Abstract
The last three years have seen an escalation in the number of sequenced bacteriophage genomes with more than five hundred now in the NCBI phage database, representing a more than three-fold increase from 2005. These span at least 70 different bacterial hosts, with two-thirds of the sequenced genomes of phages representing only eight bacterial hosts. Three key features emerge from the comparative analysis of these genomes. First, they span a very high degree of genetic diversity, suggesting early evolutionary origins. Secondly, the genome architectures are mosaic, reflecting an unusually high degree of horizontal genetic exchange in their evolution. Thirdly, phage genomes contain a very high proportion of novel genetic sequences of unknown function, and likely represent the largest reservoir of unexplored genes. With an estimated 1031 bacterial and archael viruses in the in the biosphere, our view of the virosphere will draw into sharper focus as further bacteriophage genomes are characterized.
Introduction
Because of their relatively small size and simplicity of isolation, bacteriophages were the first complete genomes to be sequenced, beginning with the 5,386bp single-stranded DNA (ssDNA) phage φX174 in 1977 [1]. The first complete sequence of a double-stranded DNA phage was that of the lambda 48,502bp genome [2], and the sequence of the 39,936bp phage T7 genome was reported shortly after [3]. The first complete sequence of a dsDNA tailed-phage genome of a virus infecting a non-Escherichia coli host was that of mycobacteriophage L5 almost ten years later [4], and the number of sequenced bacteriophage genomes has exploded since that time as DNA sequencing methodologies have advanced.
The application of epifluorescence techniques to the enumeration of phage particles in environment samples in the late 1980′s [5-7] contributed to a significant re-thinking of the roles of bacteriophages in biology and in the environment. Viral densities in ocean samples are 106 − 107 particles per milliliter, and given the size of the oceans and determinations of similar terrestrial viral populations [8], the total phage population is estimated to be around 1031 particles [9,10]. This is validated by independent estimates of 1030 bacterial cells in the biosphere [11], along with the finding that environmental viral-bacterial ratios are typically 5−10:1 [10,12]. The size of this viral population is stunning, and suggest that prokaryotic viruses are a majority of all biological forms [13]. Furthermore, experimental studies show that the population is highly dynamic, with an estimated 1023 infections/second globally [12].
The advances in bacteriophage genomics have been fueled with two primary goals. First, the abundance and turn over of the phage population poses questions as to how phage genomes are related to each other and to their hosts, and what evolutionary mechanisms shaped this population. Secondly, genomics has advanced the use of phages for development of genetic, biotechnological, and clinical tools, and a large variety of utilities and approaches have been described. In this review I will largely focus on what we have learned about the structure and content of bacteriophage genomes, and how they have evolved, focusing on developments since the review by Casjens in 2005 [14]. Several additional reviews and resources on phage genomics might be of interest [15-20].
Sequenced bacteriophage genomes
At the time of writing, the NCBI phage genome database contains 500 completely sequenced genomes, corresponding to a total of 22.3Mbp of sequence information. Although phage genomes are on average only approximately 1% the size the bacterial chromosomes, the number of sequenced bacterial genomes in the NCBI bacterial chromosome list (741) is substantially greater in number and a 100-fold larger in total sequence information (2.42Gbp). This is somewhat surprising given the high diversity of the phage population and its abundance, and it seems likely that the next few years will see a substantial shift in the ratio of sequence phage and bacterial genomes. There are a large number of resident prophages in sequenced bacterial genomes [21,22], possibly outnumbering sequenced phage genomes, although the proportion of these retaining potential for lytic growth is unclear.
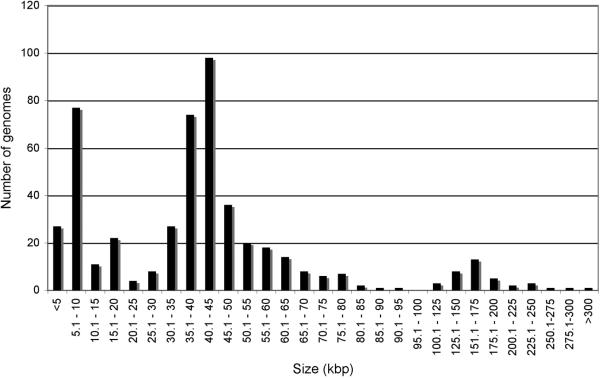
Sequenced phage genomes vary considerably in size from Leuconostoc phage L5 (2,435bp) to Pseudomonas phage 201phi2−1 (316,674b). However, there is not a uniform distribution of genome sizes across this spectrum (Fig. 1), and there is a predominance of genome sizes in three separate ranges. The largest is a peak of genomes in the 30−50kbp interval, corresponding to nearly 50% of all phages (Fig. 1). A second group (abut 20% of total) is those smaller than 10kbp, and the third group contains those in the 100−200 kbp interval (6% of the total). This overall distribution likely reflects the phage isolation methods employed and the sequencing technologies available at that time, and is not a true reflection of size distributions of phages in the environment. For example, the group of genomes smaller than 10kbp has a median size value of only 5.5kbp and many were characterized prior to automated sequencing methodologies. In contrast, the dearth of larger phage genomes (>250kbp) is not from technological availability, but because bacteriophages with larger heads and corresponding larger genomes form extremely small plaques on standard agar plates and are thus easily overlooked [23]. The use of alternative microbiological substrates and electron microscopy suggests there are many large phages in the environment that have yet to be explored.
Figure 1. Size distribution of sequenced bacteriophage genomes.
As of early July 2008, the phage database at NCBI contains 500 genomes ranging from 2.3 kbp to 316 kbp. The numbers of sequenced genomes in each size range is shown. Size intervals are 5 kbp from 2−100 kbp, and 25 kbp from 100−300 kbp.
The predominant bacteriophage morphological forms are tailed phages containing dsDNA genomes. However, few of these if any have genomes smaller than 10kbp, and the vast majority are larger than 15kbp. This is consistent with the finding that the virion structure and assembly genes typically encompass at least 15 kbp of genome space; moreover, the Siphoviruses (characterized by a long flexible non-contractile tail) all contain a tapemeasure protein gene whose length corresponds to the length of the phage tail, and ranges from 1.5 − 6 kbp in length. Therefore the majority of phages with Siphoviral morphologies have genomes longer than 20 kbp. In contrast, most of the phages with larger genomes (>125kbp) are Myoviruses (with contractile tails), and the largest Siphoviral genome is that of Bacillus phage SPBc2 (134,416 bp). The reasons for the lack of larger Siphoviruses is not clear.
Phage genome sequence diversity
The genetic diversity of the bacteriophage population is remarkable. In general, the nucleotide sequences of genomes derived from phages with non-overlapping host ranges rarely share sequence similarity, although this may not be surprising if the bacterial hosts are distantly related. For example, there is no obvious or extensive nucleotide sequence similarity between the four published genomes of Streptomyces phages and a collection of 50 mycobacteriophage genomes, even though the Mycobacterium and Streptomyces hosts both are members of the Actinomycetales. Presumably, phages of non-overlapping hosts enjoy at least a transient genetic isolation from each other.
Since bacteriophages infecting a common bacterial host are in genetic contact with each other, it is not surprising that they sometimes share common nucleotide sequences. More than 30 phage genomes have been isolated for Pseudomonas (33), Staphylococcus (48) and Mycobacterium (50) hosts, and there are many examples where phages of a common host enjoy related sequences [24-26]. Perhaps somewhat more surprising is that many phages within these groups share little or no sequence similarity, as illustrated by nucleotide sequence comparisons of mycobacteriophages and Pseudomonas phages (Fig. 2). In general, phages with different virion morphotypes have different genome organizations and greater sequence diversity; genome architecture may therefore impose constraints on genetic exchange. It is noteworthy that the high genetic diversity among phages of a common hoss coupled with the still limited number of available sequences suggests there is an abundance of new viral genome sequences as yet unidentified.
Figure 2. Nucleotide sequence relationships among phages infecting common bacterial hosts.
Bacteriophage genomes of 30 Pseudomonas and 50 mycobacteriophages were concatenated and compared using the program Gepard. The comparison illustrates two key points. First, that there are groups of genomes within each set that share significant nucleotide sequence similarity, while there is no detectable sequence similarity between the two groups (data not shown). Secondly, even within phages infecting a common host, there is enormous genetic diversity.
Phage genome architectures
Comparative analysis of phage genomes reveals some common architectural themes. In general, in phages with Siphoviral morphotypes there is clear synteny among genes encoding the virion structure and assembly functions [14]. The genes for head and tail assembly are arranged together with the head genes 5′ to the tail genes. The head genes typically include one or two terminase subunits, the portal protein, a prohead protease, a scaffold protein, and the major capsid subunit; the tail genes include the major tail subunit, two overlapping open reading frames expressed via a programmed translational frameshift [27] and the tapemeasure protein, followed by the minor tail proteins [14]. The length of the tapemeasure protein gene corresponds to the length of the phage tail, and it thus commonly the largest gene in the genome [28,29].
The syntenic relationship of the virion structure and assembly genes is conserved in phages that not only have no nucleotide sequence similarity but also in genomes in which none of the predicted protein amino acid sequences are detectably related [30], and presumably represents a relatively early feature in bacteriophage evolution. This notwithstanding, there are many architectural elaborations of this theme. For example, in the mycobacteriophages, the lysis genes are found in different locations relative to the structure and assembly genes. In some phages [e.g. L5; [4]], the lysis genes are to the left of the terminase gene, between terminase and the leftmost cos site; in other phages [e.g. TM4; [31]], the lysis genes are located to the right of the minor tail protein genes. Many phages also contain a cassette of integration functions (int, attP), which are typically located close to the center of the genome, even though genome length can vary by more than two-fold [32]. In one instance, the integration cassette appears to have relocated to a position within the tail genes [33], a process that might have occurred though use of a secondary attachment site within the genome. Comparison of mycobacteriophage genomes shows that while the structure and assembly genes typically occupy the ‘left arm’ region between a centrally located integration cassette and the leftmost cos site, the size of this region can vary by more than two fold [29]. In the largest instance (phage Omega), the gene order of the structure and assembly genes is conserved, but there are numerous additional genes of unknown function distributed between them [29].
Phages with somewhat larger genomes (>125kbp) have much less well conserved arrangements of gene functions, although there is still considerable grouping of virion structural genes. Many of these – including phages of several different hosts – are related to phage T4 and these contain conserved core groups of genes that are interrupted with hyper plastic regions [34]. The genes in these hyperplastic regions are relatively small and mostly of unknown functions, although they include a subset of virion internal proteins that are implicated in adsorption and protection from DNA modification by the host [35].
Genome mosaicism and its origins
One of the most striking features of bacteriophage genomes is their apparent mosaic structure; in essence, each genome can be considered as a unique combination of modules that are exchangeable among the population. The size of the modules, their rates of exchange, and the phage genomes carrying them all vary greatly, with phages of different virion morphology, size, and host-range all participants in an orgy of recombination [36]. This mosaicism is by no means unique to the phage population and is also prevalent in bacteria, where genes are acquired by horizontal genetic exchange (predominantly transduction, transformation and conjugation) [37]; but the extent of mosaicism in phage genomes is remarkable and has become even clearer as the number of genomes available for comparative analysis has grown.
Phage genome mosaicism can be viewed at two levels. The first is by comparison of nucleotide sequences, described initially by DNA heteroduplex mapping [38], and subsequently by sequence comparison [39]. At the resolution of DNA sequence information, precise junctions can be observed corresponding to the boundaries of two DNA segments that clearly have distinct evolutionary origins. One of the most notable features of these functions is that they predominantly correspond to the boundaries of open reading frames (Fig. 3A). There are two types of models to explain the recombination mechanisms that give rise to these patterns. The first proposes that there are short conserved boundary sequences at gene junctions that serve to target exchange events catalyze by homologous recombination, using either host- or phage-encoded recombinases [40]. Some examples of short conserved sequences have been reported [41], although their role is not clear, and this model would not seem to account for the vast majority of exchange events. A second model proposes that the recombination events are not targeted and occur randomly (or with only short sequence preferences), such that most events give rise to non-functional genomic trash [13]. Most of the progeny would not be viable, except for those that have an appropriate genome size and that retain gene functions, thus accounting for the correlation with gene borders. Productive genetic exchange may require multiple recombination events, and while the overall process is expected to occur at low frequency, this process is potentially highly creative, yielding new combinations of genes, as well as new combinations of protein domains. A recent report suggests that homeologous recombination mediated by phage-encoded RecE/T-like recombination systems may play an important role in generating mosaicism [42], and bacterial acquisition of CRISPR sequences could represent an additional mechanism [43].
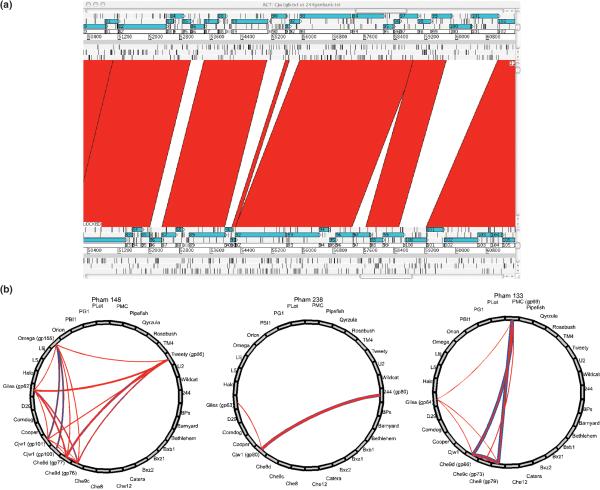
Figure 3. Phage genome mosaicism at the nucleotide and protein sequence levels.
A. Nucleotide sequence comparison of mycobacteriophages Cjw1 and 244. The two annotated genomes [29] were aligned using the Artemis Comparison Tool, and highly related sequences shown by red shading. The correspondence of nucleotide discontinuities with gene boundaries is evident. B. Phamily circles of thee consecutive genes (62−64) of mycobacteriophage Giles [24,33]. Each circle corresponds to three different Phams representing group of related proteins; and the thickness of the lines connecting each phamily constituent is related to the strengths of the relationships. Each of the three Giles genes clearly has a different evolutionary history.
Comparing the predicted amino acid sequences of phage gene products generates an alternative manifestation of mosaicism [20,24,29]. This is an informative approach since many groups of phages – including those that infect common hosts – may not share any nucleotide sequence information; protein sequence data reveals genes that share much older ancestry. Sequence comparison and phylogenetic reconstruction shows that different genes, groups of genes, or segments of genes, all share different ancestry, and thus represent modules within a mosaic genome. However, the phylogenetic comparisons can be complex because the diversity is sufficiently high that none or few of the participating genomes in the analysis are in common. One alternative method or representing these relationships is through the use of phamily circles [24] in which all of the participating genomes are represented, even those that do not contribute to that particular phylogenetic relationship (Fig, 3B). The extent of phage genomic mosaicism thwarts the simple determination of phylogenetic relationships of whole phage genomes as units of evolution, and their histories can best be considered as the totality of the evolutionary routes taken by each constituent module.
Phage metagenomics
Phage metagenomics provides an alternative view of phage genome structure that is different but complementary to whole genome sequence determination. A variety of phage metagenomic studies have been reported [44] and one of the most notable features is that they contain a high proportion of unknown sequences, consistent with the high genetic diversity in the completely sequenced genomes. These studies offer considerable insight into the geographic distribution of phages and phage types, but as yet provide relatively little insight into whole phage genome structure and evolution, because rarely do whole genome sequence emerge. However, phage metagenomics is well-suited to the ultra high throughput sequencing methodologies just now becoming available, and large scale viral metagenomics will be a crucial asset in understanding phage genomics in the next few years.
Conclusions
In summary, enormous advances have been made in phage genomics over the past few years and while we do not yet have detailed answers as to the extent of phage diversity and evolutionary pathways, the questions have become clearer and the groundwork well established. The technologies for sequence determination are no longer limiting and it is reasonable to expect a deluge of new information in the next few years. Hopefully a clearer picture will emerge of phage genomic structure along with new questions as to what functions the many newly discovered phage genes do, and how they may be exploited for biological and biotechnological ends.
Acknowledgements
I would like to thank Drs. Roger Hendrix and Jeffrey Lawrence for their collaboration in phage genomics and the many students and fellows who also have contributed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 3.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 4.Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 5.Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 6.Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol. 2005;8:444–450. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Hennes KP, Suttle CA. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol Oceanogr. 1995;40:1050–1055. [Google Scholar]
- 8.Ashelford KE, Day MJ, Fry JC. Elevated abundance of bacteriophage infecting bacteria in soil. Appl Environ Microbiol. 2003;69:285–289. doi: 10.1128/AEM.69.1.285-289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 10.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •12.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [The abundance of marine viruses, their dynamic relationships with their hosts, and their influence on biogeochemistry of the planet emphasizes the importance of dissecting their genetic relationships.] [DOI] [PubMed] [Google Scholar]
- 13.Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 14.Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–458. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Ackermann HW, Kropinski AM. Curated list of prokaryote viruses with fully sequenced genomes. Res Microbiol. 2007;158:555–566. doi: 10.1016/j.resmic.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Toussaint A, Lima-Mendez G, Leplae R. PhiGO, a phage ontology associated with the ACLAME database. Res Microbiol. 2007;158:567–571. doi: 10.1016/j.resmic.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Lima-Mendez G, Toussaint A, Leplae R. Analysis of the phage sequence space: the benefit of structured information. Virology. 2007;365:241–249. doi: 10.1016/j.virol.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 18.Prangishvili D, Garrett RA, Koonin EV. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [Genomics of archaeal viruses provides whole new perspective on phage genomics, and together with their morphological diversity suggests an important and somewhat understudied area.] [DOI] [PubMed] [Google Scholar]
- 19.Glazko G, Makarenkov V, Liu J, Mushegian A. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol Direct. 2007;2:36. doi: 10.1186/1745-6150-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Glazko G, Mushegian A. Protein repertoire of double-stranded DNA bacteriophages. Virus Res. 2006;117:68–80. doi: 10.1016/j.virusres.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Fouts DE. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- 23.Serwer P, Hayes SJ, Thomas JA, Hardies SC. Propagating the missing bacteriophages: a large bacteriophage in a new class. Virol J. 2007;4:21. doi: 10.1186/1743-422X-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [Mycobacteriophages are highly genetically diverse and their mosaicism can be seen through sequence comparison of the predicted protein sequences and grouping into Phamilies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J Bacteriol. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J. A conserved frameshift strategy in dsDNA long tailed phages [Ph.D.] University of Pittsburgh; Pittsburgh: 2000. [Google Scholar]
- 28.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 29.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 30.Casjens S, Hatfull GF, Hendrix R. Evolution of dsDNA tailed-bacteriophage genomes. Sem. Virol. 1992;3:383–397. [Google Scholar]
- 31.Ford ME, Stenstrom C, Hendrix RW, Hatfull GF. Mycobacteriophage TM4: genome structure and gene expression. Tuber Lung Dis. 1998;79:63–73. doi: 10.1054/tuld.1998.0007. [DOI] [PubMed] [Google Scholar]
- 32.Hatfull GF. Mycobacteriophages. In: Calendar R, editor. The Bacteriophages. Oxford University Press; 2006. pp. 602–620. [Google Scholar]
- 33.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J. Bacteriol. 2008;190:2172–2182. doi: 10.1128/JB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •34.Comeau AM, Bertrand C, Letarov A, Tetart F, Krisch HM. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology. 2007;362:384–396. doi: 10.1016/j.virol.2006.12.031. [Sequence comparison of a group of phages related to T4 reveals common core genes and regions of relatively small regions that are highly variable among these phage genomes.] [DOI] [PubMed] [Google Scholar]
- 35.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol. 2008;375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Retchless AC, Lawrence JG. Temporal fragmentation of speciation in bacteria. Science. 2007;317:1093–1096. doi: 10.1126/science.1144876. [DOI] [PubMed] [Google Scholar]
- 38.Highton PJ, Chang Y, Myers RJ. Evidence for the exchange of segments between genomes during the evolution of lambdoid bacteriophages. Mol Microbiol. 1990;4:1329–1340. doi: 10.1111/j.1365-2958.1990.tb00712.x. [DOI] [PubMed] [Google Scholar]
- 39.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 40.Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark AJ, Inwood W, Cloutier T, Dhillon TS. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J Mol Biol. 2001;311:657–679. doi: 10.1006/jmbi.2001.4868. [DOI] [PubMed] [Google Scholar]
- 42.Martinsohn JT, Radman M, Petit MA. The lambda red proteins promote efficient recombination between diverged sequences: implications for bacteriophage genome mosaicism. PLoS Genet. 2008;4:e1000065. doi: 10.1371/journal.pgen.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- •44.Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M, Liu H, Furlan M, Wegley L, Chau B, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [Metagenomics provides a powerful approach to exploring the diversity and biogeography of bacteriophages and offers a helpful and complementary view to that of whole genome sequencing.] [DOI] [PubMed] [Google Scholar]