Abstract
Diverse members of the Paramyxovirus family of negative-strand RNA viruses effectively suppress host innate immune responses through the actions of their V proteins. The V protein mediates interference with the interferon regulatory RNA helicase MDA5 to avoid cellular antiviral responses. Analysis of the interaction interface revealed the MDA5 helicase C domain as necessary and sufficient for association with V proteins from human parainfluenza virus type 2, parainfluenza virus type 5, measles virus, mumps virus, Hendra virus, and Nipah virus. The identified ∼130-residue region is highly homologous between MDA5 and the related antiviral helicase LGP2, but not RIG-I. Results indicate that the paramyxovirus V proteins can also associate with LGP2. The V protein interaction was found to disrupt ATP hydrolysis mediated by both MDA5 and LGP2. These findings provide a potential mechanistic basis for V protein-mediated helicase interference and identify LGP2 as a second cellular RNA helicase targeted by paramyxovirus V proteins.
Detection of pathogens by the mammalian innate immune system is mediated by pattern recognition receptors. For viruses, nucleic acids are often the trigger for innate responses that culminate in antiviral gene expression, including the production of type I interferon (IFN-α/β). Foreign nucleic acids outside the cell can be recognized by transmembrane Toll-like receptors at the cell surface or in the lumen of endocytic vesicles (9). Intracellular nucleic acids are recognized by cytoplasmic receptor proteins (24). In both cases, receptor binding to the nucleic acid ligand triggers a signal transduction cascade that activates immediate transcriptional responses, including the induction of the antiviral cytokines in the IFN family.
An important class of receptors for the detection of cytosolic nonself RNAs is represented by the proteins encoded by retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (24). These proteins share functional domains, including an amino-terminal protein interaction motif that functions in signal transduction and is homologous to the caspase activation and recruitment domain (CARD) (8) and a C-terminal DECH-box family RNA helicase domain. The helicase domains share approximately 30 to 40% overall sequence identity. Experimental and structural studies have demonstrated a third functional region at the extreme C terminus of RIG-I that forms a zinc-mediated fold and functions as a regulatory domain for the recognition and discrimination of RNA 5′ ends (6, 20). Current evidence supports a model in which binding to RNA ligand induces a change in accessibility to the CARD, allowing it to interact with downstream signaling proteins, including the mitochondrial resident IPS-1 (also identified as MAVS, VISA, and Cardiff [12, 15, 21, 28]). This association coordinates a serine kinase-mediated cascade that activates latent transcription factors, including NF-κB and IFN regulatory factor 3, culminating in the expression of IFN-β and a number of other crucial antiviral effector genes (8).
A third helicase protein, LGP2, resembles RIG-I and MDA5 in the helicase domain, also exhibiting 30 to 40% sequence identity. This overall level of similarity may underestimate the relatedness of these proteins, as they are more identical in their key helicase sequence motifs and encompassing domains (2). Significantly, LGP2 lacks the N-terminal CARD homology region. As a result, LGP2 expression is not typically associated with the ability to directly activate downstream signaling. Ectopic expression of LGP2 interferes with double-stranded RNA (dsRNA) and virus-induced antiviral signaling, while reduction of LGP2 expression by RNA interference results in enhanced IFN-β synthesis and antiviral responses. Expression of LGP2, like that of RIG-I and MDA5, is induced by virus infection, nucleic acid transfection, and IFN stimulation, suggesting it has the properties of a negative feedback inhibitor (14, 19, 20, 29).
The importance of these RNA helicase proteins in antiviral responses is validated by the phenotypes of mice harboring targeted disruptions in these genes (9). Deficiency in RIG-I leads to a widespread enhancement in replication of many RNA virus types, due to suppressed IFN biosynthesis and antiviral responses (11). Cells deficient in RIG-I exhibit a general defect in the ability to respond to foreign dsRNAs or single-stranded RNAs (ssRNAs) bearing phosphorylated 5′ ends. MDA5 deficiency showed a more specific phenotype, resulting in increased susceptibility to picornavirus infection and insensitivity to the synthetic dsRNA analog poly(I:C) (7).
LGP2 deficiency leads to a more complex phenotype (27). LGP2-deficient mice are more sensitive to IFN induction by cytosolic poly(I:C), an MDA5 ligand, and more resistant to vesicular stomatitis virus, a virus that triggers RIG-I signaling. Negative regulation of the IFN response remains intact overall, indicating that LGP2 may not be the primary negative regulator of type I IFN production. However, LGP2 deficiency results in a suppressed IFN response to infection with encephalomyocarditis virus, a picornavirus that has been demonstrated to trigger MDA5-mediated antiviral signaling. These data suggest that LGP2 executes both negative and positive regulatory functions related to RIG-I and MDA5 signaling. The disparate effects of LGP2 deficiency are difficult to interpret without a better mechanistic understanding of LGP2 functions in cellular innate antiviral immunity.
Further affirmation of the antiviral role of the helicase proteins derives from the fact that individual viruses have evolved means to evade or disrupt their activity, either by direct interference or by antagonizing the signaling intermediates. The large Paramyxovirus family of negative-strand RNA viruses is well known to evade IFN antiviral responses. Most viruses within this family encode a protein, called V, that is essential for IFN signaling evasion. In addition to suppressing the activity of signal transducer and activator of transcription (STAT) proteins to compromise IFN signal transduction, paramyxovirus V proteins can also limit the induction of IFN gene expression by interfering with the MDA5 protein (1, 3). Associations between the V protein C-terminal domain (CTD), a 60- to 70-residue zinc binding fold that is the conserved hallmark of V proteins, and the MDA5 helicase domain prevents signal transduction downstream of poly(I:C), a synthetic RNA ligand that is a known MDA5 activator (1, 3). This inhibition was observed for all tested V proteins of the large Paramyxovirus family (3). Despite the potential importance of the V protein-MDA5 interface as an antiviral target, the mechanistic basis and consequences for MDA5 interference by V proteins are poorly understood and difficult to reconcile with the lack of phenotype in MDA5-deficient mice. Recent evidence indicates that MDA5 is involved in the detection of Sendai virus-defective interfering (DI) genomes, providing plausible biological relevance to the observed V protein interference that may not have been apparent from gene disruption studies (31).
Identification of the MDA5 target recognized by paramyxovirus V proteins led to the discovery that LGP2 is also a target for V protein antagonism. Results demonstrate that the helicase C domain of both proteins functions as a V protein binding region common to both MDA5 and LGP2 but absent in RIG-I. We demonstrate that interaction with the V protein interferes with the catalytic activity of both target helicases, providing a biochemical basis for V protein helicase antagonism.
MATERIALS AND METHODS
Cells and virus.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% cosmic calf serum (HyClone). Parainfluenza virus type 5 (PIV5) was obtained from Robert Lamb (Northwestern University, Evanston, IL) and was propagated and titers were determined in Vero cells.
Plasmids, immunoprecipitations, and immunoblotting.
To construct helicase protein fragment expression vectors, PCR-amplified segments of MDA5, LGP2, or RIG-I cDNAs were subcloned into plasmid pEF-FLAG to generate an N-terminal in-frame epitope tag, unless otherwise specified in the text. All resulting plasmids were verified by DNA sequencing and expression analysis to detect the epitope-tagged protein fragment. Expression vectors for hemagglutinin (HA)-tagged paramyxovirus V proteins and the glutathione S-transferase (GST)-measles virus and mumps virus V protein fragment fusions have been described elsewhere (17, 26). For immunoprecipitation experiments 5 μg of FLAG-helicase vector and 5 μg of HA-V (or GST-V) vector were transfected by the CaPO4 method in HEK293T cells. Twenty-four hours later, cells were washed once with ice-cold phosphate-buffered saline and subsequently lysed with whole-cell extract buffer (WCEB) as described previously (26). For immunoprecipitation, lysates were prepared in WCEB and precleared with Sepharose beads. Protein complexes were purified by overnight incubation with FLAG M2 affinity resin (Sigma) and washed with WCEB. Alternatively, GST fusions were collected on glutathione Sepharose (Sigma). After elution with sodium dodecyl sulfate, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and processed for immunoblotting. For immunoblotting, proteins were separated, transferred to nitrocellulose, probed with antibodies recognizing FLAG, HA, or GST (Sigma), and visualized by enhanced chemiluminescence (NEN Life Sciences). Rabbit antiserum recognizing the PIV5 P and V proteins was generated using a GST-V protein purified from Escherichia coli (Cocalico Biologicals, Inc.) and screened by immunoblotting.
ATP hydrolysis.
For ATP hydrolysis assays FLAG-tagged helicase proteins in the presence or absence of PIV5, mumps virus, measles virus, or Nipah virus V proteins were immunoprecipitated as described above and eluted with FLAG tripeptide (Sigma). Purified protein or helicase-V protein complexes were incubated with 2 μg poly(I:C) in 50-μl ATPase reaction mixtures containing 5 mM morpholinopropanesulfonic acid (pH 6.5), 0.3 mM MgCl2, 0.2 mM dithiothreitol for 10 min at room temperature. The reaction was started by addition of 0.5 mM ATP (Sigma), 0.66 nM [γ-32P]ATP (Perkin-Elmer) and incubated for 1 h at 37°C. Aliquots (10%) of the reaction mixtures were loaded onto thin-layer chromatography plates (Sigma) and analyzed by phosphorimaging with a Molecular Dynamics Storm scanner and subsequently quantified using ImageQuant software. The remaining reaction mixture was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using anti-FLAG antibody (Sigma) and antisera detecting PIV5 V protein or HA epitope tag. For the experiment shown in Fig. 6A, below, 293T cells were transfected with 2 μg MDA5 expression plasmid by calcium phosphate precipitation and after 24 h infected with 10 PFU/ml PIV5 for 24 h prior to lysis. For the experiment shown in Fig. 6B, 293T cells were infected with 10 PFU/ml PIV5 for 24 h and then transfected with 8 μg LGP2 expression plasmid using polyethyleneimine (Polysciences) for 24 h prior to lysis.
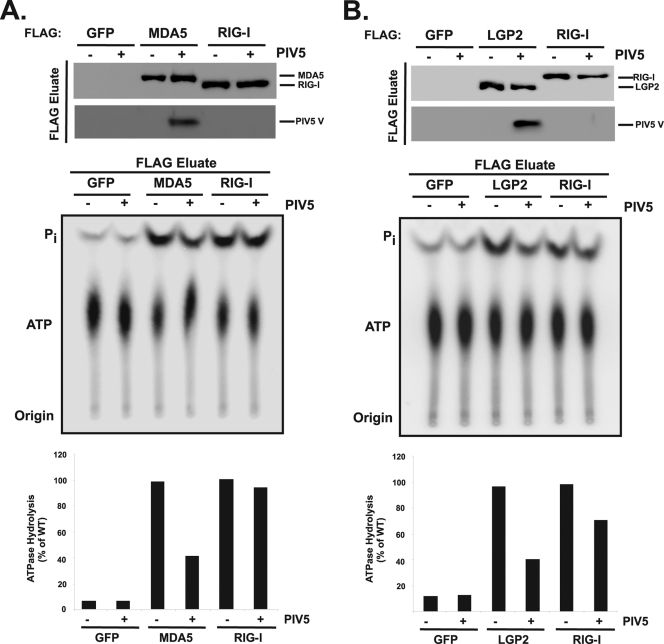
FIG. 6.
Interference with ATP hydrolysis in infected cells. Results are for an experiment similar to that shown in Fig. 5, but FLAG-tagged MDA5, LGP2, RIG-I, or control GFP were immunoaffinity purified from HEK293T cells that were also infected with PIV5 and eluates were subjected to ATP hydrolysis analysis in the presence of poly(I:C). Top panels demonstrate copurification of V protein (PIV5V) with MDA5 (A) or LGP2 (B). Middle panels demonstrate ATP hydrolysis activities by autoradiography, and bottom panels contain graphical representations of ATP hydrolysis activities, quantified by phosphorimaging analysis. Graphs plot the ratio of Pi to ATP, expressed as a percentage of the ATP hydrolysis activity without V protein.
RESULTS
LGP2 and MDA5 share a V-protein binding region in the helicase domain.
Diverse paramyxovirus V proteins are capable of disrupting IFN production by binding to the MDA5 protein through protein interactions between the V protein CTD and the MDA5 helicase domain. In contrast, the related RIG-I protein is not inhibited by V proteins (1, 3). To gain further insights into the underlying selectivity of these protein interactions, coimmunoprecipitation experiments were used to map the interaction sites. FLAG epitope-tagged MDA5, RIG-I, their N-terminal CARD or C-terminal helicase domain, or LGP2 was expressed along with HA-tagged measles virus V protein (Fig. 1A and B). Following FLAG immunoprecipitations, HA immunoblot assays confirmed that neither RIG-I nor any fragment of RIG-I was able to coprecipitate the V protein. Both MDA5 and the MDA5 helicase domain specifically bound to the V protein, but the MDA5 CARD region did not. The full-length LGP2 protein was also tested in this assay and was found to efficiently coprecipitate with the V protein (Fig. 1B).
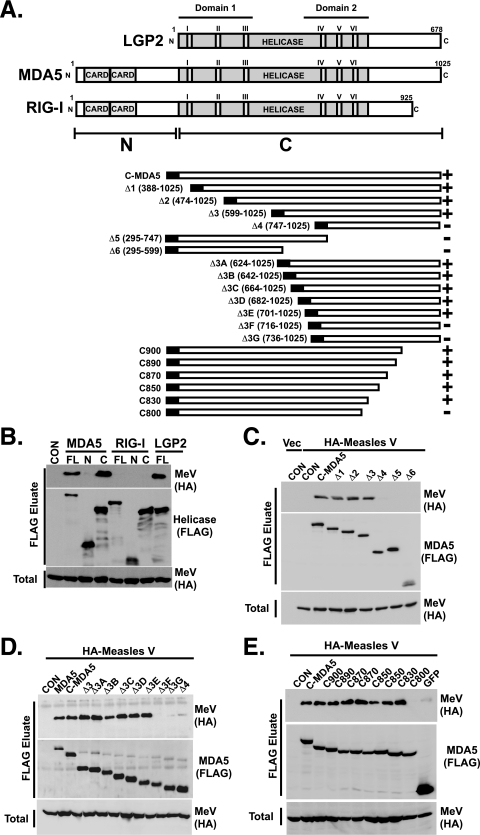
FIG. 1.
Map of V protein interaction regions of MDA5 and LGP2. (A) Diagrammatic representations of the LGP2, MDA5, and RIG-I proteins. (Top) Box diagrams illustrate the key features of the proteins, with CARD and helicase regions highlighted. Roman numerals depict the conserved helicase motifs, and boundaries of domain 1 and domain 2 are indicated. (Bottom) Illustration of MDA5 helicase domain protein fragments used for mapping the minimum V protein binding region. The black box depicts the N-terminal FLAG epitope tag, numbers refer to amino acid residues encompassed by the construct in parentheses, and + or − indicates V protein coprecipitation. (B) V protein associates with MDA5, the MDA5 helicase domain, and LGP2, but not RIG-I. FLAG-tagged helicase proteins were expressed in HEK293T cells along with HA-tagged measles virus V protein and lysates subject to FLAG immunoaffinity purification and anti-HA immunoblotting to detect protein coimmunoprecipitation. (C and D) Mapping the N-terminal boundary of the MDA5 V protein binding region. The experiment was similar to that shown in panel B, but we used serially truncated proteins as indicated. (E) Map of the C-terminal boundary of the MDA5 V protein binding region. The experiment was similar to those shown in panels B to D, but we used C-terminal truncation series as indicated. Vec, empty HA expression vector; Con, empty FLAG expression vector; FL, full-length protein; N, N-terminal CARD fragment; C, C-terminal helicase domain.
To map the minimum region required for V protein interactions, a comprehensive nested series of MDA5 fragments was constructed sequentially, designed to encompass the entire helicase domain with both N-terminal and C-terminal deletions (Fig. 1A). Coimmunoprecipitation experiments using N-terminal deletions revealed that the C-terminal half of the helicase domain was able to coprecipitate with measles virus V (Fig. 1C), ultimately defining the amino-terminal boundary between residues 701 and 716 (Fig. 1D, compare Δ3E to Δ3F). Similar analysis mapped the C-terminal boundary between residues 800 and 830 (Fig. 1E, compare C830 to C800). These results tentatively identified the V protein target as approximately 130 amino acids in length, between MDA5 residues 701 and 830, which is analogous to LGP2 residues 351 to 479.
LGP2 and MDA5 minimum V protein binding region coincides with helicase C domain.
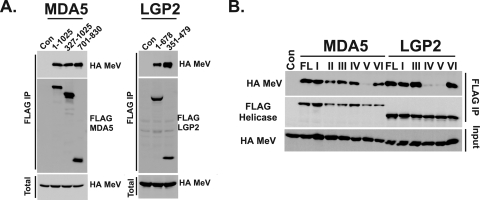
To test if these N- and C-terminal boundaries represent an independent protein domain that is sufficient for V protein interactions, the minimal MDA5 and LGP2 target regions were expressed as individual FLAG fusions and subjected to coprecipitation assays. Both MDA5701-830 and LGP2351-479 minimal target fragments copurified measles virus V protein as well as or better than the full-length proteins (Fig. 2A), demonstrating that they represent a minimal V protein binding region (MVBR) sufficient for interaction.
FIG. 2.
The MDA5 and LGP2 helicase C domains mediate the V protein interaction. (A) V protein association with defined MDA5 and LGP2 minimal targets. (Left) Full-length MDA5 (residues 1 to 1025), the MDA5 helicase domain (residues 327 to 1025), or MVBR fragment (residues 701 to 830) coprecipitate with the measles virus V protein. (Right) Full-length LGP2 (residues 1 to 678) or MVBR fragment (residues 351 to 479) coprecipitate with the measles virus V protein. (B) V protein interaction requires structural integrity of the helicase C domain. A coimmunoprecipitation assay was conducted using full-length, wild-type MDA5 or LGP2 (FL), or full-length proteins engineered with mutations to the indicated helicase conserved motifs (roman numerals).
Structural studies of RNA helicase proteins have revealed the enzymatic core to consist of two tandem RecA-like domains, referred to as domain 1 and domain 2, that are joined together by an interdomain linker region. These two domains form a cleft into which ATP can bind and present a binding surface for RNA interactions (18). The conserved amino acid sequence motifs I, II, and III are present in domain 1 while motifs IV, V, and VI are present in domain 2, which is also known as the helicase C domain (Fig. 1A). The juxtaposition of these motifs in the binding cleft coordinates RNA-stimulated ATP hydrolysis with helicase activities (5, 25). Sequence comparison and structural modeling of the MDA5 and LGP2 helicase domains reveals that the experimentally defined V protein target region boundaries coincide with the boundaries of helicase domain 2 (2, 16).
To probe the importance of the conserved helicase motifs in V protein interactions, a series of MDA5 and LGP2 proteins containing specific mutations in the conserved motifs (2) were subjected to coimmunoprecipitation assays with measles virus V protein (Fig. 2B). All of these mutations disrupt the enzymatic activity of the helicase (2). MDA5 proteins with point mutations to motifs I, II, III, or IV had little or no effect on the ability to coprecipitate with the V protein. The mutation in motif V, a nine-residue deletion, disrupted the association, while an eight-amino-acid deletion to motif VI did not (Fig. 2B). LGP2 proteins with point mutations to motifs I and III also retained the ability to bind V protein, but deletions in either motif IV (seven residues) or motif V (nine residues) resulted in loss of V protein interaction. Again, deletion of eight residues comprising the LGP2 motif VI did not disrupt interactions. The two LGP2 mutants that lost V protein association also lack the ability to function as negative regulators, which suggests disruption of protein structure and interference with protein-protein interactions (2). These results demonstrate that structural integrity of the helicase C domain is critical for V protein interaction.
MDA5 and LGP2 bind diverse V proteins via the conserved C-terminal domain.
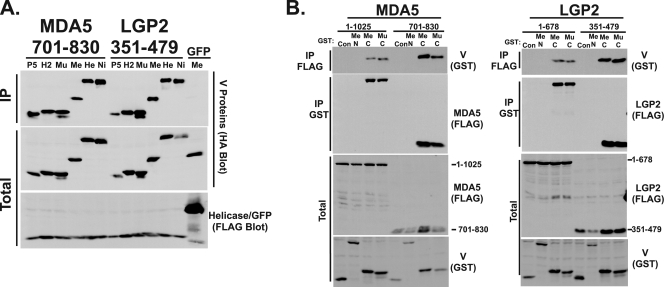
All of the MDA5 and LGP2 fragments that were capable of interacting with the measles virus V protein also coprecipitated with the Nipah virus V protein, which is only homologous in the conserved CTD region (data not shown). To demonstrate the generality of this MVBR as a target for diverse paramyxovirus V proteins, MDA5701-830 and LGP2351-479 were tested for their ability to interact with V proteins from parainfluenza virus 5, human parainfluenza virus 2, mumps virus, measles virus, Nipah virus, and Hendra virus (Fig. 3A). All of the tested V proteins were able to coprecipitate with both MDA5 and LGP2 MVBR fragments.
FIG. 3.
Targeting of MDA5 and LGP2 via the MVBR is a property of diverse paramyxovirus V proteins and is mediated by the conserved CTD. (A) Diverse paramyxovirus V proteins coprecipitate with the minimal MDA5 or LGP2 target fragments. Another coprecipitation experiment was done using V proteins from PIV5 (P5), human parainfluenza virus type 2 (H2), mumps virus (Mu), measles virus (Me), Hendra virus (He), or Nipah virus (Ni). Expression of FLAG-tagged green fluorescent protein (GFP) combined with measles virus V protein served as a control for nonspecific interactions. (B) The V protein cysteine-rich C-terminal domain is sufficient to mediate association with the LGP2 and MDA5 MVBR fragments. FLAG-tagged full-length or target fragment of MDA5 (left) or LGP2 (right) was expressed along with glutathione S-transferase fusion proteins containing the measles virus (Me) or mumps virus (Mu) C-terminal domain, or as a control the measles virus N-terminal domain. Following FLAG or GST affinity purification, samples were probed for coprecipitation with antiserum specific to GST or FLAG, as indicated.
Conversely, to verify that V proteins contact the helicase target fragments by means of their conserved CTD zinc finger regions, GST fusion proteins that express the mumps virus or measles virus CTD, or the control measles virus N-terminal domain (17), were used in coprecipitation assays (Fig. 3B). No interaction was observed between the full-length helicase or MVBR and the V protein N-terminal domain, but both measles virus and mumps virus V protein CTD fusion proteins coprecipitated with both full-length MDA5 (residues 1 to 1025) or full-length LGP2 (residues 1 to 678) and the MVBRs MDA5701-830 and LGP2351-479. It was observed that the target fragments were more efficiently coprecipitated than the respective full-length helicase proteins, suggesting that the isolated domain may be more accessible to the V protein CTD.
LGP2 and MDA5 MVBRs select V protein from PIV5-infected cells.
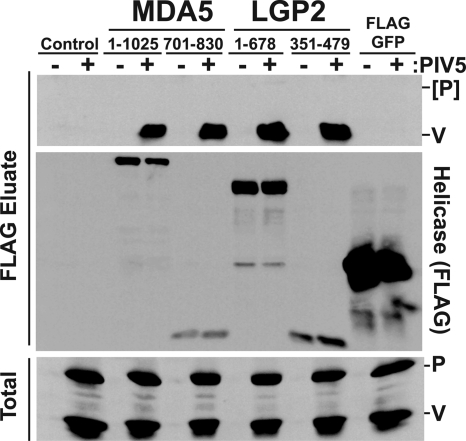
The abilities of the defined helicase target fragments to interact with physiologically expressed V protein in the context of virus infection were also tested (Fig. 4). Cells expressing the tagged full-length helicase proteins or MVBR were infected with PIV5, lysates were subjected to immunoprecipitation with FLAG immunoaffinity resin, and eluates were probed with antiserum that recognized the PIV5 P and V proteins. The V protein, but not the P protein, was specifically coprecipitated by either full-length MDA5 or LGP2 as well as with their respective minimal target regions. Together, these data define ∼130-amino-acid regions of MDA5 and LGP2 that contain the binding targets for paramyxovirus V proteins.
FIG. 4.
MVBR reacts with V protein from infected cells. Parallel plates of HEK293T cells were transfected with expression vectors for the indicated FLAG-tagged helicase proteins or fragments and then infected with 1 PFU/cell of PIV5 (+) or mock infected (−). Cell lysates were subjected to FLAG purification and immunoblotting with antiserum that recognizes the PIV5 V protein and P protein. Only V protein coprecipitated.
V protein interferes with MDA5 and LGP2 enzymatic activities.
The experimental results defined the helicase C domain as the site of V protein association with MDA5 and LGP2. MDA5 interference by V proteins has been validated with demonstration of V-mediated signaling interference leading to disruption of IFN-β promoter activity. LGP2 plays a more complex and poorly understood role, as both a negative regulator of dsRNA signaling and a positive regulator of antiviral responses (13, 14, 19, 20, 27, 29). While MDA5 and LGP2 fundamentally differ in their antiviral signaling capacities, at the catalytic level they both use RNA-dependent ATP hydrolysis to drive enzymatic activity (2, 10).
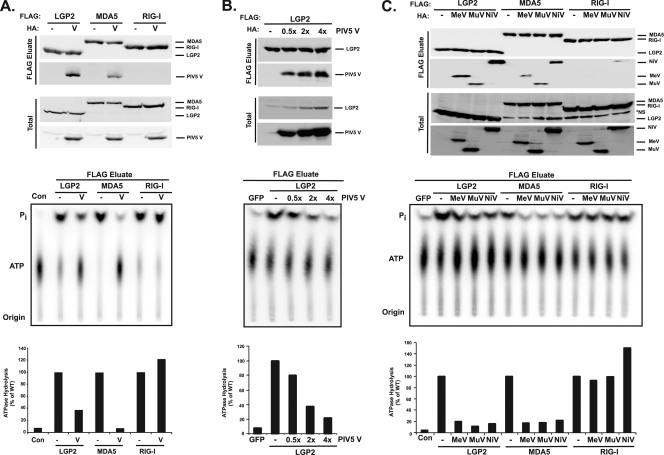
The biochemical consequences for V protein interactions with both MDA5 and LGP2 were examined at the enzymatic level with ATP hydrolysis assays (Fig. 5). Our previous studies demonstrated that FLAG-immunopurified helicases have specific ATP hydrolysis activity (2). Epitope-tagged MDA5, LGP2, and RIG-I proteins were expressed in HEK293T cells in the absence or presence of the PIV5 V protein, and lysates were subjected to immunoaffinity purification with anti-FLAG resin. Immunoblotting detected copurified V protein in complex with both MDA5 and LGP2, but not RIG-I. FLAG eluates were subjected to an ATP hydrolysis assay in the presence of poly(I:C). MDA5- and LGP2-dependent ATP hydrolysis activity was significantly diminished in the V protein-containing reactions (Fig. 5A). Although the precise stoichiometry is difficult to evaluate under these experimental conditions, the specificity of V protein-mediated inhibition of LGP2 is shown by dose-dependent interference (Fig. 5B). Similar effects on the helicase catalytic activity were observed for measles virus, mumps virus, and Nipah virus V proteins (Fig. 5C). These findings demonstrate that the V protein interferes with the enzymatic activity of both MDA5 and LGP2.
FIG. 5.
V protein interferes with ATP hydrolysis by MDA5 and LGP2. FLAG-tagged MDA5, LGP2, and RIG-I were immunoaffinity purified from HEK293T extracts with or without coexpressed V protein, and eluates were subjected to ATP hydrolysis analysis in the presence of poly(I:C). Top panels demonstrate copurification of V protein (PIV5V) with LGP2 and MDA5 but not RIG-I by immunoblotting. Middle panels demonstrate ATP hydrolysis activities by autoradiography, with positions of the origin, ATP, and free phosphate indicated. Bottom panels contain graphical representations of ATP hydrolysis activities, quantified by phosphorimaging analysis. Graphs plot the ratio of Pi to ATP, expressed as the percentage of the ATP hydrolysis activity without V protein. (A) Plasmids coding for FLAG-tagged LGP2, MDA5, or RIG-I RNA helicases were cotransfected with plasmids coding for HA-tagged PIV5 V protein or empty vector plasmid at a 1:1 ratio into HEK293T cells. Con, no-protein control; WT, wild type. (B) Four micrograms of cDNA plasmids coding for FLAG-tagged LGP2 helicase were cotransfected with empty vector (−) or 2 μg, 8 μg, or 16 μg of plasmid coding for HA-tagged PIV5 V protein. Total amounts of transfected DNA were equalized with empty vector for each reaction. (C) Plasmids coding for FLAG-tagged LGP2, MDA5, or RIG-I RNA helicases were cotransfected with empty vector control or plasmids coding for HA-tagged measles virus, mumps virus, or Nipah viurs V protein at a 1:3 ratio into HEK293T cells. *NS, nonspecific band.
To verify the ATP hydrolysis interference in a more native context, a similar experiment was carried out with physiologically expressed V protein from PIV5 infection (Fig. 6). Again, both MDA5 and LGP2, but not RIG-I, were capable of coprecipitating the V protein from the infected cells. Analysis of ATP hydrolysis in the purified and eluted material demonstrated interference with both MDA5 and LGP2, but not RIG-I, reducing the enzymatic activity of these helicases by approximately 60% of uninfected controls.
DISCUSSION
RNA helicase proteins are essential for a vast array of cellular processes, and the recent identification of RIG-I, MDA5, and LGP2 as essential for recognition of and response to viral pathogens establishes their importance for innate antiviral immunity (24). The significant role of these proteins in antiviral signaling makes them prime targets for virus-encoded host evasion and antagonism. Antagonism of MDA5 signaling is a unique property attributed to paramyxovirus V proteins and results in a disengagement of MDA5-mediated IFN-β promoter activation (1, 3). Here, LGP2 is identified as a second helicase protein targeted by paramyxovirus V proteins.
Interaction mapping has defined an MVBR that is necessary and sufficient for contact with MDA5, encompassed by residues 701 to 830. The analogous region of LGP2, encompassed by residues 351 to 479, was also found to be targeted by paramyxovirus V proteins. Sequence analysis of the identified MVBR reveals unusually high conservation between MDA5 and LGP2, with 57% identity and 78% similarity. In contrast, RIG-I is only 37% identical and 58% similar to LGP2 and 41% identical and 58% similar to MDA5 in this region, the degree of homology observed when comparing the full-length proteins.
RNA helicase catalytic activity is thought to arise from the coordinated action of two structural domains, known as domain 1 and domain 2 (5, 18). These two lobes encompass all of the conserved sequence motifs that contribute to ATP binding and hydrolysis as well as RNA interaction and unwinding. Domain 2, also referred to as the helicase C domain, represents a widely recognized structural element of diverse RNA and DNA helicase families. The boundaries of the MVBRs defined here coincide exactly with the helicase C domains. Results from ATP hydrolysis assays indicate that the V protein interaction can interfere with enzymatic activity, and disruption of the MVBR by deletion mutagenesis abrogates V protein interactions. Although the relationship between enzymatic activity of the helicase domain and signal transduction by these proteins remains poorly understood, the disruption of helicase domain structure and function provides a reasonable mechanistic basis for the observed V protein antagonism.
The boundaries of the MDA5 MVBR characterized here, residues 701 to 830, are in close agreement with those defined using yeast two-hybrid methods (4). The site of PIV5 V protein interaction with MDA5 was mapped to MDA5 residues 676 to 816. This study also observed that PIV5 V protein-mediated MDA5 interference is directed at the level of RNA-dependent MDA5 oligomerization. This is not inconsistent with the observed disruption of ATPase activity, as the detection of RNA substrate is a prerequisite for MDA5-mediated ATP hydrolysis. Prevention of RNA access by V proteins might contribute to the observed loss of enzymatic activity in the presence of V protein. However, the weak RNA binding capacity of MDA5 complicates analysis of RNA interactions, and we have not observed V-dependent loss of LGP2 RNA contact. Further work is required to determine the precise point of V protein antagonism for both MDA5 and LGP2.
MDA5 and LGP2 MVBR fragments are targeted by the V protein C-terminal domain, a zinc binding domain that exhibits high amino acid conservation among all paramyxovirus V proteins. In agreement with the generality of V-mediated MDA5 interference (1, 3), we demonstrated that the MDA5 and LGP2 MVBRs are recognized by V proteins from measles virus, PIV5, mumps virus, human parainfluenza virus type 2, Nipah virus, and Hendra virus, as well as by the isolated CTDs from measles virus and mumps virus V proteins. The expressed MVBR fragments can selectively copurify the PIV5 V protein from infected cells, demonstrating complex formation at physiologically relevant levels of V protein expression.
Mice deficient in RIG-I expression are more susceptible to infection with Sendai virus, but MDA5-deficient mice retain normal resistance. Therefore, intracellular detection of paramyxovirus infection is considered widely to be mediated primarily by RIG-I (11). As a consequence, the precise role for MDA5 in antiviral responses to paramyxovirus infection remains enigmatic. Recent studies have demonstrated a specific role for MDA5 signaling in response to Sendai virus Cantell, a strain that is known to accumulate DI genomes (31). DI genomes are known to be potent stimulators of IFN biosynthesis and antiviral responses that can activate RIG-I-dependent pathways (22, 23, 30, 32), but available evidence suggests that DI particles function as a paramyxovirus-associated molecular pattern that can stimulate dendritic cell activation via MDA5. Dendritic cells derived from MDA5-deficient mice were inefficiently activated by Sendai virus Cantell, and sole expression of Sendai virus V protein can inhibit direct early dendritic cell activation by DI particles (31). These data provide biological relevance to the reported MDA5 interference by V proteins and identify DI genomes as a potential natural MDA5 activator.
At our current level of understanding, the V protein antagonism of LGP2 remains difficult to reconcile with prior studies of LGP2 action. Characterized as a negative feedback regulator of IFN synthesis and antiviral responses, analysis of LGP2-deficient mice has revealed phenotypes related to both negative and positive regulation of IFN responses and antiviral signaling (14, 19, 20, 27, 29). Further experimentation will be needed to fully appreciate the exact roles for LGP2 in detection of and response to paramyxoviruses and the precise advantages of V-mediated interference. Considering that expression of both MDA5 and LGP2 proteins is strongly controlled by virus-induced and IFN-responsive signaling pathways, their accumulation at late times following infections or in specialized cell types may function as a modifier of initial antiviral responses. Common mechanisms for targeting MDA5 and LGP2 and a shared role in detection and response to DI RNA genomes provide an additional suggestion of collaboration between these two helicases. Together with the fact that both LGP2 deficiency and MDA5 deficiency result in greater susceptibility to picornavirus infections (11, 27), it is tempting to speculate that LGP2 might act directly or indirectly in concert with the MDA5 pathway to modulate antiviral responses.
Acknowledgments
We acknowledge Aristobolo M. Silva and members of the Horvath and Barber laboratories for critical comments and helpful discussions.
This work was supported by NIH grants AI050707 and AI073919 to C.M.H.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamming, D., and C. M. Horvath. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I and LGP2. J. Biol. Chem. 2849700-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Childs, K., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359190-200. [DOI] [PubMed] [Google Scholar]
- 4.Childs, K. S., J. Andrejeva, R. E. Randall, and S. Goodbourn. 2009. Mechanism of mda-5 inhibition by paramyxovirus V proteins. J. Virol. 831465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 36717-37. [DOI] [PubMed] [Google Scholar]
- 6.Cui, S., K. Eisenacher, A. Kirchhofer, K. Brzozka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29169-179. [DOI] [PubMed] [Google Scholar]
- 7.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 1253-56. [DOI] [PubMed] [Google Scholar]
- 9.Ishii, K. J., S. Koyama, A. Nakagawa, C. Coban, and S. Akira. 2008. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe 3352-363. [DOI] [PubMed] [Google Scholar]
- 10.Kang, D. C., R. V. Gopalkrishnan, Q. Wu, E. Jankowsky, A. M. Pyle, and P. B. Fisher. 2002. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 99637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 12.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 13.Komuro, A., D. Bamming, and C. M. Horvath. 2008. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine 43350-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komuro, A., and C. M. Horvath. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 8012332-12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 16.Nishino, T., K. Komori, D. Tsuchiya, Y. Ishino, and K. Morikawa. 2005. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure 13143-153. [DOI] [PubMed] [Google Scholar]
- 17.Ramachandran, A., J. P. Parisien, and C. M. Horvath. 2008. STAT2 is a primary target for measles virus V protein-mediated alpha/beta interferon signaling inhibition. J. Virol. 828330-8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5232-241. [DOI] [PubMed] [Google Scholar]
- 19.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 1755260-5268. [DOI] [PubMed] [Google Scholar]
- 20.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 22.Strahle, L., D. Garcin, and D. Kolakofsky. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351101-111. [DOI] [PubMed] [Google Scholar]
- 23.Strahle, L., J. B. Marq, A. Brini, S. Hausmann, D. Kolakofsky, and D. Garcin. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 8112227-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 2017-22. [DOI] [PubMed] [Google Scholar]
- 25.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8251-262. [DOI] [PubMed] [Google Scholar]
- 26.Ulane, C. M., A. Kentsis, C. D. Cruz, J. P. Parisien, K. L. Schneider, and C. M. Horvath. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 7910180-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkataraman, T., M. Valdes, R. Elsby, S. Kakuta, G. Caceres, S. Saijo, Y. Iwakura, and G. N. Barber. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 1786444-6455. [DOI] [PubMed] [Google Scholar]
- 28.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19727-740. [DOI] [PubMed] [Google Scholar]
- 29.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 1752851-2858. [DOI] [PubMed] [Google Scholar]
- 30.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269383-390. [DOI] [PubMed] [Google Scholar]
- 31.Yount, J. S., L. Gitlin, T. M. Moran, and C. B. Lopez. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 1804910-4918. [DOI] [PubMed] [Google Scholar]
- 32.Yount, J. S., T. A. Kraus, C. M. Horvath, T. M. Moran, and C. B. Lopez. 2006. A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol. 1774503-4513. [DOI] [PubMed] [Google Scholar]