Abstract
Progressive hearing loss is common in the human population, but little is known about the molecular basis. We report a new ENU-induced mouse mutant, diminuendo, with a single base change in the seed region of Mirn96. Heterozygotes show progressive loss of hearing and hair cell anomalies, while homozygotes have no cochlear responses. Most microRNAs are believed to downregulate target genes by binding to specific sites on their mRNAs, so mutation of the seed should lead to target gene upregulation. Microarray analysis revealed 96 transcripts with significantly altered expression in homozygotes; notably, Slc26a5, oncomodulin, Gfi1, Ptprq and Pitpnm1 were downregulated. Hypergeometric p-value analysis showed hundreds of genes were upregulated in mutants. Different genes, with target sites complementary to the mutant seed, were downregulated. This is the first microRNA found associated with deafness, and diminuendo represents a model for understanding and potentially moderating progressive hair cell degeneration in hearing loss more generally.
Progressive hearing loss is common in the human population. About one in 850 children are born with a significant, permanent hearing impairment, but by the age of ten this number has doubled1. Age-related hearing loss in later life has a heritability approaching 50%2, and some single genes have been identified underlying progressive hearing loss in rare families (Hereditary Hearing Loss Homepage; http://webho1.ua.ac.be/hhh/). However, for the vast majority of cases of progressive hearing loss there is no molecular diagnosis. To provide candidate genes and models for hearing loss, we established a screen for new ENU-induced deaf mouse mutants3. One such mutant recovered was diminuendo (Dmdo), inherited in a semi-dominant manner. Heterozygotes (Dmdo/+) show a progressive loss of the Preyer reflex (ear flick response to sound) between 4 and 6 weeks. Homozygotes (Dmdo/Dmdo) do not demonstrate a Preyer reflex at any age, and show head bobbing and a staggering, circling gait.
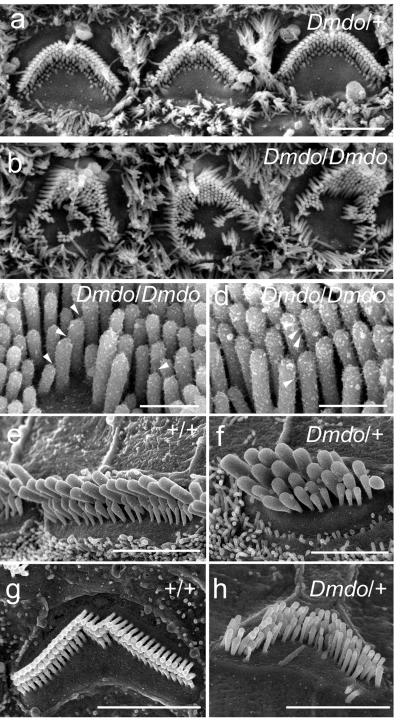
The gross structure of the middle and inner ears appeared normal in mutants so we examined the organ of Corti using scanning electron microscopy. At 4-5 days after birth, the number and arrangement of hair cells appeared normal in mutants, and heterozygotes looked similar to wildtype littermates (Fig 1a, S1a,b). However, irregular bundles and persistent clusters of ectopic stereocilia were observed in homozygotes (Fig. 1b-d) and by 7 days, homozygote hair cells displayed marked degeneration (data not shown). At four and six weeks old, very few recognisable hair cells remained in homozygotes (Fig. S1f,k). In heterozygotes many outer hair cells had degenerated in the middle and basal turns but most inner hair cells remained intact (Fig S1l-o). Remaining outer hair cell stereocilia bundles formed a loose U-shape rather than the precise V-shape of controls, and inner hair cells often showed smaller, more widely-separated bundles (Fig. 1e,f,g,h; S1e,h,j,p,q). Compound action potentials, reflecting cochlear nerve activity in response to sound, were undetectable in homozygotes at 4 weeks, and in heterozygotes thresholds were raised by around 60dB (Fig. S1r), despite the persistence of many surviving hair cells. Endocochlear potentials were within the normal range (Fig. S1s). Fused stereocilia and hair cell degeneration were evident in the vestibular systems of four week-old heterozygotes.
Figure 1. Scanning electron micrographs of diminuendo inner ear.
a, the heterozygote (Dmdo/+), and b, the homozygote (Dmdo/Dmdo) at postnatal day 5, showing irregular hair bundles and ectopic stereocilia. Scale bars = 2 μm. c, d, Stereocilia in homozygotes at P4 showing tip links (c; arrowheads) and lateral links (d; arrowheads). Scale bars = 500nm. e-h, Stereocilia bundles of inner hair cells (e, f) and outer hair cells (g, h) in heterozygotes (f, h) and wildtype littermates (e, g) at postnatal day 28. Scale bars = 3μm (e, g, h) and 2μm (f). Heterozygous outer hair cells (h) show irregular stereocilia bundles, a smaller apical surface, and are more widely spaced than in wildtypes. Inner hair cells also appear to be more widely separated, and show smaller bundles with fewer stereocilia organised in 4-5 rows (f). All Dmdo/+ stereocilia have rounded tips.
We mapped the mutant phenotype to proximal chromosome 6, between D6Mit159 and D6Mit268, a 4.96Mb interval (Fig S2a,b). We sequenced 87% of the ~900 exons within the interval (http://www.ensembl.org), and located two mutations. The first mutation was a silent C>T substitution in exon 5 of 2310005E10Rik, a member of the aldo-keto reductase family and a putative orthologue of the human gene AKR1B10. The mutated base is the third in the codon, and the amino acid, asparagine, is not affected. The variant in the Dmdo genome is identical to the equivalent wildtype human reference sequence. Normalised cDNA from the organs of Corti of three P4 Dmdo/Dmdo and +/+ sibling pairs gave bands of identical size and intensity when subjected to PCR with primers in exons 4 and 6. We concluded that this variant was unlikely to be involved in causing the hearing impairment in the diminuendo mutant (Fig S2c,d).
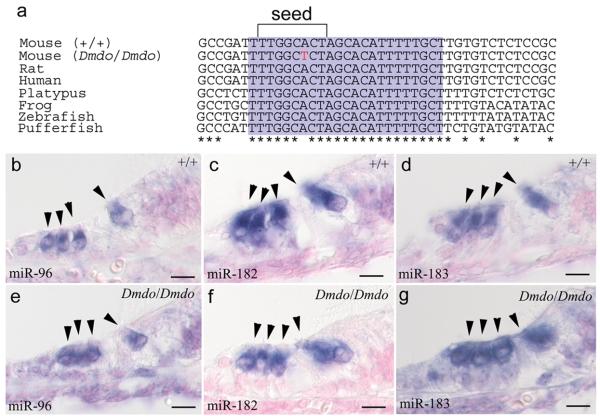
The second mutation was an A>T substitution in Mirn96 (mmu-miR-96; miR-96), a microRNA (miRNA). MicroRNAs are small non-coding RNAs that bind to specific sites in the 3′UTR of target mRNAs, inducing transcript destabilisation and translational inhibition. The mutation lies within the miRNA seed region (nucleotides 2-7), which confers binding specificity4, making this variant a strong candidate for the causative mutation. The sequence of mature miR-96 is perfectly conserved between human, mouse, rat and fish (Fig. 2a).
Figure 2. miR-96, miR-182 and miR-183 in littermates at postnatal day 5.
a, Alignment of DNA sequences from wildtype mouse, diminuendo homozygote, rat, human, platypus, frog, zebrafish and pufferfish. The mature miRNA sequence for each species is shaded purple, and the seed region critical for target binding is bracketed. The mutation, indicated by the red letter, falls within the seed region. The mature sequence is absolutely conserved between the species shown, and also between cow, dog, horse, macaque, opossum, chimpanzee, orang-utan, ground squirrel, tree shrew, mouse lemur, bushbaby, cat, armadillo, tenrec, medaka, rabbit, stickleback and tetraodon (sequences obtained from Ensembl v50; http://www.ensembl.org). b, e, expression of miR-96 in wildtype (b) and homozygote (e). c, f, expression of miR-182 in wildtype (c) and homozygote (f). d, g, expression of miR-183 in wildtype (d) and homozygote (g). No specific staining was observed using the control probe (data not shown). Probes designed against the mature miRNA sequence have been shown incapable of detecting the precursor transcript29, so these show the location of only the mature miRNA. Hair cells are marked by arrowheads. Scale bars = 10μm.
Mirn96 is one of a cluster of three miRNAs; the other two are Mirn183 and Mirn182. These miRNAs are expressed in all sensory hair cells of the inner ear and in neurons of the cochlear and vestibular ganglia in newborn mice, and are thought to be processed from a single primary transcript5. We found that all three mature miRNAs were expressed in hair cells of both wildtypes and homozygous mutants (Fig. 2b-g), suggesting the mutation does not have a major effect on processing and export of the miRNAs to the cytoplasm despite the possible interference of the mutation with stem loop formation required for cleavage of the transcript6.
We chose two complementary approaches to study the effects of the mutation. Firstly, we used the miRanda target predictor v3.07with additional filtering to produce a list of 132 genes which we annotated (Supplementary Table 1) and selected thirteen for further study (Supplementary Table 2). We used a luciferase assay with siRNAs mimicking the wildtype and mutant miR-96. The siRNAs were co-transfected into NIH 3T3 cells with a construct containing the 3′UTR of each candidate gene, either with the putative binding sites intact, or with the sites replaced by an EcoR1 site to disrupt binding. Five genes were validated as targets of miR-96: Aqp5, Celsr2, Myrip, Odf2 and Ryk (Fig. S3a,b). Quantitative RTPCR showed Aqp5 and Celsr2 were significantly upregulated in mutant cochlear tissues compared with wildtype. However, the difference in expression levels was small (Fig. S3c). We used antibodies against the validated targets and found all five were expressed in or near wildtype hair cells at P3 and P5, but there was no visible difference in diminuendo (Fig S3d-m and data not shown). However, miRNAs may have multiple small effects on the expression levels of a number of genes8 and immunohistochemical tests may not show such small effects. Therefore, we adopted a genome-wide approach to investigate the mechanism of action of the mutation.
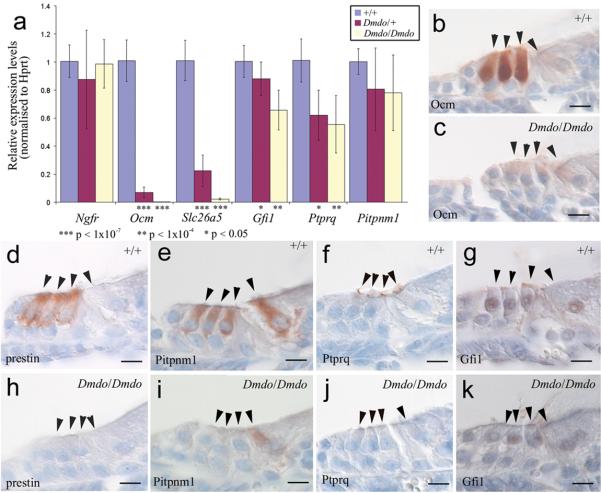
We compared gene expression of both direct and indirect targets by microarray analysis of the organ of Corti of P4 mutants and wildtypes. We retrieved 96 significantly affected transcripts (P-value<0.05); 50 genes were up-regulated and 36 down-regulated (Supplementary Table 3; the remaining 10 probes were either duplicates (6) or mapped to intergenic regions (4)). Thirteen of these so far have been confirmed by qRTPCR (Fig. S4a). Of the downregulated genes, five in particular were of interest; Slc26a5 (prestin), Ocm (oncomodulin), Pitpnm1, Gfi1 and Ptprq. None of these genes have a predicted wildtype or mutant miR-96 target site, so the downregulation is presumably a downstream effect. Prestin is a voltage-sensitive motor molecule that mediates outer hair cell length changes responsible for amplification of sound within the cochlea9. Prestin knockout mice have short hair cells and hair cell degeneration10. Oncomodulin is expressed in outer hair cells and may act as a cytosolic calcium ion buffer11. Ptprq is required for maturation of the hair bundle, and is thought to be a component of interstereocilial shaft connectors12. Gfi1 is expressed in hair cells, and knockout mice display hair cell degeneration13. The difference in expression of these genes was confirmed by qRTPCR in both heterozygotes and homozygotes (Fig. 3a) and by immunohistochemistry (Fig. 3b-k). We found no evidence of genomic changes that might account for the extreme downregulation of Ocm and Slc26a5: exon 1 of each gene amplified correctly in mutants, surrounding genes were expressed normally in the microarray, and the diminuendo phenotype showed Mendelian inheritance and mapped to the Mirn96 locus. Epigenetic downregulation of any one of these five genes could explain the hearing impairment, as three are known to lead to deafness when knocked out and the remaining two are highly expressed in sensory hair cells.
Figure 3. Ocm, Slc26a5, Pitpnm1, Ptpr1 and Gfi1 expression in diminuendo.
a, Quantitative real-time PCR on cDNA generated from normalised RNA from the organs of Corti of 4 day old littermates. Ocm, Slc26a5, Ptprq and Gfi1 are downregulated in heterozygotes and homozygotes. Error bars represent standard deviation. Quantities normalised to Hprt1 levels; Ngfr is expressed in support cells adjacent to hair cells30 and was used to assess the quantity of sensory material. Ngfr: Wildtype, n=12, mean=1.01±0.12 (s.d.); heterozygote, n=12, mean=0.89±0.35 (s.d.); homozygote, n=12, mean=0.99±0.17 (s.d.) Ocm: Wildtype, n=9, mean=1.01±0.15 (s.d.); heterozygote, n=9, mean=0.07±0.04 (s.d.); homozygote, n=9, mean=0.003±0.001 (s.d.) Slc26a5: Wildtype, n=9, mean=1.01±0.14 (s.d.); heterozygote, n=9, mean=0.22±0.11 (s.d.); homozygote, n=9, mean=0.02±0.01 (s.d.) Gfi1: Wildtype, n=9, mean=1.01±0.12 (s.d.); heterozygote, n=9, mean=0.88±0.12 (s.d.); homozygote, n=9, mean=0.66±0.14 (s.d.) Ptprq: Wildtype, n=9, mean=1.01±0.15 (s.d.); heterozygote, n=9, mean=0.62±0.18 (s.d.); homozygote, n=9, mean=0.56±0.20 (s.d.) Pitpnm1: Wildtype n=8, mean=1.00±0.09 (s.d.); heterozygote, n=9, mean=0.80±0.29 (s.d.); homozygote, n=9, mean=0.78±0.27 (s.d.). Three animals were used for each genotype and DNA from each was run in triplicate. T-tests: Ngfr heterozygote p=0.25 (Welch's t-test), homozygote p=0.75 (Student's t-test); Ocm heterozygote p=1.51×10−8 (Welch's t-test), homozygote p=3.46×10−8 (Welch's t-test); Slc26a5 heterozygote p=7.73×10−10 (Student's t-test), homozygote p=3.37×10−8 (Welch's t-test); Gfi1 heterozygote p=0.038 (Student's t-test), homozygote p=3.39×10−5 (Student's t-test); Ptprq heterozygote p=1.37×10−4 (Student's t-test), homozygote p=6.46×10−5 (Student's t-test); Pitpnm1 heterozygote p=0.084 (Welch's t-test), homozygote p=0.35 (Student's t-test); α=0.05. b-k, location of oncomodulin (b, c), prestin (d, h), Pitpnm1 (e, i), Ptprq (f, j) and Gfi1 (g, k) in 5-day old wildtype (b, d-g) and homozygote (c, h-k) littermates. Scale bars = 10μm.
We asked whether the striking downregulation of oncomodulin and prestin was a generic feature of degenerating hair cells by looking at immunostaining intensity in nine other mouse mutants which exhibit early hair cell degeneration: headbanger and shaker14626SB (Myo7a)14,15, Snell's waltzer (Myo6) 16, headturner (Jag1)17, beethoven and deafness (Tmc1) 18,19, oblivion (Atp2b2) 20, catweasel (Six1)21 and whirler (Whrn)22. Oncomodulin and prestin showed hair cell labelling in mutants as strong as in the littermate controls (Fig. S5a-p and data not shown), suggesting that the reduction of expression in diminuendo was a specific feature. Furthermore, other markers of hair cells and supporting cells of the organ of Corti showed normal immunostaining intensity in diminuendo mutants at P0, P3 and P5, including Myo7a, Cdkn1b (p27Kip1), Sox2 and Jag1 (Fig S5q-y and data not shown).
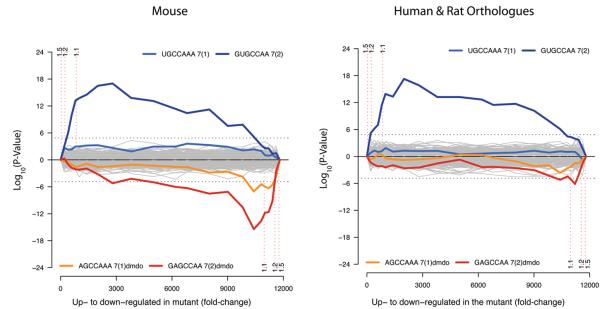
We next searched for wider miRNA effects on the mRNA profile of diminuendo using Sylamer23. Analysis of all miRNA heptamers shows that the heptamer complementary to the seed region of miR-96 (GUGCCAA) is greatly enriched in the 3′UTRs of hundreds of genes upregulated in diminuendo homozygotes (Fig. 4). This indicates that miR-96 normally modulates expression of a broad range of target genes, and that it affects mRNA levels rather than affecting translation alone. Among the most downregulated genes, the heptamer complementary to the mutant miR-96 is enriched (GAGCCAA, Fig. 4), indicating that mutant miR-96 influences expression of newly-acquired target genes. We analysed conservation of these signals. Wildtype seed matches are enriched in 3′UTRs of human and rat orthologues of the most upregulated mouse genes (Fig 4), suggesting that these sites are conserved and may be functional. However, enrichment of mutant miR-96 binding sites in human and rat orthologues of downregulated genes is barely above significance threshold (dotted line). We also employed a simple log ratio analysis of seed match biases in gene sets at different cutoffs (Fig S4b-j). These results agreed with the Sylamer analysis.
Figure 4. Microarray analysis showing enrichment and depletion of heptamers in 3′UTRs.
a, Microarray analysis showing enrichment and depletion of heptamers in 3′UTRs using Sylamer23. The x-axis represents the sorted gene list from most up-regulated (left) to most down-regulated (right). The y-axis shows the hypergeometric significance for enrichment or depletion of heptamers in 3′UTRs at leading parts of the gene list. Positive values indicate enrichment (−log10(P-value)) and negative values depletion (log10(P-value)). Heptamers that are depleted in the initial part are accordingly enriched in the complementary part with the same P-value, as a consequence of the hypergeometric distribution. For each miRNA (including the diminuendo mutant miR-96) the two heptamers matching the 5′ seed region, starting at positions 1 and 2, were considered. Each heptamer was tested at regularly placed rank cutoffs in the gene list. The P-value indicates the significance of the enrichment or depletion of the heptamer in the set of 3′UTRs in the initial part of the gene list when compared to the 3′UTRs in the complementary set. The horizontal dotted lines represent an E-value threshold (P-value corrected for multiple testing) of 0.01. Vertical dotted lines indicate Fold Change cutoffs of >1.5, >1.2, and >1.1, and the parts of the gene lists defined by these cutoffs. b, The same analysis as in (a), where each 3′UTR has been replaced by the concatenation of its orthologous 3′UTRs in Human and Rat. The seed match for the wild type miR-96 shows similar enrichment as compared with the analysis in (a). In contrast, the enrichment of the miR-96 diminuendo mutant binding sites in the down-regulated genes is barely above background (dotted line).
To elucidate the link between the mutated microRNA and the abnormally-expressed genes revealed by the microarray analysis, we examined the 500bp upstream of the top 356 upregulated genes and the top 425 downregulated genes to find any transcription factor motifs that were enriched in the affected genes (Fig S4k-n). Several interesting motifs were found, including a Gfi1-like motif found among the upregulated genes; a binding site for Mitf, known to be regulated by miR-96 24; and targets associated with control of Notch signalling, such as Pou2f125, Rbpj26, and bHLH transcription factors27. Any one of the transcription factor binding motifs discovered could be involved in linking the mutation in miR-96 to the misregulation of genes directly required for hair cell development and survival. The large number of genes whose expression is affected by miR-96 and the complexity of their interactions suggests that there may not be a simple mechanism that explains the effects of the mutation, but rather a combination of many small effects that act in concert to cause hair cell dysfunction.
The diminuendo mutant shows progressive hearing impairment in heterozygotes and profound deafness in homozygotes associated with hair cell defects. Although hearing impairment is often thought to be caused by hair cell loss, this and previous studies suggest that the degeneration is instead a correlate or consequence of a prior dysfunction of the hair cells. The mutation in the seed region of miR-96 is highly likely to cause the hearing impairment in diminuendo mutants because it co-segregates with the phenotype, it occurs in the seed region of the miRNA known to be critical for target recognition, miR-96 is expressed specifically in the cells most affected by the mutation, Sylamer analysis indicated that the mutation has a direct effect on expression of many genes as well as indirect effects, and we found no other plausible mutation despite resequencing the vast majority of the coding sequence within the non-recombinant region of chromosome 6. In addition, the finding of two different single base changes in the seed region of miR-96 in humans with progressive hearing loss in the accompanying report28 provides critical support for our proposal that the single base change in miR-96 is the causative mutation behind the diminuendo phenotype, and furthermore suggests that the phenotype results from a lack of repression of normal targets even though we show a gain of repression of novel targets. Although the link between the direct targets of the miRNA and the phenotype are not clear, we have shown that several genes known to be important for hair cell function are specifically downregulated in the diminuendo mutant and any one could account for the hair cell dysfunction. This is the first ENU-induced mutation found in a miRNA and the first miRNA found to be associated with deafness. Understanding the mechanism by which miR-96 leads to progressive hearing loss will give us clues to help develop therapies to ameliorate the effects of progressive deafness, whatever the trigger.
Methods Summary
The diminuendo mutant was recovered from a screen for new dominantly-inherited mutations using N-ethyl-N-nitrosurea (ENU) as a mutagen. Electron microscopy was carried out on inner ears of mice at 4 and 5 days old, and four and six weeks old, using the OTOTO method. Compound action potentials and endocochlear potentials were recorded using standard techniques. The mutation was localised using a backcross and genome scan, followed by sequencing of the exons within the non-recombinant region.
RNA Extraction
The organs of Corti of four-day-old mice were dissected and stored at −20°C in RNAlater stabilisation reagent (QIAgen, cat. no. 76106). RNA was extracted using QIAshredder columns (QIAgen, cat. no. 79654) and the RNeasy mini kit (QIAgen, cat. no. 74104), following the manufacturer's instructions.
Expression Analysis
Pups were collected on the day they were born, three, or five days after birth. Animals were dissected in ice cold PBS, fixed for two days in 10% formalin at 4°C, embedded in paraffin and cut into 8μm sections. Immunohistochemistry and in situ hybridisations were carried out using the Ventana Discovery machines and reagents according to the manufacturer's instructions. From each animal, at least five sections were used per probe or antibody. Quantitative RT-PCR was carried out on cDNA from normalised organ of Corti RNA, using reagents from Applied Biosystems.
Supplementary Material
Acknowledgements
We thank Peter Ellis, Keith James and Robert Andrews for assistance with the microarrays, Huw Williams for help with the quantitative RT-PCR, Antony Rodriguez for helpful discussions on miRNAs, Amy Taylor for help with bioinformatics, Sarah Rose, Michael Drummond, Kelvin Hawker and Angela Lucas for help with mapping the mutation, Fei Zhu for her work on Gfi1 and Ptprq, Rosalind Lacey and Dawn Savage for animal care, Aziz El-Amraoui and Christine Petit for the anti-MyRIP antibody, Guy Richardson for the anti-Ptprq antibody, and Agnieszka Rzadzinska for high resolution scanning electron microscopy analysis. This work was supported by Deafness Research UK, the Wellcome Trust, Medical Research Council (UK), EC (CT-97-2715, LSHG-CT-2004-512063), Spanish Ministerio de Ciencia y Tecnología (SAF2005-06355), Spanish Fondo de Investigaciones Sanitarias (FIS PI-051942; G03/203, CP03/00014, PI08/0045), NGFN (Germany), and charitable donations from individuals affected by deafness.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests. The authors declare no competing financial interests.
References
- 1.Fortnum HM, Summerfield AQ, Marshall DH, Davis AC, Bamford JM. Prevalence of permanent childhood hearing impairment in the United Kingdom and implications for universal neonatal hearing screening: questionnaire based ascertainment study. Bmj. 2001;323:536–40. doi: 10.1136/bmj.323.7312.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gates GA, Couropmitree NN, Myers RH. Genetic associations in age-related hearing thresholds. Arch Otolaryngol Head Neck Surg. 1999;125:654–9. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- 3.Hrabe de Angelis M, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nature Genetics. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Research. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–31. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 7.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dallos P, et al. Prestin-based outer hair cell motility is necessary for mammalian cochlear amplification. Neuron. 2008;58:333–339. doi: 10.1016/j.neuron.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liberman MC, et al. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature. 2002;419:300–304. doi: 10.1038/nature01059. [DOI] [PubMed] [Google Scholar]
- 11.Sakaguchi N, Henzl MT, Thalmann I, Thalmann R, Schulte BA. Oncomodulin is expressed exclusively by outer hair cells in the organ of Corti. Journal of Histochemistry & Cytochemistry. 1998;46:29–39. doi: 10.1177/002215549804600105. [DOI] [PubMed] [Google Scholar]
- 12.Goodyear RJ, et al. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci. 2003;23:9208–9219. doi: 10.1523/JNEUROSCI.23-27-09208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallis D, et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- 14.Holme RH, Steel KP. Stereocilia defects in waltzer (Cdh23), shaker1 (Myo7a) and double waltzer/shaker1 mutant mice. Hear. Res. 2002;169:13–23. doi: 10.1016/s0378-5955(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes CR, et al. A Myo7a mutation cosegregates with stereocilia defects and low-frequency hearing impairment. Mammalian Genome. 2004;15:686–697. doi: 10.1007/s00335-004-2344-x. [DOI] [PubMed] [Google Scholar]
- 16.Self T, et al. Role of myosin VI in the differentiation of cochlear hair cells. Developmental Biology. 1999;214:331–341. doi: 10.1006/dbio.1999.9424. [DOI] [PubMed] [Google Scholar]
- 17.Kiernan AE, et al. The Notch ligand Jagged1 is required for inner ear sensory development; Proceedings of the National Academy of Sciences of the United States of America; 2001. pp. 3873–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock GR, Steel KP. Inner ear pathology in the deafness mutant mouse. Acta oto-laryngologica. 1983;96:39–47. doi: 10.3109/00016488309132873. [DOI] [PubMed] [Google Scholar]
- 19.Vreugde S, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nature Genetics. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 20.Spiden SL, et al. The Novel Mouse Mutation Oblivion Inactivates the PMCA2 Pump and Causes Progressive Hearing Loss. PLoS Genetics. 2008 doi: 10.1371/journal.pgen.1000238. In Press, Accepted Manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosman EA, Quint E, Fuchs H, de Angelis MH, Steel KP. Catweasel mice: A novel role for Six1 in sensory patch development and a model for Branchio-Oto-Renal syndrome. Developmental Biology. 2009 doi: 10.1016/j.ydbio.2009.01.030. In Press, Accepted Manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mburu P, et al. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nature Genetics. 2003;34:421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- 23.van Dongen S, Abreu-Goodger C, Enright AJ. Fast assessment of microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008 doi: 10.1038/nmeth.1267. In Press, Accepted Manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 25.Kiyota T, Kato A, Altmann CR, Kato Y. The POU homeobox protein Oct-1 regulates radial glia formation downstream of Notch signaling. Dev Biol. 2008;315:579–592. doi: 10.1016/j.ydbio.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, et al. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 27.Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- 28.Mencia A, et al. Mutations in the seed region of the human miR-96 are responsible for non-syndromic progressive hearing loss. Nature Genetics. 2008 doi: 10.1038/ng.355. In Press, Accepted Manuscript. [DOI] [PubMed] [Google Scholar]
- 29.Obernosterer G, Leuschner PJF, Alenius M. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller KL, Jacques BE, Kelley MW. Fibroblast growth factor signaling regulates pillar cell development in the organ of Corti. Journal of Neuroscience. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.