Abstract
Polo-like kinase 1 (Plk1) overexpression is associated with tumorigenesis by an unknown mechanism. Likewise, Plk1 was suggested to act as a negative regulator of tumor suppressor p53, but the mechanism remains to be determined. Herein, we have identified topoisomerase I-binding protein (Topors), a p53-binding protein, as a Plk1 target. We show that Plk1 phosphorylates Topors on Ser718 in vivo. Significantly, expression of a Plk1-unphosphorylatable Topors mutant (S718A) leads to a dramatic accumulation of p53 through inhibition of p53 degradation. Topors is an ubiquitin and small ubiquitin-like modifier ubiquitin-protein isopeptide ligase (SUMO E3) ligase. Plk1-mediated phosphorylation of Topors inhibits Topors-mediated sumoylation of p53, whereas p53 ubiquitination is enhanced, leading to p53 degradation. These results demonstrate that Plk1 modulates Topors activity in suppressing p53 function and identify a likely mechanism for the tumorigenic potential of Plk1.
Polo-like kinase-1 (Plk1)3 has multiple functions required for cell cycle progression, and overexpression of Plk1 is observed in various types of human tumors (1, 2). Thus, Plk1 has been proposed as a novel diagnostic marker for cancers. Accumulating evidence suggests that Plk1 negatively regulates the function of the tumor suppressor p53, whose loss-of-function mutations have been observed in nearly 50% of human tumors (1). In our earlier studies, we were the first to demonstrate that Plk1 depletion results in increased p53 level in HeLa cells (3) and that human cells with different levels of p53 respond to Plk1 depletion differently (4). Subsequently, it was shown that Plk1 directly binds to the DNA-binding domain of p53 through its N-terminal kinase domain and inhibits the transactivation as well as the proapoptotic function of p53 (5). Although it has been suggested that Plk1 might regulate p53 through direct phosphorylation (5), our repeated efforts to prove p53 as a direct target of Plk1 have been unsuccessful.
Topors was discovered in a screen searching for proteins that bind to DNA topoisomerase I (6) and was also identified as a p53-binding protein (7). Although Topors is widely expressed in normal human tissues, its expression is decreased or undetectable in colon, lung, and brain adenocarcinomas, indicating that it might function as a tumor suppressor (8). Topors contains an N-terminal C3HC4-type RING domain that is closely related in sequence to the RING domains of known E3 ligases (see Fig. 1A) and is the first example of a protein that has both ubiquitin and SUMO-1 E3 ligase activity. Topors functions as an E3 ubiquitin ligase for p53 and NKX3.1, and Topors-mediated ubiquitination leads to the degradation of these proteins (9, 10). Substrates of the SUMO-1 E3 ligase activity of Topors include DNA topoisomerase I and p53 (11, 12). In contrast to ubiquitination-induced protein degradation, Topors-induced p53 sumoylation is accompanied by an increase in the level of p53 protein (11). Taken together, these studies indicate that Topors functions both as an ubiquitin and as a SUMO-1 E3 ligase for p53. Therefore, it is likely that the effects of Topors on p53 depend on cellular context (10).
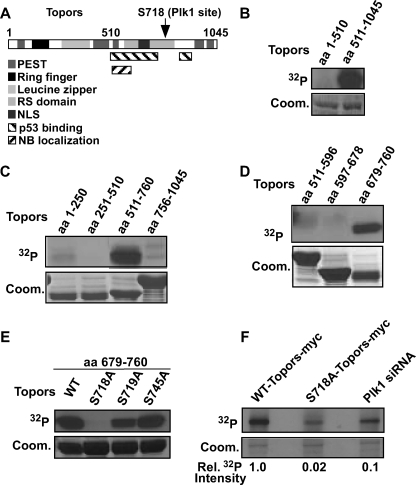
FIGURE 1.
Plk1 phosphorylates Topors at Ser718in vitro and in vivo. A, schematic representation of the domain structure of Topors. Two separate regions encoding putative p53-binding domains are aa 456–731 and 854–916. Amino acid residues in the putative Ring finger motif are shown in a black box. PEST, sequences rich in Pro, Glu, Ser, and Thr; RS domain, Arg- and Ser-rich domain; NLS, nuclear localization sequence; NB, nuclear bodies. B, purified Plk1 was incubated with purified GST-Topors (aa 1–510) or GST-Topors (aa 511–1045) for 30 min at 30 °C in the presence of [γ-32P]ATP (32P). Reaction mixtures were resolved by SDS-PAGE followed by autoradiography. Coom., Coomassie Blue. C and D, Plk1 phosphorylates Topors (aa 679–760). Purified Plk1 was incubated with purified GST-Topors fragments (aa 1–250, 251–510, 511–760, 756–1045, 511–596, 597–678, and 679–760). Kinase assays were performed as described in B. E, Ser718 of Topors is a Plk1 phosphorylation site in vitro. Purified Plk1 was incubated with the indicated serine to alanine Topors (aa 679–760) mutants and analyzed as in B. F, Topors is phosphorylated in vivo at Ser718 by Plk1. HEK293T cells were transfected with WT-Topors-Myc (lanes 1 and 3) or S718A-Topors-Myc (lane 2) and depleted of Plk1 by using double-stranded RNA targeting Plk1 (lane 3). After overnight incubation, cells were treated with nocodazole for 10 h and metabolically labeled with [32P]orthophosphate. Phosphoproteins were immunoprecipitated with anti-Myc antibodies, resolved by SDS-PAGE, and subjected to autoradiography. Relative 32P (Rel. 32P) incorporations of Topors are indicated on the bottom.
In this study, we provide evidence that Plk1 phosphorylates Topors on Ser718. Significantly, we demonstrate that the Plk1-mediated phosphorylation of Topors results in reduced sumoylation of p53, whereas the ubiquitination activity toward p53 is increased, thereby facilitating p53 degradation.
EXPERIMENTAL PROCEDURES
Cell culture, transfections, kinase assays, immunoblotting, and metabolic labeling are described in the supplemental Experimental Procedures.
RESULTS
Physical Interaction of Plk1 with Topors
Plk1 expression is elevated in various human tumors including hepatitis B virus-mediated hepatocellular carcinoma (13). The X protein (pX) of hepatitis B virus is implicated in hepatocellular carcinoma development (14). Recent studies have demonstrated the Plk1 is involved in pX-induced hepatocyte transformation using an in vitro pX-expressing cellular model (Studach et al. (28)). Furthermore, Topors was identified as a gene whose depletion via a small interfering RNA library screen rescued pX-expressing cells from DNA damage-induced apoptosis.4 Based on these findings, we explored the possibility of potential direct connection between Plk1 and Topors.
To test whether Topors is a binding partner of Plk1, a co-immunoprecipitation experiment was performed. As shown in supplemental Fig. 1A, the Plk1 protein was specifically pulled down by the anti-GFP antibody from lysates of cells expressing GFP-Topors, indicating an association between Topors and Plk1. To examine the interaction between the relevant endogenous proteins, HeLa cells were treated with hydroxyurea or nocodazole, and total cell lysates were subjected to anti-Topors IP followed by anti-Plk1 Western blot. As shown in supplemental Fig. 1B, Plk1 co-immunoprecipitates with Topors using lysates from cells synchronized in S and M phases.
Plk1 Phosphorylates Topors-Ser718 Both in Vitro and in Vivo
Considering the in vivo interaction between Topors and Plk1, we investigated whether Plk1 phosphorylates Topors. We performed in vitro Plk1 kinase assays, employing affinity-purified GST-Plk1 expressed in baculovirus-infected Hi5 insect cells and affinity-purified GST-Topors, the N-terminal (amino acids 1–510) or C-terminal fragments (aa 511–1045), in the presence of [γ-32P]ATP. As shown in Fig. 1B, C-terminal Topors, but not N-terminal Topors, was phosphorylated by Plk1 efficiently, indicating that C-terminal Topors is a Plk1 substrate.
To identify the phosphorylation site(s), four shorter Topors fragments (aa 1–250, 251–510, 511–760, and 756–1045) were purified and subjected to Plk1 kinase reactions. Only the fragment containing amino acids 511–760 yielded a strong phosphorylation signal (Fig. 1C). Moreover, a similar experiment with three additional non-overlapping Topors fragments (aa 511–596, 597–678, and 679–760) revealed that that only the fragment spanning amino acids 679–760 was phosphorylated by Plk1 (Fig. 1D). Next, to map the potential phosphorylation site(s) for Plk1, we mutated every serine/threonine in the fragment spanning aa 679–760 into alanine. When compared with the phosphorylation level of WT-Topors (aa 679–760), the phosphorylation level of S718A-Topors was completely abolished, indicating that Ser718 is the phosphorylation site for Plk1 (Fig. 1E). Significantly, the sequence context of the Plk1 phosphorylation site that we have mapped in Topors is 717ESSY720. Earlier studies by others have determined that Plk1 prefers a negatively charged residue in the nearby sequence (15–17). Thus, the sequence context of the Plk1 phosphorylation site in Topors (717ESSY720) matches this rule.
To examine whether Plk1-dependent phosphorylation of Topors occurs in vivo, metabolic labeling experiments were performed. HEK293T cells were transfected with Myc-Topors (WT or S718A), retransfected with double-stranded RNA to deplete Plk1, treated with nocodazole, and metabolically labeled with [32P]orthophosphate. Topors was immunoprecipitated with an anti-Myc antibody, and the level of Topors phosphorylation was determined (Fig. 1F). The mutation S718A significantly reduced Topors phosphorylation, suggesting that Ser718 is the major site phosphorylated in vivo under these conditions. Moreover, depletion of Plk1 also reduced the phosphorylation of Topors, indicating that Plk1 does phosphorylate Topors in vivo (Fig. 1F). Moreover, the C-terminal fragment of Topors was expressed and analyzed in vivo. As shown in supplemental Fig. 2, overexpressed GFP-Topors, both in WT and in the S718A mutant, localizes in punctate nuclear regions associated with promyelocytic leukemia nuclear bodies (18). In contrast, GFP-Topors-aa 679–760, both in the WT and in the S718A mutant, is detected throughout the cell with the same localization pattern as GFP (supplemental Fig. 2). Metabolic labeling experiments indicated that this fragment of Topors is not phosphorylated in vivo (supplemental Fig. 2B), likely due to the mislocalization of the protein fragment (supplemental Fig. 2A).
Plk1-dependent Phosphorylation of Topors Negatively Regulates the Protein Level of p53
To explore the role of Plk1-mediated phosphorylation of Topors, U2OS cells were generated that stably express Myc-Topors (WT or S718A). As shown in Fig. 2A, the expression level of Myc-Topors (WT or S718A) is similar in the stably transfected cells.
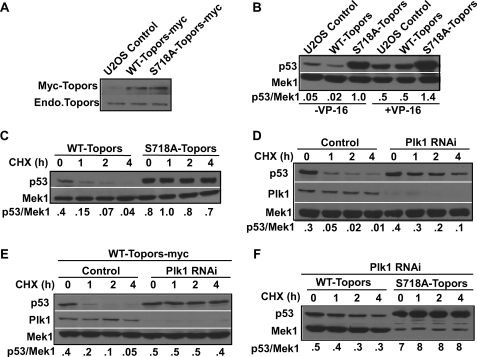
FIGURE 2.
The protein level of p53 is negatively regulated by Plk1-dependent phosphorylation of Topors. A, Western blots of extracts of control U2OS cells or U2OS cells stably expressing Myc-Topors (WT or S718A) using Myc (top) or Topors (bottom) antibodies. Endo., endogenous. B, U2OS cells stably expressing Myc-Topors (WT or S718A) were treated with or without 200 μm VP-16 for 18 h and analyzed by anti-p53 immunoblot. C, U2OS cells stably expressing Myc-Topors (WT or S718A) were treated with 75 μg ml−1 of cycloheximide (CHX) for the indicated time course and analyzed as in B. The ratios between p53 and Mek1 are indicated on the bottom. D, U2OS cells were transfected with pBS/U6-Plk1 to deplete Plk1, treated with cycloheximide as indicated, and harvested for anti-p53 immunoblot. For control samples, cells were transfected with pBS/U6 empty vector only. RNAi, RNA interference. E, U2OS cells stably expressing WT-Topors-Myc were depleted of Plk1 as in D, incubated overnight, and treated with cycloheximide for the indicated times. F, U2OS cells stably expressing Myc-Topors (WT or S718A) were depleted of Plk1, incubated overnight, and treated with cycloheximide for the indicated times. The amount of lysate loaded in the right four lanes was 10% of that in the left four lanes, as indicated by Mek1 Western blotting.
Because Topors is a dual ubiquitin and SUMO E3 ligase, modulating the stability of p53 (9, 11), we asked whether the Plk1-mediated phosphorylation of Topors regulates p53 stability. To address this question, U2OS cells stably expressing Myc-Topors (WT or S718A) were subjected to an anti-p53 Western blot. Consistent with a previous report (9), expression of WT-Topors led to a slight reduction in the protein level of p53. In striking contrast, the level of p53 protein was significantly elevated in cells expressing S718A-Topors (Fig. 2B, left three lanes). Considering that p53 is stabilized in response to DNA damage, similar experiments were performed in the presence of VP-16, a topoisomerase II inhibitor, used to induce DNA double strand breaks (19). In the presence of VP-16, the p53 level of cells expressing S718A-Topors was still significantly higher than that of cells expressing WT-Topors, suggesting that Plk1 phosphorylation of Topors negatively regulates p53 stability in a DNA damage-independent manner (Fig. 2B, right three lanes). Consistent with the above observation, p21, whose transcription is under control of p53, is clearly elevated in cells expressing S718A-Topors (supplemental Fig. 3A).
The increased level of p53 protein in U2OS cells expressing S718A-Topors could be due to either an increased level of protein translation or a reduced rate of protein degradation. To distinguish between these two possibilities, we quantified the half-life of p53 after treatment with cycloheximide, a protein translation inhibitor (20). First, U2OS cells stably expressing Topors (WT or S781A) were treated with cycloheximide for different times and analyzed by an anti-p53 Western blot. Cycloheximide treatment resulted in a gradual decrease in the level of p53 in cells expressing WT-Topors but not in cells expressing S718A-Topors, suggesting that the S718A mutation prevents p53 degradation (Fig. 2C and supplemental Fig. 4A). This result agrees with our earlier observations that Plk1 depletion leads to an elevation in the level of p53 in HeLa cells (3).
To examine whether this is a general phenomenon or a cell type-dependent event, U2OS cells were depleted of Plk1 by RNA interference (3), treated with cycloheximide, and analyzed by an anti-p53 Western blot. As indicated, the half-life of p53 increased from less than 20 min in control U2OS cells to about 2 h following Plk1 depletion (Fig. 2D and supplemental Fig. 4B). These results indicate that p53 stabilization in response to Plk1 depletion is likely to be a general phenomenon. To further confirm these observations, we analyzed p53 degradation in U2OS cells stably expressing WT-Topors. In these cells, Plk1 depletion also strongly prevented p53 degradation (Fig. 2E and supplemental Fig. 4C) in a pattern almost identical to that in S718A-Topors-expressing cells (Fig. 2C, right four lanes), confirming that Plk1-mediated phosphorylation of Topors promotes p53 degradation. Finally, we compared the stability of p53 in cells expressing different forms of Topors (WT or S718A) after Plk1 depletion. As expected, although p53 degradation was almost completely blocked in S178A-Topors-expressing cells, p53 was also significantly stabilized in cells expressing WT-Topors (Fig. 2F and supplemental Fig. 4D). Because the level of p53 immunostaining in S178A-Topors-expressing cells is very pronounced, only 10% of lysate was loaded in Fig. 2F, right four lanes, resulting in the low level of Mek1 used as our internal control.
To determine the half-life of p53 in cells expressing different forms of Topors (WT or S718A) by quantification, we needed to control p53 Western signals of cells expressing S718A-Topors in a liner range. For that purpose, we deliberately underloaded the S718A samples (Fig. 2F and supplemental Fig. 3B). The amount of lysate loaded in the right four lanes (S718A samples) was only 10% of that in the left four lanes (WT samples), as indicated by Mek1 Western blotting. Therefore, the apparent low Mek1 levels in cells expressing S718A-Topors is not a phenotype (Fig. 2F and supplemental Fig. 3B) because the Mek1 levels of cells expressing S718A-Topors were the same as those of cells expressing WT-Topors if the same amount of lysates was loaded (Fig. 2C).
Plk1-mediated Phosphorylation of Topors Modulates Sumoylation Versus Ubiquitination of p53
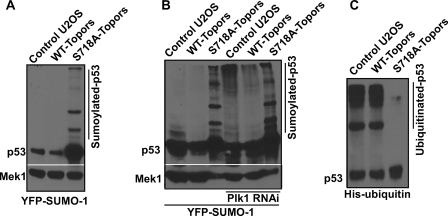
Considering that Topors functions both as a ubiquitin and as a SUMO-1 E3 ligase for p53, we asked whether the Plk1-dependent phosphorylation of Topors is involved in the sumoylation or ubiquitination of p53. Toward this end, U2OS cells stably expressing Topors (WT or S718A) were transfected with YFP-SUMO-1 and analyzed by an anti-p53 Western blot. As indicated in Fig. 3A, multiple SUMO-1-modified forms of p53 were detected in lysates from cells expressing S718A-Topors. By contrast, sumoylated forms of p53 were absent in WT-Topors-expressing or non-transfected U2OS cells, suggesting that Plk1-mediated phosphorylation of Topors inhibits its sumoylation activity toward p53. This is not due to a loading difference because sumoylated forms of p53 can be easily detected even with only 5% of the lysate from S718A-Topors-expressing cells (supplemental Fig. 5A). The increased level of p53 sumoylation in cells expressing S718A-Topors was also confirmed by anti-SUMO-1 Western blotting (supplemental Fig. 5B). To provide more evidence that Plk1 activity regulates sumoylation of p53, U2OS cells stably expressing Topors (WT or S718A) were co-transfected with YFP-SUMO-1 and pBS/U6-Plk1, the vector used to deplete Plk1 previously (3). When compared with cells with a normal Plk1 level, both untransfected cells and WT-Topors-expressing cells accumulated sumoylated forms of p53 upon Plk1 depletion (Fig. 3B). Based on these observations, we conclude that Plk1-mediated phosphorylation of Topors at Ser718 negatively regulates sumoylation of p53 in mammalian cells.
FIGURE 3.
Plk1-mediated phosphorylation of Topors suppresses sumoylation but increases ubiquitination of p53. A, U2OS cells stably expressing Myc-Topors (WT or S718A) were transfected with YFP-SUMO-1, incubated for 1 day, and analyzed by anti-p53 Western blotting. B, U2OS cells stably expressing Myc-Topors (WT or S718A) were co-transfected with YFP-SUMO-1 and pBS/U6-Plk1 at a ratio of 4:3, incubated for 1 day, and analyzed as in A. For cells expressing S718A-Topors (lanes 3 and 6), only 10% of lysate was loaded when compared with the rest of samples. C, U2OS or U2OS cells expressing Myc-Topors (WT or S718A) were transfected with His-ubiquitin, incubated for 1 day, and harvested. Lysates were subjected to anti-p53 IP followed by anti-p53 Western blotting.
In addition, we examined whether Plk1 phosphorylation of Topors also regulates the ubiquitination of p53. Accordingly, U2OS cells stably expressing Topors (WT or S718A) were transfected with His-ubiquitin and subjected to an anti-p53 IP followed by an anti-p53 Western blot. As shown in Fig. 3C and supplemental Fig. 5C, strong ubiquitin-modified forms of p53 were observed in both control and WT-Topors-expressing cells, whereas ubiquitinated forms p53 were absent in S718A-Topors-expressing cells. These results demonstrate that phosphorylation of Topors at Ser718 is required for ubiquitination of p53. Taken together, our data strongly support the notion that Plk1-mediated phosphorylation of Topors suppresses the sumoylation of p53, inducing instead p53 ubiquitination, which results in p53 degradation.
DISCUSSION
Phosphorylation of Topors was reported in a screen for nuclear phosphorylated proteins in HeLa cells (21). Recently, mass spectrometry was used to identify four Topors phosphorylation sites (22). Ser718 was not identified as a residue to be phosphorylated in this approach, likely due to an incomplete coverage of Topors fragments by enzymatic digestion of arginine/serine-rich regions (22). It is likely that multiple kinases act together to regulate Topors functions. In the case of Plk1, it has been established that a priming kinase phosphorylates the substrate to create a docking site for the C-terminal Polo box domain of Plk1 and that the recruited Plk1 phosphorylates the substrate to regulate its function (16, 23). Being a part of a serine-proline dipeptide, Ser98 is a likely cyclin-dependent kinase site (24). Whether cyclin-dependent kinase serves as a priming kinase for Ser98 to regulate Plk1-mediated phosphorylation of Topors at Ser718 is a likely possibility.
Although SUMO-1 is thought to be mono-conjugated to its substrate, a recent publication provides the first definite demonstration of the assembly of a poly-SUMO-1 chain on a protein substrate at a single SUMO-1 acceptor site (25). Therefore, it is possible that the multiple sumoylated p53 bands we observed in Fig. 3A are due to the formation of the poly-SUMO-1 chain on p53. Alternatively, because at least two sumoylation sites (Lys26 and Lys302) have been identified for Drosophila p53 (26), additional sumoylation sites may also exist for human p53, in addition to the already established site Lys386 (27). Possible multiple sumoylation sites can also be a reason for the multiple bands we observed in Fig. 3A. The mechanism by which the Plk1 phosphorylation of Ser718 inhibits the sumoylation activity of Topors while enhancing the ubiquitination activity is currently unknown. Several possibilities exist, including affecting ubiquitin carrier protein (E2) enzyme interaction, substrate binding, cellular localization, or protein stability (22). These possibilities are currently under investigation.
Supplementary Material
Acknowledgments
We thank Dr. R. Erikson for support and the critical review of this manuscript. We thank Drs. H. Koseki, S. Weger, and A. Saleem for various Topors constructs. We thank S. Liu, B. Song, and G. Weber for helpful discussions.
This work was supported, in whole or in part, by the National Institutes of Health Howard Temin Award K01 CA114401 through the NCI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and supplemental Figs. S1–S5.
W. H. Wang and O. M. Andrisani, unpublished data.
- Plk1
- Polo-like kinase 1
- Topors
- topoisomerase I-binding protein
- SUMO
- small ubiquitin-like modifier
- E3
- ubiquitin- protein isopeptide ligase
- pX
- X protein
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- aa
- amino acids
- WT
- wild type
- IP
- immunoprecipitation.
REFERENCES
- 1.Eckerdt F., Yuan J., Strebhardt K. (2005) Oncogene 24, 267–276 [DOI] [PubMed] [Google Scholar]
- 2.Takai N., Hamanaka R., Yoshimatsu J., Miyakawa I. (2005) Oncogene 24, 287–291 [DOI] [PubMed] [Google Scholar]
- 3.Liu X., Erikson R. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X., Lei M., Erikson R. L. (2006) Mol. Cell. Biol. 26, 2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ando K., Ozaki T., Yamamoto H., Furuya K., Hosoda M., Hayashi S., Fukuzawa M., Nakagawara A. (2004) J. Biol. Chem. 279, 25549–25561 [DOI] [PubMed] [Google Scholar]
- 6.Haluska P., Jr., Saleem A., Rasheed Z., Ahmed F., Su E. W., Liu L. F., Rubin E. H. (1999) Nucleic Acids Res. 27, 2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou R., Wen H., Ao S. Z. (1999) Gene 235, 93–101 [DOI] [PubMed] [Google Scholar]
- 8.Saleem A., Dutta J., Malegaonkar D., Rasheed F., Rasheed Z., Rajendra R., Marshall H., Luo M., Li H., Rubin E. H. (2004) Oncogene 23, 5293–5300 [DOI] [PubMed] [Google Scholar]
- 9.Rajendra R., Malegaonkar D., Pungaliya P., Marshall H., Rasheed Z., Brownell J., Liu L. F., Lutzker S., Saleem A., Rubin E. H. (2004) J. Biol. Chem. 279, 36440–36444 [DOI] [PubMed] [Google Scholar]
- 10.Guan B., Pungaliya P., Li X., Uquillas C., Mutton L. N., Rubin E. H., Bieberich C. J. (2008) J. Biol. Chem. 283, 4834–4840 [DOI] [PubMed] [Google Scholar]
- 11.Weger S., Hammer E., Heilbronn R. (2005) FEBS Lett. 579, 5007–5012 [DOI] [PubMed] [Google Scholar]
- 12.Hammer E., Heilbronn R., Weger S. (2007) FEBS Lett. 581, 5418–5424 [DOI] [PubMed] [Google Scholar]
- 13.Chen X., Cheung S. T., So S., Fan S. T., Barry C., Higgins J., Lai K. M., Ji J., Dudoit S., Ng I. O., Van De Rijn M., Botstein D., Brown P. O. (2002) Mol. Biol. Cell 13, 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrisani O. M., Barnabas S. (1999) Int. J. Oncol. 15, 373–379 [DOI] [PubMed] [Google Scholar]
- 15.Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E. (2003) J. Biol. Chem. 278, 25277–25280 [DOI] [PubMed] [Google Scholar]
- 16.Wu Z. Q., Liu X. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z. Q., Yang X., Weber G., Liu X. (2008) J. Biol. Chem. 283, 25503–25513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasheed Z. A., Saleem A., Ravee Y., Pandolfi P. P., Rubin E. H. (2002) Exp. Cell Res. 277, 152–160 [DOI] [PubMed] [Google Scholar]
- 19.Mok T. S., Wong H., Zee B., Yu K. H., Leung T. W., Lee T. W., Yim A., Chan A. T., Yeo W., Chak K., Johnson P. (2002) Cancer 95, 1511–1519 [DOI] [PubMed] [Google Scholar]
- 20.Takano Y. S., Harmon B. V., Kerr J. F. (1991) J. Pathol. 163, 329–336 [DOI] [PubMed] [Google Scholar]
- 21.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H. J., Zheng H., Kulkarni D., Kerrigan J., Pungaliya P., Saleem A., Rubin E. H. (2008) Biochemistry 47, 13887–13896 [DOI] [PubMed] [Google Scholar]
- 23.Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 24.Songyang Z., Blechner S., Hoagland N., Hoekstra M. F., Piwnica-Worms H., Cantley L. C. (1994) Curr. Biol. 4, 973–982 [DOI] [PubMed] [Google Scholar]
- 25.Yang M., Hsu C. T., Ting C. Y., Liu L. F., Hwang J. (2006) J. Biol. Chem. 281, 8264–8274 [DOI] [PubMed] [Google Scholar]
- 26.Mauri F., McNamee L. M., Lunardi A., Chiacchiera F., Del Sal G., Brodsky M. H., Collavin L. (2008) J. Biol. Chem. 283, 20848–20856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson I. R., Irwin M. S. (2006) Neoplasia 8, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studach L. L., Rakotomalala L., Wang W. H., Hullinger R. L., Cairo S., Buendia M. A., Andrisani O. M. (2009) Hepatology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.