Abstract
Thiol-dependent redox systems are involved in regulation of diverse biological processes, such as response to stress, signal transduction, and protein folding. The thiol-based redox control is provided by mechanistically similar, but structurally distinct families of enzymes known as thiol oxidoreductases. Many such enzymes have been characterized, but identities and functions of the entire sets of thiol oxidoreductases in organisms are not known. Extreme sequence and structural divergence makes identification of these proteins difficult. Thiol oxidoreductases contain a redox-active cysteine residue, or its functional analog selenocysteine, in their active sites. Here, we describe computational methods for in silico prediction of thiol oxidoreductases in nucleotide and protein sequence databases and identification of their redox-active cysteines. We discuss different functional categories of cysteine residues, describe methods for discrimination between catalytic and noncatalytic and between redox and non-redox cysteine residues and highlight unique properties of the redox-active cysteines based on evolutionary conservation, secondary and three-dimensional structures, and sporadic replacement of cysteines with catalytically superior selenocysteine residues.
Keywords: cysteine, redox, selenocysteine, thiol oxidoreductase, thioredoxin
INTRODUCTION
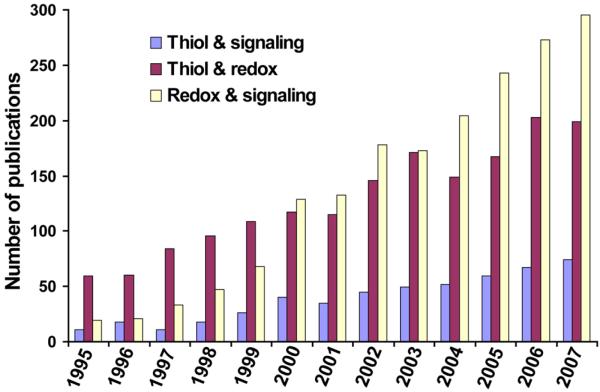
The past ten years have seen a growing evidence for the key roles of thiol-dependent redox processes in cellular redox regulation. This research has revealed molecular mechanisms by which thiol-based redox systems protect cells against oxidative stress, mediate signal transduction, support protein folding and participate in many other physiological processes. The roles of thiol redox control in cancer and aging-related diseases (Matés et al., 2008), such as Parkinson's disease (Andres-Mateos et al., 2007; Wassef et al., 2007), Alzheimer's disease (Calabrese et al., 2006; Newman et al., 2007) and in other pathophysiological processes has also been described. A growing interest in thiol-dependent processes can be illustrated by the number of PubMed entries in the past 12 years (Fig. 1).
Fig. 1.
Publications involving thiol oxidoreductases and their functions. Articles were counted by their year of their publication in PubMed using indicated keywords.
The thiol-based redox control in cells is provided primarily by thiol oxidoreductases, a diverse group of enzymes which utilize their redox-active cysteine (Cys) residues for redox catalysis (Fig. 2). Many such proteins have a thioredoxin fold (Martin, 1995), but other folds are also abundantly present. Identification and characterization of thiol oxidoreductases is challenging because of high divergence of protein families that represent these enzymes and a variety of folds that were adapted by this protein group. Specific functions are known for some thiol oxidoreductases, whereas many other proteins are described only by their general participation in redox processes. Several independent approaches have been advanced for prediction of thiol oxidoreductases in sequence and structure databases. For example, one is based on the identification of CxxC (two Cys separated by two residues) motifs in the context of protein secondary structure (Fomenko and Gladyshev, 2002; 2003). Another is based on the analyses of structural features of these enzymes (Fetrow and Skolnick, 1998; Fetrow et al., 1998). An additional method searches for sporadic Cys/selenocysteine (Sec) pairs in homologous sequences (Fomenko et al., 2007). It this review, we discuss functional categories of Cys in proteins and unique features of catalytic redox-active Cys that support their identification and functional characterization.
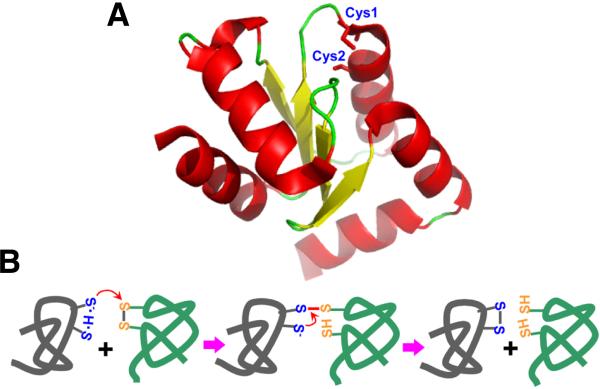
Fig. 2.
Thiol oxidoreductases and their catalytic mechanisms. (A) A 3D-structure of a thiol oxidoreductase (human glutaredoxin 2, accession code 2FLS). The attacking catalytic Cys is indicated as Cys1 and the resolving Cys as Cys2. Both Cys residues are located near the protein surface and are accessible for protein-protein interactions. (B) Catalytic mechanism of a representative thiol oxidoreductase, thioredoxin. The attacking Cys is shown as thiolate, stabilized by hydrogen bond with the resolving Cys. A transient intermolecular disulfide bond is formed with the target protein during the reaction. The resolving Cys in the thiol oxidoreductase attacks this intermolecular disulfide forming a disulfide with the catalytic Cys, releasing the reduced substrate.
Functional categories of cysteine residues
Among the 20 common amino acids in proteins, Cys is one of two least abundant yet the most conserved residue that is frequently present in functionally important sites (i.e., catalytic, regulatory, cofactor binding, etc.) in proteins. These properties are likely due to higher reactivity of Cys compared to other amino acids. Organisms living in harsh environments show a tendency for reduced use of Cys (Beeby et al., 2005). Cys residues serve numerous functions that can be arbitrarily defined as follows:
1. Catalytic redox-active Cys residues
These Cys are present in the active sites of thiol oxidoreductases and are directly involved in catalysis. Such Cys participate in oxidation, reduction and isomerization of disulfide bonds as well as in other reactions that change the redox state of these residues. These Cys residues are highly conserved in protein sequences.
2. Regulatory Cys
These Cys regulate or modulate protein activity by changing their redox state, but themselves they are not catalytic. Such residues are present in transcription factors, for example, in OxyR and Yap1 (Azevedo et al., 2003; Delaunay et al., 2002; Wood et al., 2004), kinases (Rhee et al., 2000; Veal et al., 2004), phosphatases (Juarez et al., 2008; Salmeen et al. 2003), chaperone Hsp33 (Ilbert et al., 2007; Jakob et al., 2000), mitochondrial branched chain aminotransferase (Conway et al., 2004) and many other proteins. Redox regulation involving these regulatory Cys residues may involve reversible intramolecular and intermolecular disulfide bonds as well as glutathionylation, S-nitrosylation (Dalle-Donne et al., 2007; 2008; Sun et al., 2006) are other Cys modifications.
3. Structural Cys
These Cys residues participate in protein structure and folding through formation of stable intramolecular and intermolecular disulfide bonds. Such Cys are present in numerous proteins, many of which are secreted, membrane-bound or endoplasmic reticulum (ER)-resident proteins.
4. Metal-coordinating Cys
This is a large and diverse group of highly conserved Cys residues that coordinate metal ions. Metal-binding Cys frequently occur in the form of CxxC motifs (Gladyshev et al., 2004). Because of that, discrimination between redox-active and metal-binding Cys by sequence and structure analysis is challenging as both frequently utilize such motifs.
5. Catalytic non-redox Cys
These residues participate in catalysis but do not change their redox state in the reaction. Catalytic non-redox Cys are typically highly conserved, serve as nucleophiles and occur in diverse protein families. Examples of enzymes with such Cys residues are glyceraldehyde-3-phosphate dehydrogenase and various Cys proteases (Fermani et al., 2007).
6. Cys as sites of posttranslational modifications
Reactivity of Cys residues is also utilized for targeting proteins to membranes and as sites of posttranslational modifications. For example, a C-terminal Cys can form a thioether bond with farnesyl and geranylgeranyl groups, leading to protein lipidation. Such modifications may influence biological activities of proteins, attach proteins to membranes or engage them in protein-protein interactions (Zhang and Casey, 1996).
It should be mentioned, however, that in some cases a Cys can fall into more than one of the above categories: for example, the catalytic non-redox Cys in glyceraldehyde-3-phosphate dehydrogenase also undergoes regulation by reactive oxygen species and thiols (Fermani et al., 2007; Hook and Harding, 1997). Finally, there are Cys residues without any specific function. These Cys show low conservation in protein sequences and their abundance may be influenced by environment (Beeby et al., 2005). They share some features such as low exposure of the sulfur atom, no proximity to other Cys residues and high pKa (often higher than 9), which are consistent with their lower reactivity (Sanchez et al., 2008).
Among the six functional categories of Cys residues, our focus has been on the catalytic redox-active Cys, which are the functional sites in thiol oxidoreductases. Since different folds are utilized in these enzymes, their defining feature must be a conserved catalytic redox Cys. However, other functional Cys also exhibit strong conservation. Thus, while query proteins can be initially screened for Cys conservation, identification of thiol oxidoreductases should also involve procedures for specific recognition of these enzymes based on unique features of their Cys residues as well as specific discrimination of these enzymes from proteins with other Cys functions.
Conservation of protein sequences can be determined by collecting homologous sequences and building multiple sequence alignments. The most popular sequence analysis and alignment tools are a set of BLAST programs (Altschul et al., 1997). These programs exist as stand-alone tools for most computer platforms and systems and as web-versions, for example, at http://www.ncbi.nlm.nih.gov/BLAST/. Conservation of a Cys in a protein sequence would suggest a possibility that this residue may have a functional role, but would not tell what that function might be. We further discuss the application of computational tools with regard to Cys classification with focus on catalytic redox-active Cys residues.
Regulatory cysteines
This Cys class is common in regulatory and metabolic proteins. Signaling mechanisms involving regulatory Cys are based on disulfide bond formation, S-nitrosylation, glutathionylation or other types of Cys modifications (Biswas et al., 2006). Regulatory Cys are generally conserved; however, their conservation level is not as high as that of metal-coordinating or catalytic Cys residues. Most data on the identity and function of regulatory Cys have been derived from experimental work. Only several specific features, such as acid-base motifs and high frequency of charged amino acids in Cys-flanking regions, have been described, but they are insufficiently specific. For example, while these features seem to better recognize S-nitrosylated Cys (Greco et al., 2006), they fail with other common Cys modifications such as the oxidation of Cys thiol to sulfenic acid (Salsbury et al., 2008).
Neither structure nor sequence determinants have been reported for regulatory Cys. Regulatory redox Cys are often direct targets for thiol oxidoreductases and these interactions influence cellular pathways. Computational identification of regulatory Cys is one of the major challenges in redox biology, but as of now, low reliability of experimental data and low conservation preclude in silico prediction of proteins with such residues.
Structural cysteines
Intramolecular disulfide bond formation is one of the major mechanisms of protein structure stabilization. It is often used in secreted proteins and those located in oxidizing environments, such as bacterial periplasm and eukaryotic ER, and is much less frequent in reducing environments (e.g., cytosol, nucleus and mitochondrial matrix). However, in some thermophilic bacteria, structure stabilization through disulfide bonds is used even for intracellular proteins (Pedone et al., 2008). Structural disulfide bonds are formed between two specific Cys residues in a process of oxidative folding involving specialized machinery (Coller et al., 2002; Tu et al., 2004). This class of conserved Cys can be characterized computationally through protein structure analysis. A current version of PDB (http://www.rcsb.org) contains a redundant set of 51,663 protein structures (as of June 2008). An interesting approach for automated screening of structural databases for intermolecular and intramolecular disulfide bonds is based on the analysis of distance between any two sulfur atoms of Cys residues in the same protein (Beeby et al., 2005). This distance-based approach has been implemented via PDB advanced search engine (with the user-defined Cys-Cys distance), and thus can be effectively employed by the user to directly scan the database. A growing flow of protein structures from structural genomics projects should help improve functional analysis of disulfide bonds in proteins.
Metal-binding cysteines
Metal-binding Cys residues are found in structurally and evolutionary distinct groups of proteins which are present in all branches of life. Zinc, iron, copper, nickel, cobalt, manganese, selenium, molybdenum, calcium and perhaps other metals and metalloids could be coordinated by Cys residues in proteins. Metals in proteins can function as structure-stabilizing elements or be involved directly in catalysis, function or regulation as cofactors. Metal-binding proteins can be identified using conserved protein domain and pattern identification tools, for example, PROSITE (Gattiker et al., 2002; Hulo et al., 2008) profiles and patterns (http://ca.expasy.org/prosite/). A current version of PROSITE database contains 1316 patterns and 801 profiles. Information about amino acids which are involved in metal coordination is based on prior experimental data, such as protein structures, and the results of site-directed mutagenesis experiments. The growth of sequence and structure databases provides opportunities for better definition of metalloproteomes in organisms (Zhang et al., 2008a; 2008b). Some metal-binding Cys are sensitive to oxidation and may release metal and in turn form disulfide bonds (Giles et al., 2003; Ilbert et al., 2007), but at present there are no reliable criteria for the identification of such redox-regulated metal-binding proteins. Cys and histidines are most frequently involved in metal coordination and the presence of proximal conserved Cys, when in combination with other conserved Cys and histidines, may help predict metal binding sites in proteins. Metal-coordinating Cys are frequently present in the form of conserved CxxC motifs, but these motifs are also typical of thiol oxidoreductases. Nevertheless, the presence of several proximal conserved CxxC motifs in protein sequences almost always indicates a metal-binding rather than redox function. Attempts have been made to collect metal-binding proteins and organize the data in the form of database, e.g., http://metallo.scripps.edu (Castagnetto et al., 2002).
Catalytic non-redox cysteines
At present, this broad group of evolutionary conserved proteins with conserved nucleophilic Cys could be predicted only by sequence or structure similarity to functionally characterized proteins. Perhaps, approaches that analyze enzyme active sites and their structural environments may help detect these proteins, but this has not been tried thus far.
Catalytic redox-active cysteines
Catalytic redox-active Cys are functional, evolutionary conserved residues. Although in silico prediction of Cys classes in general is difficult, several methods have been developed for prediction of thiol oxidoreductases, with the key method searching for Cys/Sec pairs in homologous sequences.
Sporadic cysteine/selenocysteine pairs in proteins
Sec is known as the 21st amino acid in the genetic code and differs from Cys by a single atom (i.e., Se versus S) (Hatfield and Gladyshev, 2002; Stadtman, 1996; Wessjohann et al., 2007). Sec is inserted cotranslationally in protein sequences in response to a stop codon, UGA, when a specific RNA hairpin structure, SECIS element, is present in selenoprotein genes. Replacement of S with Se (i.e., Cys with Sec) may significantly improve catalytic properties of thiol oxidoreductases (Bock et al., 1991; Jacob et al., 2003; Kim and Gladyshev, 2005; Kim et al., 2006). In contrast to the many functions of Cys, Sec is always (at least in functionally characterized selenoproteins) located in the active sites of redox proteins and serves as the catalytic group in these enzymes (Fomenko et al., 2007). In addition, most selenoproteins have close Cys-containing homologs, whose catalytic Cys are highly conserved. These observations were used to develop a method for high-throughput identification of catalytic redox Cys in protein sequences by searching for sporadic Cys/Sec pairs in homologous sequences.
Table 1 shows 41 selenoproteins families which have at least one Cys homolog (note that several protein families, such as PDI, DsbA, DsbC, DsbG, and DsbE are included in a large thioredoxin superfamily). Most of these Cys-containing proteins have previously been described as thiol oxidoreductases. Only six thiol oxidoreductases (some belonging to the same family), including DsbB, dihydrolipoamide dehydrogenase, glutathione reductase, Erv1, Erv2, and Ero1 do not have known Sec-containing homologs. Thus, occurrence of selenoprotein homologs and location of Sec in these proteins may in itself be indicative of a redox function of the corresponding Cys in Cys-containing proteins. A method that employs this observation is thus not dependent on sequence motifs, structure or origin of sequences. It first identifies unique Cys/Sec pairs flanked by homologous sequences within a pool of translated nucleotide sequences. These pairs then serve as seeds for sequence analysis at the level of protein families and subfamilies. As shown in Table 1, application of this method identifies the majority of known proteins containing catalytic redox-active Cys, while filtering out proteins in which conserved Cys are involved in other functions (i.e., non-redox catalysis, regulation, structural disulfides, posttranslational modifications, and metal binding) (Fomenko et al., 2007; Fomenko and Gladyshev, unpublished). It should be noted that for oxidoreductases containing multiple conserved Cys, identity of the attacking nucleophilic Cys could be determined by the location of Sec in the selenoprotein homolog.
Table 1.
Protein families identified by searching for Cys/Sec pairs in homologous sequences in nucleotide and protein sequence databases
| Protein | Comments | Protein family | Bacteria Cys/Sec | Archaea Cys/Sec | Eukaryota Cys/Sec |
|---|---|---|---|---|---|
| Thioredoxin-fold proteins | |||||
| Deiodinase (includes thyroid hormone deiodinases 1, 2 and 3 and bacterial homologs) | Reductive deiodination of thyroid hormones (in animals). | CDD1392 | +/+ | −/− | −/+ |
| Unknown function (in bacteria) | |||||
| Glutathione peroxidase (also includes phospholipid hydroperoxide glutathione peroxidase and other homologs) | Reduction of hydroperoxides, UxxT motif | CDD10260 | +/+ | −/− | +/+ |
| CDD25459 | |||||
| Peroxiredoxin (Prx) | Reduction of hydroperoxides, TxxU motif | Derived | +/+ | +/− | +/− |
| Thioredoxin (includes thioredoxin-like proteins, protein disulfide isomerases, DsbA, DsbC, DsbG, DsbE and other homologs) | Reduction, formation or isomerization of disulfide bonds, UxxC motif | Derived | +/+ | +/− | +/+ |
| Glutaredoxin (includes glutaredoxin-like proteins) | Reduction of intramolecular disulfides and mixed disulfide bonds involving gluathione, UxxC, UxxS motifs | Derived | +/+ | +/− | +/− |
| Arsenate reductase | Reduction of arsenate, UxxS motif | CDD17412 | +/+ | −/− | −/− |
| CDD11108 | |||||
| SelU homologs | Function not known, CxxU motif | − | −/− | −/− | +/+ |
| SelT homologs | Function not known, CxxU motif | − | −/− | −/− | +/+ |
| Sep15 (includes Fep15 and SelM) | Function not known, CxxC, CxU/U motif | − | −/− | −/− | +/+ |
| Rdx (includes SelW, SelV, SelT and SelH homologs) | Function not known, CxxU motif | CDD16464 | +/+ | +/− | +/+ |
| CDD12854 | |||||
| Non-thioredoxin fold proteins | |||||
| Methionine-S-sulfoxide reductase (MsrA) | Reduction of methionine-S-sulfoxides | CDD25795 | +/+ | +/− | +/+ |
| Methionine-R-sulfoxide reductase (MsrB) | Reduction of methionine-R-sulfoxides | CDD25798 | +/− | +/− | +/+ |
| Animal thioredoxin reductase (TR) | Reduction of thioredoxins, glutaredoxins and some biofactors | CDD10363 | −/− | −/− | +/+ |
| Bacterial thioredoxin reductase | Reduction of thioredoxins, glutaredoxins and some biofactors | PRK10262 | +/+ | +/− | +/− |
| CMD and AhpD domain-containing proteins | Oxidoreduction | CDD25931 | +/+ | +/− | +/− |
| CDD10469 | |||||
| CDD11836 | |||||
| OsmC-like protein | Thiol peroxidase | CDD25924 | +/+ | +/− | −/− |
| CDD11475 | |||||
| CDD11476 | |||||
| Formylmethanofuran dehydrogenase | Oxidation of formylmethanofuran | CDD29457 | −/− | +/+ | −/− |
| F420-reducing hydrogenase alpha subunit | Hydrogen oxidation or proton reduction | CDD12595 | +/+ | +/+ | −/− |
| F420-reducing hydrogenase, delta subunit | Hydrogen oxidation or proton reduction | CDD10549 | +/+ | +/+ | −/− |
| Methylviologen-reducing hydrogenase | Hydrogen oxidation or proton reduction | − | +/+ | +/+ | −/− |
| NADH oxidoreductase | Possible electron transport | CDD13801 | +/+ | +/− | −/− |
| Formate dehydrogenase alpha chain (FDH) | Oxidation of formate to carbon dioxide | CDD29449 | +/+ | +/+ | −/− |
| Glycine reductase selenoprotein B | Glycine reductase | CDD12612 | +/+ | −/− | −/− |
| SelJ homologs | Possible ADP-ribosylation | − | +/− | −/− | +/+ |
| SelK homologs | Function not known | − | −/− | −/− | +/+ |
| SelS homologs | Translocation of misfolded proteins from the ER to cytosol | − | −/− | −/− | +/+ |
| SelO homologs | Function not known | CDD3203 | +/− | −/− | +/+ |
| A subfamily of selenophosphate synthetase (SPS, SelD) homologs | Synthesis of selenophosphate | CDD10578 | +/+ | +/+ | +/+ |
| A subfamily of SAM-dependent methyltransferases (arsenic methyltransferase) | Arsenic detoxification | CDD10371 | +/+ | +/− | +/− |
| HesB-like protein | Biosynthesis of iron-sulfur clusters | CDD23223 | +/+ | +/+ | −/− |
| Heterodisulfide reductase | Reduction of disulfides/sulfur metabolism | CDD10867 | +/+ | +/+ | −/− |
| Molybdopterin biosynthesis MoeB family proteins | Possible reduction of a disulfide between MoaD and a rhodanese | CDD30111 | +/+ | +/− | +/− |
| CDD10349 | |||||
| CDD30117 | |||||
| A subfamily of glutathione S-transferase homologs | Possible glutathione-dependent oxidoreducase | CDD10495 | +/+ | −/− | +/− |
| Rhodanese-related sulfurtransferase superfamily | Possible redox functions | CDD01448 | +/+ | +/− | +/− |
| CDD01444 | |||||
| CDD01524 | |||||
| CDD00158 | |||||
| DsrE-like protein | Sulfur oxidation/reduction | CDD15459 | +/+ | +/− | −/− |
| CDD11267 | |||||
| Periplasmic substrate-binding protein | Possible iron transport/reduction, CxxU motif | CDD29747 | +/+ | −/− | −/− |
| 5′-nucleotidase/2′,3′-cyclic phosphodiesterase | Possible cyclic phosphodiesterase, CxU motif | CDD10605 | +/+ | −/− | −/− |
| Proline reductase PrdB | Amino acid metabolism | CDD27462 | +/+ | −/− | −/− |
| Ferredoxin-thioredoxin reductase | Ferredoxin-thioredoxin reductase catalytic subunit, CxU motif | CL01977 | +/+ | +/− | +/− |
| Formylmethanofuran dehydrogenase subunit fmdB | Putative regulatory protein, CxxU motif | CD02761 | +/+ | +/− | −/− |
| Gamma-interferon inducible thiol reductase | Lysosomal thiol reductase, UxxC motif | pfam03227 | −/− | −/− | +/+ |
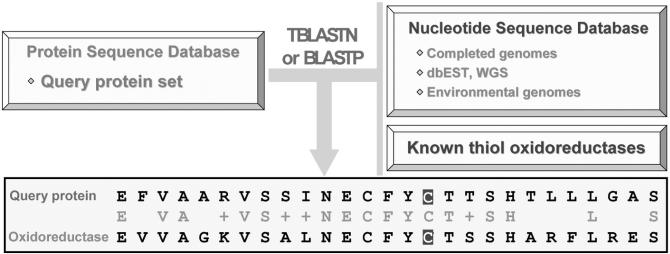
In a real search, a query Cys-containing sequence is analyzed against available nucleotide databases using TBLASTN. This program translates nucleotide sequences in six open reading frames and they are further searched for similarity to proteins in a query protein database, such as NCBI non-redundant database. The output is analyzed for Cys residues corresponding to TGA codons in the translated nucleotide sequences. This analysis is accompanied by searches for SECIS elements (thus avoiding sequencing errors that result in in-frame TGA codons) (Kryukov et al., 2003; Zhang and Glodyshev, 2005). A publicly available SECISearch (genomics.unl.edu/SECISearch.html) is used for the identification of eukaryotic SECIS elements, and a separate tool exists for bacterial structures (http://genomics.unl.edu/bSECISearch/). The presence of both a TGA codon aligning with Cys and a SECIS element in the TGA-containing nucleotide sequence could be used as a strong evidence for the occurrence of a functional Sec and also indicates the corresponding redox-active Cys. Eukaryotic selenoprotein genes contain SECIS elements in the 3′ untranslated regions and prediction of such SECIS elements could be complicated for large genes. A requirement for the occurrence of multiple Sec-containing proteins homologous to a candidate thiol oxidoreductase can further decrease false predictions. A general scheme for the identification of thiol oxidoreductases by similarity of query proteins to selenoproteins as well as to known thiol oxidoreductases is shown in Fig. 3. This combined method detects all thiol oxidoreductases currently known (Table 1). Since most thiol oxidoreductase families have Sec-containing homologs, it is likely that most thiol oxidoreductases which are currently unknown also have homologous selenoproteins. Indeed, there are many proteins with no known function, whose Cys residues align with Sec in selenoproteins, and we predict that many of such proteins are thiol oxidoreductases. Moreover, we expect further growth in the predictive power of the method with an increased output of genomics and metagenomics projects.
Fig. 3.
High-throughput identification of catalytic redox-active Cys residues. The scheme illustrates the method for prediction of redox-active Cys residues by homology to sporadic Sec-containing proteins and known thiol oxidoreductases using TBLAST and BLASTP searches.
Cysteine-based redox motifs
Many thiol-oxidoreductases contain a conserved CxxC motif or motifs derived from it (Chivers et al., 1996; 1997). Typically, the first Cys (attacking residue) occurs in the form of a reactive thiolate and the second Cys (resolving residue) in the redox motif stabilizes the first Cys through hydrogen bond. Properties of the CxxC motif have been studied in detail (Iqbalsyah et al., 2006), which suggested structural and chemical-physical determinants behind its high reactivity (i.e., favorable pKa and redox potentials). The CxxC motif is often present in thioredoxins, glutaredoxins, protein disulfide isomerases and numerous other thiol oxidoreductases. Sometimes, a resolving Cys in the redox motif may be replaced with serine or threonine, which could also stabilize the thiolate. In rare cases, a resolving Cys could be replaced by other amino acids (Fomenko and Gladyshev, 2002; 2003). Most frequently, the attacking Cys is located in the N-terminal position of the redox motif e.g., in thioredoxins, glutaredoxins, glutathione peroxidases, and arsenate reductases. However, a C-terminal position of Cys in the redox motif may also be functional (e.g., in peroxiredoxins).
Secondary structure context of redox-active cysteines
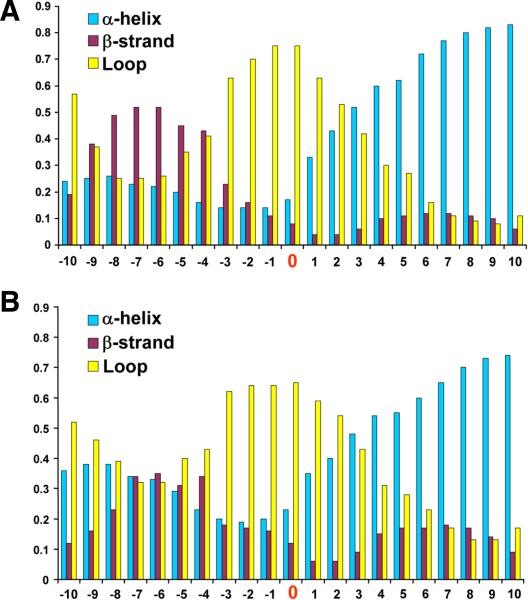
Thiol oxidoreductases are members of diverse protein families which evolved independently, and protein sequence similarity tools alone could not be used for the identification of new thiol oxidoreductases. However, comparison of secondary structure context of redox-active Cys for the majority of known thiol oxidoreductases revealed that β-strands are frequent upstream of the redox-active Cys, and β-helices are frequent downstream of these residues. Redox-active Cys themselves are most frequent in loops (Fig. 4A).
Fig. 4.
Secondary structure context of redox-active Cys residues. (A) Frequency of secondary structures was determined for ten residues upstream and ten residues downstream of the catalytic redox-active Cys (position zero) in the ten-protein sets representing 27 protein families. (B) Secondary structure context was determined for 17 non-thioredoxin fold protein families, each represented by ten proteins. In each case, β-strand is most abundant upstream of the redox-active Cys, α-helix downstream of this residue and the redox active Cys itself is most often located in loops.
A thioredoxin-fold superfamily represents approximately half (if not more) of known thiol oxidoreductases, and therefore it is of interest to determine if the observed secondary structure context of catalytic Cys also holds for other protein folds. Fig. 4B shows the occurrence of secondary structure elements in non-thioredoxin fold thiol oxidoreductases, and the general trend is similar to that for thioredoxins. The downstream α-helix might stabilize the reactive thiolate and could be used as an additional predictor of oxidoreductase function (Kortemme and Creighton, 1995). In contrast, most metal-binding CxxC motifs lack this property (Fomenko and Gladyshev, unpublished). Detailed structure-based analysis of the CxxC motif in thiol oxidoreductases has been reported that pointed to strong local secondary structure effects with positive influence on the reactivity of catalytic Cys (Iqbalsyah et al., 2006; Moutevelis and Warwicker, 2004).
Recent progress in structural bioinformatics led to increased accuracy of secondary structure analysis. There are numerous secondary structure prediction tools available on the web, such as at http://ca.expasy.org/tools/#secondary. For example, PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) is an effective secondary structure prediction method, which is based on sequence alignment outputs obtained from PSI-BLAST (Jones, 1999). The use of more than one method for secondary structure prediction is encouraged and often will result in more accurate prediction. For example, simultaneous use of PSIPRED and PROFSEC (http://www.predictprotein.org/) is routinely carried out by some automated methods in the CAFASP blind tests (http://www.cs.bgu.ac.il/~dfischer/CAFASP5/). In addition, along with the secondary structure predictive tools, independent methods specifically evaluating disordered regions in proteins, such as DISOPRED (http://bioinf.cs.ucl.ac.uk/disopred/) could be employed to exclude highly unstructured regions (Jones and Ward, 2003) which, for the reasons described before, are less likely to contain a catalytic redox-active Cys.
Structure modeling
Redox-active Cys in thiol oxidoreductases should be accessible for interactions with redox partners. This consideration may result in improved accuracy of predictions when searches are carried out at the level of protein structures. Protein structure could be modeled based on sequence similarity to proteins with already known structures. One popular 3D structure prediction tool is Structure Prediction Meta Server (http://bioinfo.pl/meta), which combines various independent methods for homology modeling, fold recognition and local structure prediction. This server is indeed particularly useful when analyzing proteins sharing low similarity with known structures. In simpler cases, a faster yet effective homology modeling methods such as Swiss model (http://swissmodel.expasy.org/SWISS-MODEL.html) or MODELER (http://www.salilab.org/modeller/) could be used for structure prediction.
Structure modeling may help filter out structural Cys and, often, metal-coordinating Cys. One of the structural methods, Fuzzy Functional Forms (FFF), was used for prediction of thioredoxin-fold proteins (Fetrow et al., 2001). This method employs three-dimensional structure information to identify functional sites in proteins. A similar structural approach has been recently applied to active site functional profiling for the identification of common properties of catalytic sites (Cammer et al., 2003) and redox-active Cys residues (Salsbury et al., 2008).
Comparative sequence analysis of thiol-based oxidoreductases
As discussed above, thiol oxidoreductases could be predicted by sequence similarity to known thiol oxidoreductases and selenoproteins (Fig. 3). In addition, comparative genomics may help in annotation and functional characterization of thiol oxidoreductases. STRING (http://string.embl.de/) (von Mering et al., 2007) is a search tool for functional associations of proteins; it features a precalculated database containing over 1,5 million proteins from 373 species. This tool combines information from gene neighborhoods in completely sequenced genomes, domain fusion and protein co-occurrence in phylogenetic profiles and can be used for prediction of pathways in which thiol oxidoreductases are involved, prediction of their interacting partners and substrates, and for the identification of evolutionary relationships among different thiol oxidoreductases.
CONCLUDING REMARKS
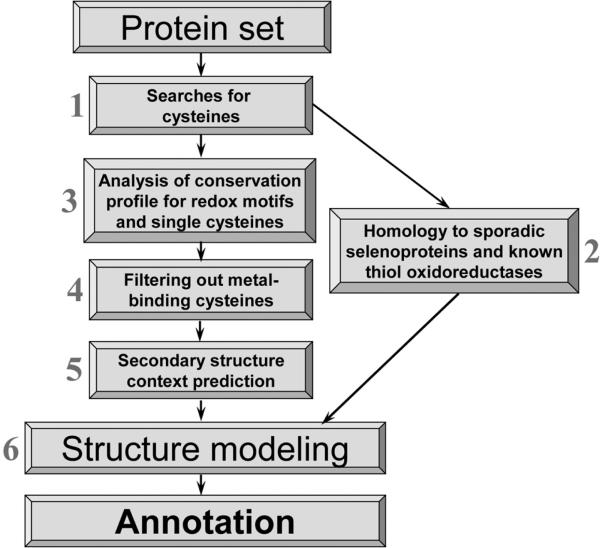
In this review, we analyzed various groups of functional Cys residues and described computational methods that could predict specifically thiol oxidoreductases and their catalytic redox-active Cys. These proteins are directly involved in thiol-based redox regulation and act as key components of thiol networks in cells. High divergence and lack of evolutionary relationships among many thiol oxidoreductase families make traditional methods of protein function prediction inefficient for these proteins. An overall procedure for prediction for redox-active Cys is shown in Fig. 5. Most steps in the procedure can be performed automatically. Query proteins may be viewed as candidate thiol oxidoreductases if they pass step 2 or steps 3−6. Significant progress in genomics and metagenomics projects during the last several years resulted in a dramatic growth of sequence databases, and we therefore expect further improvements in the predictive power of the described methods.
Fig. 5.
Overall procedure for the identification of thiol oxidorductases. The algorithm consists of six steps. A protein is viewed as a candidate thiol oxidoreductase if Cys/Sec pair is identified (step 2) or the protein passes all other steps (steps 3−6). In the latter case, proteins are identified whose Cys residues are conserved, not involved in metal binding, contain a downstream α-helix, and are accessible for interactions with redox partners.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, Ko HS, Sasaki M, Ischiropoulos H, Przedborski S, et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. USA. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo D, Tacnet F, Delaunay A, Rodrigues-Pousada C, Toledano MB. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 2003;35:889–900. doi: 10.1016/s0891-5849(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Beeby M, O'Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, Yeates TO. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005;3:e309. doi: 10.1371/journal.pbio.0030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem. Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Bock A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: An expansion of the genetic code. Trends Biochem. Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Sultana R, Scapagnini G, Guagliano E, Sapienza M, Bella R, Kanski J, Pennisi G, Mancuso C, Stella AM, et al. Nitrosative stress, cellular stress response, and thiol homeostasis in patients with Alzheimer's disease. Antioxid. Redox Signal. 2006;8:1975–1986. doi: 10.1089/ars.2006.8.1975. [DOI] [PubMed] [Google Scholar]
- Cammer SA, Hoffman BT, Speir JA, Canady MA, Nelson MR, Knutson S, Gallina M, Baxter SM, Fetrow JS. Structure-based active site profiles for genome analysis and functional family subclassification. J. Mol. Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Pique ME. MDB: the metalloprotein database and browser at the scripps research institute. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- Collet JF, Bardwell JC. Oxidative protein folding in bacteria. Mol. Microbiol. 2002;44:1–8. doi: 10.1046/j.1365-2958.2002.02851.x. [DOI] [PubMed] [Google Scholar]
- Conway ME, Poole LB, Hutson SM. Roles for cysteine residues in the regulatory CXXC motif of human mitochondrial branched chain aminotransferase enzyme. Biochemistry. 2004;43:7356–7364. doi: 10.1021/bi0498050. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. S-glutathionylation in protein redox regulation. Free Radic. Biol. Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Milzani A, Gagliano N, Colombo R, Giustarini D, Rossi R. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxid. Redox Signal. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redoxtransducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Fermani S, Sparla F, Falini G, Martelli PL, Casadio R, Pupillo P, Ripamonti A, Trost P. Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. USA. 2007;104:11109–11114. doi: 10.1073/pnas.0611636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetrow JS, Skolnick J. Method for prediction of protein function from sequence using the sequence-to-structure-to-function paradigm with application to glutaredoxins/thioredoxins and T1 ribonucleases. J. Mol. Biol. 1998;281:949–968. doi: 10.1006/jmbi.1998.1993. [DOI] [PubMed] [Google Scholar]
- Fetrow JS, Godzik A, Skolnick J. Functional analysis of the Escherichia coli genome using the sequence-to-structure-to-function paradigm: identification of proteins exhibiting the glutaredoxin/thioredoxin disulfide oxidoreductase activity. J. Mol. Biol. 1998;282:703–711. doi: 10.1006/jmbi.1998.2061. [DOI] [PubMed] [Google Scholar]
- Fetrow JS, Siew N, Di Gennaro JA, Martinez-Yamout M, Dyson HJ, Skolnick J. Genomic-scale comparison of sequence- and structure-based methods of function prediction: Does structure provide additional insight? Protein Sci. 2001;10:1005–1014. doi: 10.1110/ps.49201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko DE, Gladyshev VN. CxxS: fold-inependent redox motif revealed by genome-wide searches for thiol/disulfide oxidoreductase function. Protein Sci. 2002;11:2285–2296. doi: 10.1110/ps.0218302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko DE, Gladyshev VN. Identity and functions of CxxC-derived motifs. Biochemistry. 2003;42:11214–11225. doi: 10.1021/bi034459s. [DOI] [PubMed] [Google Scholar]
- Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-Throughput Identification of Catalytic Redox-Active Cysteine Residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinform. 2002;1:107–108. [PubMed] [Google Scholar]
- Giles NM, Watts AB, Giles GI, Fry FH, Littlechild JA, Jacob C. Metal and redox modulation of cysteine protein function. Chem. Biol. 2003;10:677–693. doi: 10.1016/s1074-5521(03)00174-1. [DOI] [PubMed] [Google Scholar]
- Gladyshev VN, Kryukov GV, Fomenko DE, Hatfield DL. Identification of trace element-containing proteins in genomic databases. Annu. Rev. Nutr. 2004;24:579–596. doi: 10.1146/annurev.nutr.24.012003.132241. [DOI] [PubMed] [Google Scholar]
- Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook DW, Harding JJ. Inactivation of glyceraldehyde 3-phosphate dehydrogenase by sugars, prednisolone-21-hemisuccinate, cyanate and other small molecules. Biochim. Biophys. Acta. 1997;1362:232–242. doi: 10.1016/s0925-4439(97)00084-7. [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Giles GI, Giles NM, Sies H. Sulfur and selenium: The role of oxidation state in protein structure and function. Angew. Chem. Int. Ed. 2003;42:4742–4758. doi: 10.1002/anie.200300573. [DOI] [PubMed] [Google Scholar]
- Jakob U, Eser M, Bardwell JC. Redox switch of hsp33 has a novel zinc-binding motif. J. Biol. Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrixes. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53:573–578. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- Juarez JC, Manuia M, Burnett ME, Betancourt O, Boivin B, Shaw DE, Tonks NK, Mazar AP, Doñate F. Su-peroxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. USA. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbalsyah TM, Moutevelis E, Warwicker J, Errington N, Doig AJ. The CXXC motif at the N terminus of an alpha-helical peptide. Protein Sci. 2006;15:1945–1950. doi: 10.1110/ps.062271506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model alpha-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- Martin JL. Thioredoxin - fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Matés JM, Segura JA, Alonso FJ, Márquez J. Intracellular redox status and oxidative stress: implications for cell proliferation, apoptosis, and carcinogenesis. Arch. Toxicol. 2008;82:273–299. doi: 10.1007/s00204-008-0304-z. [DOI] [PubMed] [Google Scholar]
- Moutevelis E, Warwicker J. Prediction of pKa and redox properties in the thioredoxin superfamily. Protein Sci. 2004;13:2744–2752. doi: 10.1110/ps.04804504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, Klein JB, Turner DM, Butterfield DA. An increase in S-glutathionylated proteins in the Alzheimer's disease inferior parietal lobule, a proteomics approach. J. Neurosci. Res. 2007;85:1506–1514. doi: 10.1002/jnr.21275. [DOI] [PubMed] [Google Scholar]
- Pedone E, Limauro D, Bartolucci S. The machinery for oxidative protein folding in thermophiles. Antioxid Redox Signal. 2008;10:157–169. doi: 10.1089/ars.2007.1855. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE. 2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- Ridge PG, Zhang Y, Gladyshev VN. Comparative genomic analyses of copper transporters and cupropro-teomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS ONE. 2008;3:e1378. doi: 10.1371/journal.pone.0001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- Salsbury FR, Jr., Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Riddle M, Woo J, Momand J. Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Protein Sci. 2008;17:473–481. doi: 10.1110/ps.073252408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman TC. Selenocysteine. Annu. Rev. Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Sun J, Steenbergen C, Murphy E. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal EA, Findlay VJ, Day AM, Bozonet SM, Evans JM, Quinn J, Morgan BA. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol. Cell. 2004;15:129–139. doi: 10.1016/j.molcel.2004.06.021. [DOI] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. STRING 7-recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassef R, Haenold R, Hansel A, Brot N, Heinemann SH, Hoshi T. Methionine sulfoxide reductase A and a dietary supplement S-methyl-L-cysteine prevent Parkinson's-like symptoms. J. Neurosci. 2007;27:12808–12816. doi: 10.1523/JNEUROSCI.0322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessjohann LA, Schneider A, Abbas M, Brandt W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 2007;388:997–1006. doi: 10.1515/BC.2007.138. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- Wood MJ, Storz G, Tjandra N. Structural basis for redox regulation of Yap1 transcription factor localization. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. An algorithm for identifycation of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. Trends in selenium utilization in marine microbial world revealed through the analysis of the global ocean sampling (GOS) project. PLoS Genet. 2008a;4:e1000095. doi: 10.1371/journal.pgen.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008b;379:881–899. doi: 10.1016/j.jmb.2008.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]