater effect (Fig. 1A).
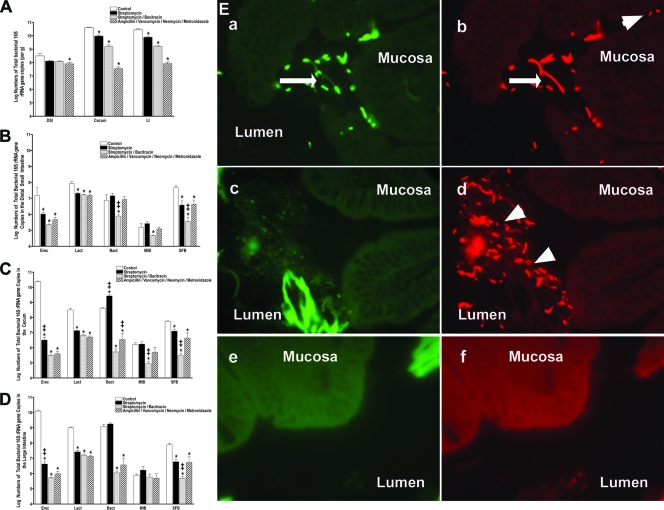
FIG. 1.
Impact of antibiotic regimens on intestinal bacterial colonization. Five-week-old female mice were given antibiotics, as indicated, in their drinking water for 7 days (n = 5 mice per group). Total bacterial genomic DNA was isolated from the DSI, cecum, and LI of each mouse and analyzed by qPCR for total bacterial numbers (A) and the abundance of specific bacterial groups (per gram) in the in the DSI (B), cecum (C), and LI (D). In the DSI, cecum, and LI, antibiotic treatment groups are all significantly different from untreated controls for E. rectale-C. coccoides (Erec), Lactobacillus (Lact), and SFB (P < 0.05). * indicates statistically significant differences from the control mice (P < 0.05); ‡ indicates statistically significant differences from all other treatment groups (P < 0.05). (E) FISH using a mixture of a Texas Red universal bacterial probe to show all bacteria (b, d, and f) and a FAM-SFB-specific bacterial probe to show SFB (a, c, and e) was performed on the terminal ileum of control (a and b), streptomycin-treated (c and d), and streptomycin-bacitracin-treated (e and f) mice and visualized by fluorescence microscopy. These images are representative of each animal studied (n = 5 mice per group). Arrows point to SFB. Arrowheads point to non-SFB commensal bacteria. The mucosal surface and lumen are labeled.
Alterations in microbiota composition are antibiotic and site specific.
Not only do variations in antibiotic treatment result in distinct changes in total bacterial numbers, with losses ranging from 0.5 to 3 logs of bacteria, the different antibiotic regimens also had distinct effects on the dominant bacterial groups present in the intestinal tract. Characterization of the intestinal microbiome in control mice demonstrates that the microbiota composition is site dependent with respect to the abundance of dominant bacterial groups. The colonization of the small intestine differs significantly from that of the cecum and distal colon. While present at a much lower abundance in the DSI, the most prominent bacterial group present in the cecum and LI is the E. rectale-C. coccoides group. The E. rectale-C. coccoides group is a member of the firmicute class of bacteria, and in the cecum and LI, it outnumbers all the other intestinal bacterial groups combined by 10- to 100-fold (Fig. 1B to D). All antibiotics tested had significant effects on this bacterial group in all segments of the GI tract, and losses of this group account for the majority of bacteria eliminated by antibiotics. The antibiotic effects on specific groups were consistent throughout the gut (Fig. 1B to D). For example, streptomycin-bacitracin had profound effects on the E. rectale-C. coccoides group, SFB, and Bacteroides in all segments of the gut, while streptomycin alone had a minimal impact on Bacteroides bacteria in any part of the gut. In addition to the E. rectale-C. coccoides group, SFB bacteria were also notably sensitive to antibiotic treatment. The SFB bacterial group was previously identified in a variety of animal hosts including mice (35), rabbits (15), and chickens (22). In the small intestine, SFB can be found to be in direct contact with the epithelium, where it has been ascribed a protective role in the prevention of enteric infection (15). While SFB are present and adherent to the DSI epithelium in untreated mice, all of the antibiotic regimens used eliminated SFB from direct contact with the DSI epithelium (Fig. 1E).
Incomplete recovery of the intestinal microbiome after antibiotic treatment.
The combination of streptomycin and bacitracin, while not absorbed systemically, is effective at eliminating a large percentage of the bacteria colonizing the intestinal tract. Mice were treated with this antibiotic regimen, and the recovery of their intestinal microbiome was monitored over 21 days by aerobic and anaerobic culture and qPCR. As noted above, prior to the withdrawal of antibiotics (day 0), neither aerobes (Fig. 2A) nor anaerobes (Fig. 2B) could be cultured from the LIs.
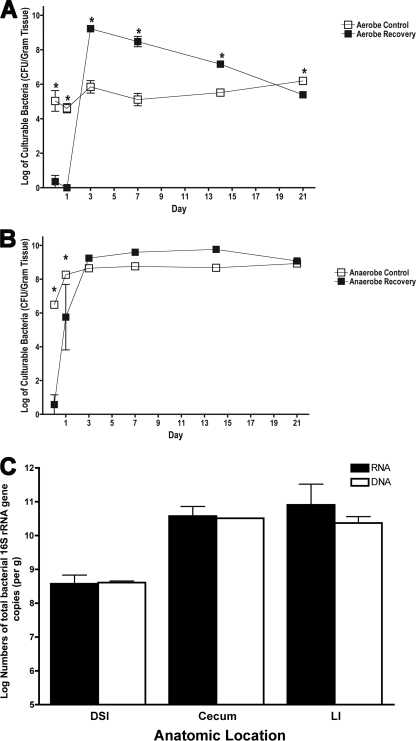
FIG. 2.
Time course of recovery of intestinal colonization. (A and B) Parallel groups of mice (n = 5 mice per group) were given untreated drinking water (open symbols) or bacitracin-streptomycin in drinking water (filled symbols) for 1 week. The antibiotics were withdrawn, and the biome was allowed to recover for 1, 3, 7, 14, or 21 days. At each time point, the LI bacteria were cultured for aerobic (A) and anaerobic (B) bacteria. * indicates statistically significant differences from the control mice (P < 0.001). To determine whether qPCR from genomic DNA represented live bacteria in the GI tract, analysis and quantification of 16S copies were performed and compared for bacterial DNA and RNA. Total bacterial RNA and genomic DNA were isolated from the DSI, cecum, and LI of adult unchallenged mice (n = 3). (C) Total 16S gene copies were quantified from genomic DNA (white bars) by qPCR and from RNA (black bars) by RT-qPCR.
Despite the elimination of culturable bacteria, the antibiotic treatments did not eliminate bacteria, as determined by quantification by molecular methods (Fig. 1A and 3A). In addition, the abundance of culturable bacteria from the LIs of untreated mice (Fig. 2A and B) is at least 1 log less than that measured by qPCR. The discordance between the quantity of culturable bacteria and that measured by molecular methods is consistent with previously reported observations, which have demonstrated that the majority of bacteria present in the GI tract are not culturable by available methods (40). Other aspects that must be considered include the fact that qPCR measures 16S gene copy numbers, which vary between bacterial species. Previous work suggested that qPCR measurements and culture measurements correlate well when individual species are measured (4). One additional concern is whether the difference is due to picking up dead bacteria by 16S qPCR. To address this concern, bacterial RNA was isolated from selected parallel samples and analyzed by RT-qPCR. Bacterial RNA is less stable than bacterial DNA and provides a good measure of live bacteria. The total bacterial abundance was unchanged when measured by this approach (Fig. 2C).
FIG. 3.
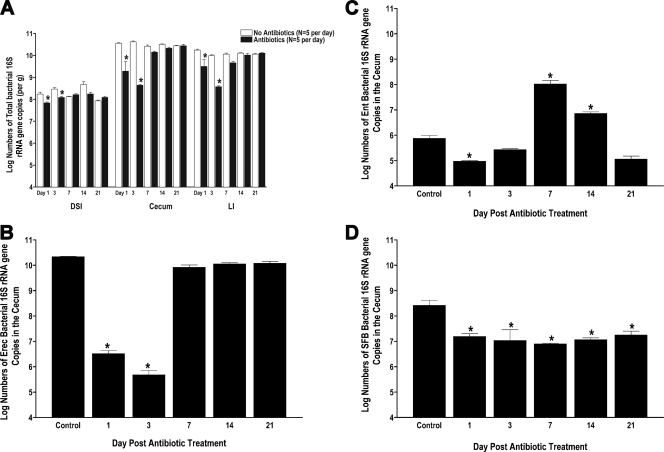
Incomplete recovery of the intestinal microbiome after antibiotic treatment. Parallel groups of mice (n = 5 mice per group) were given untreated drinking water or bacitracin-streptomycin in drinking water for 1 week. The antibiotics were withdrawn, and the biome was allowed to recover for 1, 3, 7, 14, or 21 days. Total bacterial genomic DNA was isolated from the DSI, cecum, and LI of each mouse. qPCR was performed to quantify numbers of total bacteria (A), the E. rectale-C. coccoides group (Erec) (B), the Enterobacteriaceae (Ent) (C), and SFB (D) per gram. * indicates statistically significant differences from control mice (P < 0.001).
Aerobic bacteria could not be detected 1 day after withdrawal of antibiotics but showed dramatic increases by 3 days, overgrowing the normal aerobic balance by 3 logs (Fig. 2A). This aerobic overgrowth reversed slowly and ultimately dropped slightly below levels seen in untreated mice after 3 weeks. The anaerobic bacteria showed some recovery 1 day after antibiotic withdrawal and then slightly exceeded the numbers found in untreated mice but not significantly (Fig. 2B). By 3 days after antibiotic withdrawal, the numbers of anaerobic bacteria found in antibiotic-treated and control mice were not statistically different.
Total bacterial numbers determined by qPCR were also significantly decreased compared to those for controls at 1 and 3 days after withdrawal of antibiotics (Fig. 3A). After 7 days, total numbers of bacteria had recovered (Fig. 3A), in parallel with the recovery of the dominant E. rectale-C. coccoides bacterial group (Fig. 3B). Unlike the E. rectale-C. coccoides group, the Enterobacteriaceae are present in very low numbers in the murine gut. This group expands within 7 days after antibiotic treatment and persists at elevated levels, but by 3 weeks, it is reduced back to baseline levels (Fig. 3C). Other bacterial groups examined do not show complete recovery as quickly. Bacteroides groups are highly variable during recovery from antibiotics (not shown); however, SFB levels stay consistently low and had not recovered after 3 weeks (Fig. 3D). Although the abundance and distribution of bacterial groups vary between the DSI and the rest of the lower GI tract, the recovery of specific bacterial groups is temporally consistent throughout each section of the intestine. For example, levels of the Enterobacteriaceae peak on day 7 (Fig. 3C and not shown) and return to baseline levels by day 21 in all segments of the GI tract. Overall, despite the fact that recovery is not complete after 3 weeks, the intestinal microbiota composition gravitated back toward that of untreated mice over time.
Salmonella invasion correlated with microbial abundance and recovery of the E. rectale-C. coccoides population.
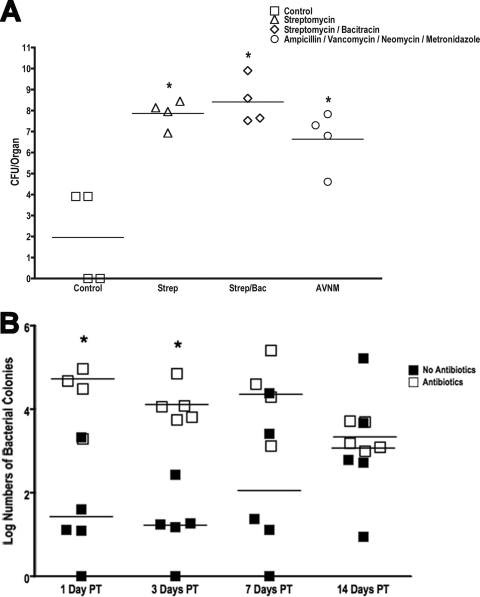
To determine whether the extent of changes in the microbiota increased host susceptibility to bacterial translocation, mice treated with either streptomycin, streptomycin-bacitracin, or AVNM were challenged with Salmonella. Salmonella burden in the livers and spleens of infected mice was analyzed. Mice not treated with antibiotics showed low-level Salmonella translocation into the spleen and liver 3 days postinfection (Fig. 4A), while pretreatment with antibiotic regimens resulted in significantly increased levels of invasion of Salmonella from the lumen to the systemic circulation, as shown previously with streptomycin treatment (30). The extent of invasion varied between antibiotic treatments but not significantly. Surprisingly, pretreatment with streptomycin-bacitracin or AVNM, although effective at eliminating large numbers of colonizing bacteria, did not promote significantly more Salmonella invasion than did streptomycin treatment alone (Fig. 4A). While this suggests that the absolute quantity of biota loss did not correlate with susceptibility to pathogen translocation, it is possible that there is a dose-dependent relationship between bacterial abundance and translocation (36) but that even the mildest antibiotic treatment used in this study was too high a dose to detect dose dependence. Correlation is evident between Salmonella translocation and the loss of several firmicute groups in every section of the GI tract, including the E. rectale-C. coccoides group, Lactobacillus sp., and SFB. Following the above-described time course for the recovery of the intestinal microbiome, antibiotics were withdrawn; groups of mice were allowed to recover for 1, 3, 7, 14, and 21 days before being orally challenged with Salmonella; and indices of infection were compared to those of non-antibiotic-treated control groups. Increased susceptibility to pathogen translocation disappeared by 7 days after antibiotic withdrawal (Fig. 4B), in direct relation to the recovery of both the E. rectale-C. coccoides (Fig. 3B) and Lactobacillus (not shown) groups. Since the recovery of the E. rectale-C. coccoides group resulted in the recovery of total bacterial numbers, it is also possible that the restoration of total bacterial abundance rather than the specific bacterial group is the critical factor in the prevention of bacterial translocation.
FIG. 4.
Effect of antibiotic regimens on Salmonella translocation. Parallel groups of mice (n = 4 mice per group) were given untreated drinking water or the indicated regimens of antibiotics for 1 week. The antibiotics were withdrawn, and the mice were inoculated with Salmonella (108 CFU/mouse) by intragastric gavage and sacrificed after 3 days. (A) The livers were isolated, homogenized, and analyzed by dilution plating onto selective agar to determine the bacterial burden and extent of translocation. Each symbol represents one mouse. The horizontal bars indicate the means of data from the four mice per group (P = 0.0055 by Wilcoxon rank-sum test). Data for all antibiotic treatment groups are significantly different from those of the controls. Tukey's multiple comparison shows that the values for antibiotic treatment groups are significantly greater than those for the control group, but they are not significantly different from each other. (B) Parallel groups of mice (n = 5 mice per group) were given untreated drinking water or bacitracin-streptomycin (Strep/Bac) in drinking water for 1 week. The antibiotics were withdrawn, and the biome was allowed to recover for 1, 3, 7, 14, or 21 days before oral infection with 108 CFU of Salmonella (open squares). Mice that were not treated with antibiotics were used for control infections (closed squares). Salmonella translocation into the liver (per gram) was determined 3 days postinoculation by dilution plating onto selective agar. Each symbol represents one mouse. The horizontal bar indicates the mean data from five mice per group. * indicates statistically significant differences (P < 0.05). PT, posttreatment.
Antibiotic disruption of the intestinal microbiota results in increased Salmonella colonization.
After antibiotic treatment, mice were given Salmonella by oral gavage, and the intestinal microbiota was examined for Salmonella abundance in the intestinal lumen (Fig. 5A). The level of Salmonella colonization of control mice was relatively low, consistent with previously reported findings (4). Antibiotic treatment of the mice resulted in increased levels of Salmonella colonization compared to that for control mice (Fig. 5A). Although the change in total bacterial quantity varied significantly between antibiotic treatments, the intestinal Salmonella burden was consistent between treatment groups, at approximately 106 16S Salmonella gene copies per gram of intestine in the DSI and 109 16S Salmonella gene copies per gram in the cecum and LI, demonstrating that there is no direct relationship between the extent of biota loss and the abundance of Salmonella colonization. As previously noted (4), during an enteric infection, Salmonella does not outcompete or replace the native biota. This suggests that the change in the total bacterial numbers may not be the most important factor in Salmonella colonization, but it is also possible that the antibiotic treatments used exceeded the range where this could be determined. To distinguish between these possibilities, intestinal colonization by Salmonella was examined with mice challenged during recovery from antibiotics. Three weeks after antibiotic withdrawal, several of the parameters of the intestinal microbiome showed recovery. This recovery included recovery of total bacterial numbers, several dominant bacterial groups, and the anaerobic population and near recovery of the aerobic population. Nevertheless, animals challenged orally with Salmonella remained susceptible to increased pathogen colonization of the intestinal tract (Fig. 5B) and again appeared to colonize to a fixed set point of 109 16S gene copies per gram in the cecum and LI, representing approximately 1% of the total bacterial population at these sites.
FIG. 5.
Antibiotic treatment enhances Salmonella colonization of the intestinal tract. (A) Mice (n = 5 mice per group) were treated with different regimens of antibiotics for 1 week, followed by Salmonella inoculation by intragastric gavage, and sacrificed after 3 days. Total bacterial genomic DNA was isolated from the DSI, cecum, and LI and analyzed for total Salmonella burden by qPCR. * indicates statistically significant differences from control mice (P < 0.05); ‡ indicates statistically significant differences from all other treatment groups (P < 0.05). (B) Untreated control mice (white bars) and antibiotic-treated mice (black bars) were allowed to recover from treatment with bacitracin-streptomycin for 1, 3, 7, 14, or 21 days prior to challenge with Salmonella by oral gavage. Mice were sacrificed after 3 days, and total bacterial genomic DNA was isolated from each segment of the GI tract and analyzed for total Salmonella burden (per gram) by qPCR (n = 5 mice per group). * indicates statistically significant differences from control mice (P < 0.05). In the cecum and LI, there were no statistically significant differences in Salmonella colonization among antibiotic-treated mice at any time point.
Alteration of the intestinal biota results in increased intestinal mucosal inflammation.
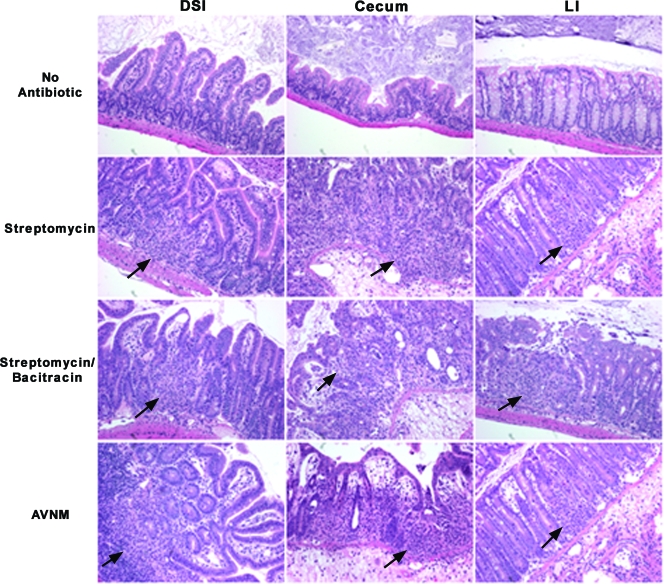
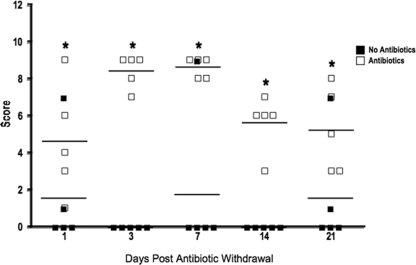
Previous work by our laboratory (4) demonstrated that Salmonella infection of mice not treated with antibiotics results in acute inflammation of the DSI. Work by other laboratories has shown that streptomycin treatment followed by Salmonella infection results in profound mucosal inflammation in the ceca of mice (5). To determine the effect of microbiota disruption on the development of intestinal inflammation, the intestinal tissues of antibiotic-treated Salmonella-infected mice were analyzed. In control mice, low-level focal inflammation was noted in the DSI (4), cecum, and LI (Fig. 6). Antibiotic treatment increased inflammation minimally in the DSI, but significantly more inflammation was observed in the cecum and LI (Fig. 6). This is consistent with previous work by Barthel et al. in the characterization of a streptomycin pretreatment mouse model for Salmonella colitis (5). Cecal inflammation involved the entire mucosal surface and was characterized by edema, extensive neutrophil influx into the lamina propria and the lumen with evidence of crypt abscesses and crypt destruction, and mucosal erosion and ulceration (Fig. 6). Again, although the antibiotics had diverse effects on the intestinal biota, the extent of these changes was not directly related to the extent of inflammation. There were minimal differences between the levels of inflammation in streptomycin-treated mice (mild changes in the biota) and those in AVNM-treated mice (profound changes in the biota). Again, this finding was confirmed by antibiotic recovery experiments. Similar to the colonization results, the extent of cecal inflammation in response to Salmonella infection remains statistically greater in mice treated by antibiotics than in non-antibiotic-treated controls even when mice are allowed to recover from antibiotic treatment for 3 weeks before oral Salmonella infection (Fig. 7).
FIG. 6.
Antibiotic pretreatment enhances the development of acute colitis. Mice (n = 5 mice per group) were treated with different regimens of antibiotics for 1 week, followed by Salmonella inoculation by oral gavage, and were sacrificed after 3 days. Hematoxylin- and eosin-stained sections of intestinal tissue from the DSI, cecum, and LI of representative mice were examined for evidence of inflammation and colitis. All images were taken at a ×20 magnification. Arrows point to areas of inflammation with neutrophil infiltrates.
FIG. 7.
Persistent susceptibility to colitis during antibiotic recovery. Mice (n = 5 mice per group) were allowed to recover from treatment with bacitracin-streptomycin (open squares) for 1, 3, 7, 14, or 21 days prior to challenge with Salmonella by oral gavage and were sacrificed after 3 days. Control mice were not pretreated with antibiotics (closed squares). Hematoxylin- and eosin-stained sections of intestinal tissue from the DSI, cecum, and LI of each mouse (n = 5) were examined for evidence of inflammation and colitis. Cecal sections were blindly scored using histological criteria for the extent of epithelial hyperplasia, edema, and acute inflammation.
DISCUSSION
Antibiotic use carries both benefit and risk. Antibiotic therapy is critical for the treatment of life-threatening infection, but misuse of antibiotics leads to the development of antibiotic resistance in common pathogens. Even routine and appropriate use of antibiotics may have a detrimental impact on the host microbial ecosystem, which is important for host mucosal protection (10, 21). In this study, we investigated how the use of oral antibiotics perturbs the intestinal microbial ecosystem in mice and the impact of that disruption on host susceptibility to a common enteric pathogen, Salmonella enterica serovar Typhimurium.
Using molecular methods to identify and quantify bacterial numbers, a more-complete understanding of the impact of antibiotics on the intestinal microbial ecosystem can be determined. While antibiotic treatment resulted in the loss of culturable bacteria, none of the antibiotic combinations tested was able to sterilize the gut. Since antibiotic treatment was limited to 1 week, it is reasonable to consider that the treatments would not be able to eliminate the slower-growing organisms. Nevertheless, complete elimination of the bacterial component of the intestinal microbiota by antibiotics is difficult to achieve and cannot be determined by culture-based methods. The various antibiotic regimens used resulted in changes in the abundance and composition of the intestinal microbiome that were antibiotic specific. The firmicute class of bacteria (including the E. rectale-C. coccoides group, Lactobacillus sp., and SFB) appears to be the most susceptible to all of the antibiotics used. This class of bacteria is also the most susceptible to disruption by diarrheal illness (4).
After antibiotic treatment, the intestinal biome gravitates over time to that of untreated mice. Using bacterial culture analysis, after the initial elimination of culturable bacteria, anaerobes recover to the levels found in untreated mice within 3 days. Levels of aerobic bacteria expand dramatically but by 3 weeks closely approximate the levels found in untreated mice. Molecular analysis reveals that total bacterial numbers rapidly recover, driven by the swift recovery of the most dominant bacterial group, the E. rectale-C. coccoides group, members of the Firmicutes. Another member of the Firmicutes, the lactobacilli, also rapidly recovers, while other groups represented at a lower abundance, such as SFB, do not.
If increased susceptibility to Salmonella infection was correlated with the quantity of colonizing bacteria eliminated by antibiotics, one would predict that streptomycin-treated mice would show less translocation and intestinal colonization by Salmonella and milder enteritis than would streptomycin-bacitracin- or AVNM-treated mice, but this was not evident in this study. Oral Salmonella challenge of antibiotic-treated mice resulted in comparable increases in intestinal Salmonella colonization, enteritis, and invasion irrespective of the antibiotic combinations used. Antibiotic recovery experiments allowed a more-complete dissection of specific aspects of Salmonella enteritis, including susceptibility to pathogen invasion, intestinal colonization, and mucosal inflammation.
Interestingly, over the time course of biome recovery from antibiotics, mice regained resistance to Salmonella translocation rapidly, but susceptibility to increased Salmonella colonization and local mucosal inflammation persisted. This suggests that invasion is mediated by different pathogen-commensal-host interactions than colonization. The prevention of Salmonella translocation is associated with a recovery of total bacterial numbers, which is driven by the recovery of the dominant firmicute populations (the E. rectale-C. coccoides group and Lactobacillus sp.).
The presence of an intact commensal biota could be preventing Salmonella invasion by effectively competing with Salmonella for attachment sites and nutrition, reducing the numbers of luminal Salmonella cells available for invasion. This would account for the association between biota recovery and resistance to translocation. However, commensal interactions with the host may also have an important role in the prevention of translocation. Several innate antimicrobial effectors produced by the intestinal epithelium are induced by intestinal colonization, including angiogenin 4 (19), RegIIIγ (9), and intestinal alkaline phosphatase (7), all of which have dual roles in intestinal homeostasis and host defense. The reduction in levels of intestinal bacteria by antibiotic treatment can result in decreased levels of expression of these effectors (44), allowing increased pathogen translocation.
Specific bacterial species may also play a role in host protection. SFB, for example, are highly susceptible to both exogenous antibiotics, as shown here, and endogenous antimicrobial peptide activity (our unpublished observations). This organism is both immune stimulatory (38, 42) and highly immune responsive (20), with increased numbers found in immunoglobulin A-deficient mice (41). The absence of SFB has been associated with increased susceptibility to enteric pathogens (15) and enteritis. While SFB abundance does not correlate with pathogen translocation, it may be critical for the increased susceptibility of animals to Salmonella colonization and enteritis. It is possible that SFB have a directly protective effect by interacting with Salmonella and preventing pathogen interactions with the mucosa. SFB may also be acting through the stimulation of the mucosally associated lymphoid tissue, resulting in more-effective mucosal host responses to the invading pathogen. Additional work, using gnotobiotic mice and controlling for the presence of this unculturable bacterial group, is needed to address this issue.
The finding that both extremely minimal disruptions in the intestinal biome, like those present 3 weeks after antibiotic withdrawal, and extremely profound disruption, such as that with AVNM treatment, result in similar luminal colonization by Salmonella was both unexpected and intriguing. It is unlikely that the intestinal biota is responsible for restricting the growth of Salmonella in these circumstances since it varies so profoundly between treatments. One explanation is that the limitation in Salmonella colonization is a result of innate host immune responses. Another possibility is that this is a colonization set point due to quorum sensing by Salmonella. Additional work will be required to distinguish these processes and determine whether this is specific to Salmonella or common to other enteric pathogens.
It is evident that the intestinal microbial ecosystem serves an important but incompletely defined role in mucosal protection. In this study, we have demonstrated that although antibiotics cannot sterilize the intestinal tract, they can have a profound impact on intestinal colonization. The murine intestinal microbiome recovers from antibiotic disruption to closely recapitulate that of untreated mice, supporting the hypothesis that attributes of the host select for a core microbiome, as previously suggested using germfree models (32). Despite the rapid recovery of several measurable parameters of the biome, residual subtle alterations in bacterial composition can persist and result in profoundly enhanced susceptibility to bacterial enteritis. In conclusion, the host drives the selection of a core biome in which bacterial quantity and composition contribute to intestinal colonization resistance. Minimal disruption of this complex balance by antibiotics can result in prolonged harmful effects on the ability of the host to resist infection.
Acknowledgments
We thank Jessica Martiny, Kevin Nallon, and Rebecca Bark for technical assistance and Lynn Gruman for assistance and advice with histology. We thank Aniko Szabo and Scott Jackson, Division of Biostatistics, the Medical College of Wisconsin, for statistical analysis of the data. We thank Joe Barbieri for helpful discussions and critically reading the manuscript.
This work was supported by Public Health Service grant AI057757 (N.H.S.) from the National Institutes of Health.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 20 April 2009.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 561919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 3071915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55541-555. [DOI] [PubMed] [Google Scholar]
- 4.Barman, M., D. Unold, K. Shifley, E. Amir, K. Hung, N. Bos, and N. Salzman. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett, J. G., N. Moon, T. W. Chang, N. Taylor, and A. B. Onderdonk. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75778-782. [PubMed] [Google Scholar]
- 7.Bates, J. M., J. Akerlund, E. Mittge, and K. Guillemin. 2007. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohnhoff, M., and C. P. Miller. 1962. Enhanced susceptibility to Salmonella infection in streptomycin treated mice. J. Infect. Dis. 111117-127. [DOI] [PubMed] [Google Scholar]
- 9.Cash, H. L., C. V. Whitham, C. L. Behrendt, and L. V. Hooper. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 3131126-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dore, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst. Appl. Microbiol. 2165-71. [DOI] [PubMed] [Google Scholar]
- 12.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 3081635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 643336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson, B. E. 1982. The physiological importance of the colonic microflora. Scand. J. Gastroenterol. Suppl. 77117-131. [PubMed] [Google Scholar]
- 15.Heczko, U., A. Abe, and B. B. Finlay. 2000. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli 0103 in Rabbits. J. Infect. Dis. 1811027-1033. [DOI] [PubMed] [Google Scholar]
- 16.Hill, G. B. 1978. Effects of storage in an anaerobic transport system on bacteria in known polymicrobial mixtures and in clinical specimens. J. Clin. Microbiol. 8680-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 2921115-1118. [DOI] [PubMed] [Google Scholar]
- 18.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22283-307. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, L. V., T. S. Stappenbeck, C. V. Hong, and J. I. Gordon. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4269-273. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, H. Q., N. A. Bos, and J. J. Cebra. 2001. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 693611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49375-390. [DOI] [PubMed] [Google Scholar]
- 22.Klassen, H., J. Koopman, M. Van den Brink, M. Bakker, F. Poelma, and A. Beynen. 1993. Intestinal segmented filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27141-150. [DOI] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 24.Lin, C. K., and H. Y. Tsen. 1996. Use of two 16S DNA targeted oligonucleotides as PCR primers for the specific detection of Salmonella in foods. J. Appl. Bacteriol. 80659-666. [DOI] [PubMed] [Google Scholar]
- 25.Lupp, C., M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor, and B. B. Finlay. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2119-129. [DOI] [PubMed] [Google Scholar]
- 26.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induce during infection and plays a role in acid tolerance. Mol. Microbiol. 34836-849. [DOI] [PubMed] [Google Scholar]
- 27.Miller, C. P., and M. Bohnhoff. 1963. Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin treatment. J. Infect. Dis. 11359-66. [DOI] [PubMed] [Google Scholar]
- 28.Murray, P. R. 1978. Growth of clinical isolates of anaerobic bacteria on agar media: effects of media composition, storage conditions, and reduction under anaerobic conditions. J. Clin. Microbiol. 8708-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Hara, A. M., and F. Shanahan. 2006. The gut flora as a forgotten organ. EMBO Rep. 7688-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Que, J. U., and D. J. Hentges. 1985. Effect of streptomycin administration on colonization resistance in mice. Infect. Immun. 48169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118229-241. [DOI] [PubMed] [Google Scholar]
- 32.Rawls, J. F., M. A. Mahowald, R. E. Ley, and J. I. Gordon. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 971166-1177. [DOI] [PubMed] [Google Scholar]
- 34.Salzman, N. H., M. M. Chou, H. de Jong, L. Liu, E. M. Porter, and Y. Paterson. 2003. Enteric salmonella infection inhibits paneth cell antimicrobial peptide expression. Infect. Immun. 711109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman, N. H., H. de Jong, Y. Paterson, H. J. M. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 1483651-3660. [DOI] [PubMed] [Google Scholar]
- 36.Sekirov, I., N. M. Tam, M. Jogova, M. L. Robertson, Y. Li, C. Lupp, and B. B. Finlay. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 764726-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snel, J., P. P. Heinen, H. J. Blok, R. J. Carman, A. J. Duncan, P. C. Allen, and M. D. Collins. 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus.” Int. J. Syst. Bacteriol. 45780-782. [DOI] [PubMed] [Google Scholar]
- 38.Snel, J., C. C. Hermsen, H. J. Smits, N. A. Bos, W. M. C. Eling, J. J. Cebra, and P. J. Heidt. 1998. Interactions between gut-associated lymphoid tissue and colonization levels of indigenous, segmented, filamentous bacteria in the small intestine of mice. Can. J. Microbiol. 441177-1182. [DOI] [PubMed] [Google Scholar]
- 39.Stecher, B., R. Robbiani, A. W. Walker, A. M. Westendorf, M. Barthei, M. Kremer, S. Chaffron, A. J. Macpherson, J. Buer, J. Parkhill, G. Dougan, C. von Mering, and W. D. Hardt. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 654799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, K., B. Meek, Y. Doi, M. Muramatsu, T. Chiba, T. Honjo, and S. Fagarasan. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 1011981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talham, G. L., H. Q. Jiang, N. A. Bos, and J. J. Cebra. 1999. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 671992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaishnava, S., C. L. Behrendt, A. S. Ismail, L. Eckmann, and L. V. Hooper. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA 10520858-20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells, C. L., R. P. Jechorek, and S. L. Erlandsen. 1990. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J. Infect. Dis. 16282-90. [DOI] [PubMed] [Google Scholar]
- 46.Wells, C. L., M. A. Maddaus, R. P. Jechorek, and R. L. Simmons. 1988. Role of intestinal anaerobic bacteria in colonization resistance. Eur. J. Clin. Microbiol. Infect. Dis. 7107-113. [DOI] [PubMed] [Google Scholar]
- 47.Wells, C. L., M. A. Maddaus, C. M. Reynolds, R. P. Jechorek, and R. L. Simmons. 1987. Role of anaerobic flora in the translocation of aerobic and facultatively anaerobic intestinal bacteria. Infect. Immun. 552689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wostmann, B. S. 1981. The germfree animal in nutritional studies. Annu. Rev. Nutr. 1257-279. [DOI] [PubMed] [Google Scholar]