Abstract
The Leishmania tarentolae mitochondrial ribosome (Lmr) is a minimal ribosomal RNA (rRNA)-containing ribosome. We have obtained a cryo-EM map of the Lmr. The map reveals several features that have not been seen in previously-determined structures of eubacterial or eukaryotic (cytoplasmic or organellar) ribosomes to our knowledge. Comparisons of the Lmr map with X-ray crystallographic and cryo-EM maps of the eubacterial ribosomes and a cryo-EM map of the mammalian mitochondrial ribosome show that (i) the overall structure of the Lmr is considerably more porous, (ii) the topology of the intersubunit space is significantly different, with fewer intersubunit bridges, but more tunnels, and (iii) several of the functionally-important rRNA regions, including the α-sarcin-ricin loop, have different relative positions within the structure. Furthermore, the major portions of the mRNA channel, the tRNA passage, and the nascent polypeptide exit tunnel contain Lmr-specific proteins, suggesting that the mechanisms for mRNA recruitment, tRNA interaction, and exiting of the nascent polypeptide in Lmr must differ markedly from the mechanisms deduced for ribosomes in other organisms. Our study identifies certain structural features that are characteristic solely of mitochondrial ribosomes and other features that are characteristic of both mitochondrial and chloroplast ribosomes (i.e., organellar ribosomes).
Keywords: cryo-EM, Leishmania tarentolae, organellar ribosome
A number of trypanosomatid protists of the genus Leishmania are responsible for severe diseases affecting millions of people worldwide. Trypanosomatids possess a single large kinetoplast mitochondrion (1). Several trypanosomatid mitochondrial genes are posttranscriptionally modified through RNA editing (2–4). Although mitochondrial translation in Leishmania, as in other organisms, is required for synthesis of several indispensable components of the respiratory chain by mitochondrial ribosomes (mitoribosomes), the process of translation is quite distinct from that in the mammalian system (5). In addition, the mechanism by which the fully-edited translatable mRNA is selected by the Leishmania mitochondrial ribosome (Lmr) to preclude translation of the immature preedited templates is unknown (5–10).
Ribosomal RNAs (rRNAs) encoded in the kinetoplast genome are extremely small (11, 12). The small subunit (SSU) of the Lmr contains a 610-nt-long 9S rRNA and the large subunit (LSU) of the Lmr contains a 1,173-nt-long 12S rRNA. Overall, the Leishmania mitochondrial rRNAs (Lm-rRNAs) have ≈61% and 28% fewer nucleotides as compared respectively with their eubacterial and mammalian mitochondrial counterparts. However, although secondary and tertiary structures are very well established for eubacterial rRNAs (13, 14), a significant portion of the secondary structures for the Lm-rRNAs (≈66% of the 9S and ≈34% of the 12S) could not be predicted by comparative sequence analyses (11, 12, 15). The dramatically less robust RNA structural scaffold could affect Lmr's stability, SSU–LSU interactions, and other properties relevant to the process of translation. It is likely that a small Lm-rRNA and a tight association of Lmr to the mitochondrial membranes were responsible for the difficulties in isolation of the trypanosomatid mitoribosomes for many decades. Recently, the Lmr was isolated from Leishmania tarentolae as a 50S protein-rich complex, and its 2 subunits were characterized as ≈28–30S SSU and ≈40S LSU particles (16, 17). These studies also revealed an unusual 45S SSU* complex, representing dimers of SSU in association with an additional protein complex.
Here, we present a 3D cryo-EM map of the 50S Lmr. Our analysis reveals that several missing eubacterial rRNA segments are structurally replaced with proteins in the Lmr structure. The intersubunit space (ISS), which accommodates the interactions of tRNAs and translational factors, is significantly remodeled in the Lmr, as compared with the ISS topology in all known structures of ribosomes (13, 14, 18–24). However, the overall morphology of the Lmr is remarkably conserved, suggesting the existence of tight structural and functional constraints on this evolutionary divergent form of the ribosome.
Results and Discussion
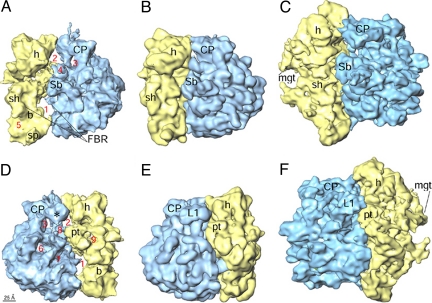
The cryo-EM map of the Lmr shows most of the landmark features of a typical ribosome (18) (Fig. 1 A and D and Fig. S1). However, a dramatic difference, with respect to all other ribosome structures (18–24), is high porosity of the Lmr. The Lmr (≈245 Å in diameter) is smaller than its eubacterial (≈260 Å; Fig. 1 B and E) and mammalian mitochondrial (≈320 Å; Fig. 1 C and F) counterparts. However, the ISS of the Lmr is larger as compared with its counterparts in other known ribosome structures, and it connects to solvent through several newly-discovered tunnels. In addition, there is a large gap between the 2 subunits in the lower body region. To interpret the cryo-EM map at the molecular level, we used (i) the conserved segments of rRNA and proteins from the X-ray crystallographic structures of eubacterial and archaebacterial ribosomes (13, 14, 22, 23), and (ii) the homology models of the mammalian mitoribosome (25, 26). Finally, the program Mfold (27), along with the cryo-EM envelope constraints, was used to model structures of Lm-rRNA segments that were not available from X-ray or homology models.
Fig. 1.
Cryo-EM map of the Lmr (A and D) and its side-by-side comparison with the maps of an eubacterial (E. coli) 70S ribosome (18) (B and E) and a mammalian (Bos taurus) 55S mitoribosome (20) (C and F). Maps are shown from the L7/L12-stalk (A–C) and the protein L1 (D–F) sides. The SSU (yellow) and LSU (blue) are depicted. Numbers (1–9 in A and D) point to a few obvious differences in the Lmr: 1, gap caused by absence of major portion of SSU rRNA helix 44 (see Fig. 2A); 2 and 3, larger gaps between the head (h) of SSU and central protuberance (CP) of LSU; 4, an extended structure emerging from the CP; and 5–9, gaps caused by absence of various rRNA segments in the Lmr. Landmarks of the SSU are: b, body; mgt, mRNA gate (in C and F); pt, platform; sh, shoulder; sp, spur. Landmarks of the LSU are: L1, L1 protein (corresponding density is marked by * in D); Sb, L7/L12 stalk base; FBR, factor binding region. The scale bar is shown in D.
Structure of the SSU.
The overall morphology of the Lmr SSU is more similar to a eubacterial SSU than it is to the mammalian mitochondrial SSU (Figs. 1 and 2). The head, platform, and body of the SSU are immediately recognizable in our map. Considering that >60% of eubacterial rRNA segments are absent in the 9S Lm-rRNA, structural similarity of Lmr SSU to eubacterial SSU is quite remarkable. We use the secondary structure of eubacterial SSU rRNA (13) as the template to describe the 9S Lm-rRNA and its helices (Fig. 2A and Fig. S2). The central core of the SSU rRNA is largely conserved, whereas the majority of peripheral helices of the eubacterial SSU rRNA are absent in 9S Lm-rRNA. However, we find that a significant portion (≈61%) of missing eubacterial rRNA segments are positionally replaced by densities corresponding to proteins.
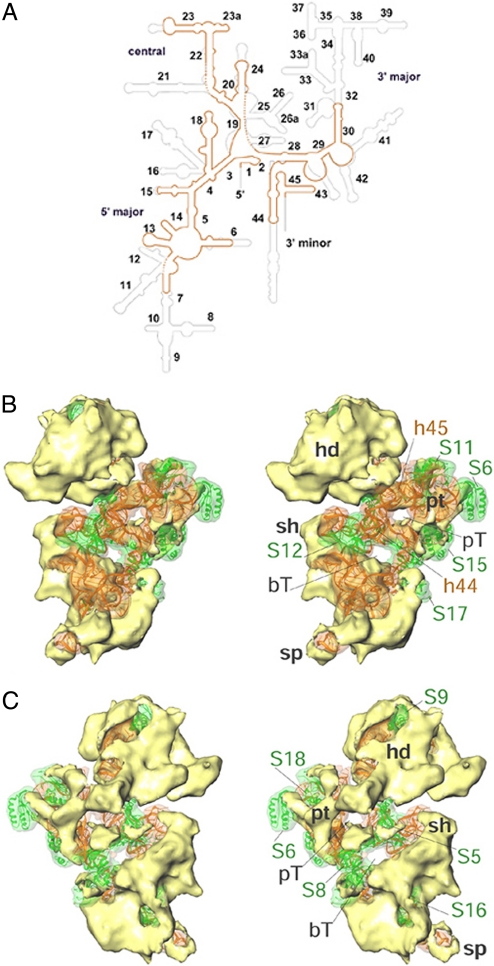
Fig. 2.
Structural analysis of the SSU. (A) Secondary-structure line diagram of the 9S Lm-rRNA (orange), superimposed on that of the bacterial 16S rRNA (13) (gray). RNA regions absent in the Lmr thus appear gray. RNA helices are identified by the adjacent numbers. Dashed lines correspond to unassigned segments of the 9S Lm-rRNA. (B) Stereo representation of the fitting of the conserved domains (ribbons) into the cryo-EM map of the SSU, shown from its interface side. The conserved components (orange, rRNA; green, proteins) are shown as translucent surfaces, and the Lmr-specific proteins (yellow) are shown as solid surfaces. (C) Same as B but shown from a solvent-side. Landmarks: numbers prefixed with S represent the SSU proteins; numbers prefixed with h identify the 9S rRNA helices; hd, head; bT, body tunnel; pT, platform tunnel. All other landmarks are as in Fig. 1.
The 9S Lm-rRNA lacks 24 of the 45 eubacterial rRNA helices (Fig. 2A). These are helices h8–h12, h16, h17, h21, h25, h26, h26a, and h31–h43. Helices h8–h10, which form the base of the eubacterial SSU, are replaced by proteins in the Lmr. Furthermore, a significant amount of additional protein mass is found in the base portion of the Lmr SSU. Given that this extra protein mass is also present in mammalian mitochondrial (20) and chloroplast (24) ribosomes, a protein-rich base thus seems to be a characteristic feature of organellar SSUs. Densities for h11, h12, and h27 are absent in the Lmr map. Through the absence of h12 (in the body) and h27 (near the body-platform junction), 2 tunnel-like features (bT and pT in Fig. 2 B and C) are created between SSU's interface and solvent sides.
The 3′ major domain of the SSU rRNA, which forms the head of the SSU (13), is dramatically smaller in Lmr than the eubacterial ribosome (Fig. 2A). Because of the absence of eubacterial helices h35–h39, another tunnel is formed between the top and bottom of the SSU head. Nevertheless, the overall architecture of the SSU head between eubacterial and Lmr is surprisingly well conserved, indicating that the majority of missing rRNA segments in the head must be replaced by proteins in Lmr. This observation suggests that the overall shape of the SSU head is critical for ribosome function.
The h44 is very small in the Lmr. Only its top portion, which constitutes the decoding site, is retained (Fig. 2A and Fig. S3). The missing segment of h44 creates a large gap between the lower body of 2 Lmr subunits (Fig. 1). Because of the absence of a portion of h24 and h27, and segments of loops between h3 and h19 and between h19 and h20 (see Fig. 2A) that stack behind h44 and h45 in the eubacterial structure (ref. 13 and Fig. S4), h44 and h45 shift closer to the platform in the Lmr.
Of the 20 eubacterial SSU proteins, 10 have homologues in the Lmr SSU. These are proteins S5, S6, S8, S9, S11, S12, and S15–S18 (16, 17, 28). Most of the bacterial homologs have long N-terminal or C-terminal extensions in the Lmr. Atomic structure of conserved portions of homologous bacterial proteins could be readily fitted into corresponding cryo-EM densities (Fig. 2 B and C). After the fitting of all segments of the rRNAs and all of the bacterial protein homologues, a significant proportion of densities in the cryo-EM map remains unexplained. These unexplained densities correspond to Lmr-specific proteins and extensions of homologous bacterial proteins and positionally replace a significant proportion (≈52%) of missing eubacterial proteins (such as S2–S4, S7, S10, S13, S19, and S20) (Fig. S3). However, ≈33% of total SSU protein mass acquires new quaternary positions in the Lmr.
Structure of the LSU.
Typical morphological features of a LSU, such as the central protuberance (CP) and 2 stalk-like features on the either side of CP, are immediately recognizable (Figs. 1 and 3) in our map. We discern 7 tunnels that connect the interface side of LSU with the solvent [e.g., tunnel near CP (CPT) in Fig. 3C]. In addition, a protein mass emerging from the CP extends into ISS. This structural feature appears to be characteristic of mitoribosomes; a similar mass was observed in the structure of a mammalian mitoribosome (20) and was referred to as the peptidyl (P)-site finger. However, the mass in the Lmr map is much more extended and spans both the aminoacylated (A)- and P-site regions; accordingly, we term it the A- and P-site finger (APSF; in Fig. 3B).
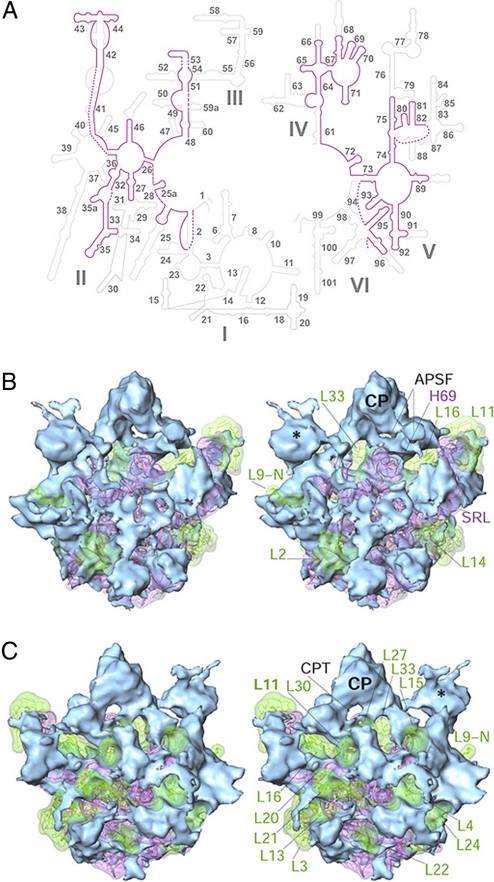
Fig. 3.
Structural analysis of the LSU. (A) Secondary-structure diagram of the 12S Lm-rRNA (purple), superimposed on that of the bacterial 23S rRNA (14) (gray). Roman numerals identify the 6 domains of the rRNA, and helices are identified by numbers. Dashed lines correspond to unassigned segments of the 12S Lm-rRNA. (B) Stereo representation of the fitting of conserved domains (ribbons) into the cryo-EM map of the LSU, shown from its interface side. The conserved components (pinkish purple, rRNA; green, proteins) are shown as translucent surfaces, and the Lmr-specific proteins (blue) are shown as solid surfaces. (C) Same as B, but shown from the solvent side. Landmarks: numbers prefixed with L identify the LSU proteins; numbers prefixed with H identify the LSU rRNA helices; APSF, A- and P-site finger; CPT, CP tunnel; L9-N, N-terminal domain of L9. All other landmarks are as in Fig. 1.
We use the bacterial LSU rRNA secondary structure (14) as the template to describe the 12S Lm-rRNA secondary structure and its helices (Fig. 3A and Fig. S5). A majority of the peripheral helices of the eubacterial LSU rRNA are absent in 12S Lm-rRNA; however, a significant proportion (≈50%) of the missing bacterial rRNA segments are positionally replaced by Lmr-specific proteins. Conserved segments from the mammalian mitoribosome LSU rRNA model (26) fitted extremely well into our cryo-EM map, with slight modification of the loop that connects helices H43 and H44 [in order to distinguish from rRNA helices of the SSU (h), all helices of LSU rRNA are identified with a prefix “H”] to the rest of the Lm-rRNA. Similarly, regions of helices H50 and H51 were modified to produce an optimum fit into the cryo-EM map.
H69 forms the central intersubunit bridge, B2a, in all ribosomes (18, 29), and plays pivotal roles in ribosome function. It provides a platform for tRNA binding at both the A and P sites of the ribosome (29) and undergoes conformational changes during the translational elongation cycle (30) and ribosome recycling (31). Unexpectedly, the H69 is partially disordered in our map, suggesting that this rRNA segment is much more flexible in the Lmr. The high flexibility of H69 could be an important regulatory feature in Leishmania, such that H69 becomes structured during protein synthesis or in the presence of binding ligands, such as tRNAs.
Densities corresponding to most of the eubacterial LSU rRNA domains V [which includes the peptidyltransferase center (PTC)] and VI [which includes the universally-conserved α-sarcin/ricin stem-loop (SRL)], are readily identifiable in our cryo-EM map. Although most of domain V is intact, the masses corresponding to helices H89 and H91 are smaller than in the eubacterial ribosome, supporting the fact that both of these helices are very small in Lm-rRNA. We find that the mass corresponding to the SRL has significantly shifted (by ≈30 Å) toward the PTC, thus decreasing the distance between the SRL (the GTPase center) and the PTC in the Lmr, as compared with the separation in the eubacterial ribosome. This shift in SRL position is apparently enabled by the absence of a neighboring mass caused by truncation in helices H89 and H91 (Fig. S6).
Of the 34 eubacterial LSU ribosomal proteins, 11 were found to have homologues in the Lmr LSU (16, 17), whereas 21 were found in a more exhaustive analysis of a closely-related organism Trypanosome brucei mitochondrial ribosome (28). These were proteins L2–L4, L9, L11–L17, L20–L24, L27–L30, and L33. Nineteen of these homologues are significantly larger than their eubacterial counterparts. Although most of eubacterial homologues can be fitted into the cryo-EM density (Fig. 3 B and C), some (e.g., L16, L30, and L33) have shifted with respect to their position in the eubacterial ribosome. The bacterial homologue of protein L1 appears to be absent in the L. major and T. brucei genome databases (www.genedb.org/genedb). However, because we see a density feature corresponding to L1 in our map (* in Figs. 1 D and 3 B and C), it is likely that L1 is substituted by a Lmr-specific protein. Moreover, the eubacterial L1-binding segment of the rRNA (H76–H78) is absent in Leishmania, and so is the density corresponding to H76–H78 in our map. In our map, the L1 feature is connected to rest of LSU through density masses that protrude from the side of CP and H75. Approximately 37% of missing bacterial proteins are positionally replaced by Lmr-specific proteins in LSU (Fig. S7). However, ≈41% of total LSU protein masses acquire new quaternary positions in the Lmr.
Intersubunit Bridges.
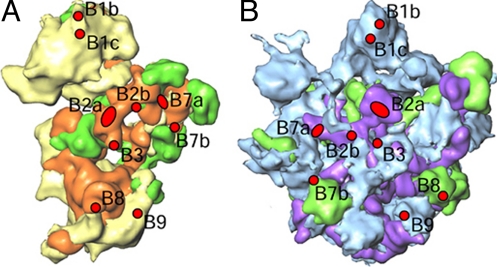
The 2 subunits of the Lmr are held together by 9 bridges (Fig. 4), significantly fewer than the 13 bridges in eubacterial ribosomes (18, 29) and the 15 bridges in mammalian mitochondrial ribosomes (20). We describe the Lmr bridges by using existing nomenclature (18, 20, 29). Components of eubacterial bridges B2c (h27), B5 (h44), and B6 (h44) are absent in the 9S Lm-rRNA, and components of bridges B1a (H38), B4 (H34), B5 (H62), and B6 (H62) are absent in the 12S Lm-rRNA. Indeed, density features corresponding to these 5 bacterial bridges (B1a, B2c, B4, B5, and B6) are absent in the Lmr. Although components of bacterial bridges B2a (h44 and H69), B2b (h24), B7a (h23 and H68), and B3 (H71) have slightly shifted (by ≈8–12 Å) in position, other bridges have retained their overall positions but have undergone change in composition caused by the absence of bacterial protein homologues (e.g., S13 and L5). Some of these bridges are replaced by Lmr-specific protein–protein bridges, such as bridges B1b and B1c. Another bridge, positionally analogous to B9 of mammalian mitoribosome (20), is formed by the Lmr-specific proteins and appears to be a feature specific to mitoribosomes. Overall, the 2 subunits are held together by 3 RNA–RNA bridges (B2a, B2b, and B7a), 3 RNA–protein bridges (B3, B7b, and B8), and 3 protein–protein bridges (B1b, B1c, and B9). The presence of fewer RNA–RNA bridges in the Lmr (3, vs. 5 in eubacteria) could explain the lack of sensitivity of the Lmr to changes in Mg2+ concentration (16).
Fig. 4.
Locations of intersubunit bridges. The SSU (A) and LSU (B) are shown from their interface sides. Bridges have been marked on both subunits as red ellipses and circles. Color codes for the SSU and LSU components are the same as in Figs. 2 and 3.
The mRNA Channel and the tRNA Binding Sites.
The mRNA channel of the Lmr is highly protein rich (Fig. 5B; and see ref. 32 for the eubacterial mRNA channel). Both the entrance and the exit of the channel possess Lmr-specific proteins, and the surface of the channel along the SSU head is lined throughout by Lmr-specific proteins. However, the topology of the mRNA entrance (Fig. S8) is quite divergent from that in the mammalian mitoribosome (20). The mRNA entrance is also much wider in the Lmr, potentially to allow interaction of the mRNA with both the Lmr itself and the translational activator proteins in Leishmania.
Fig. 5.
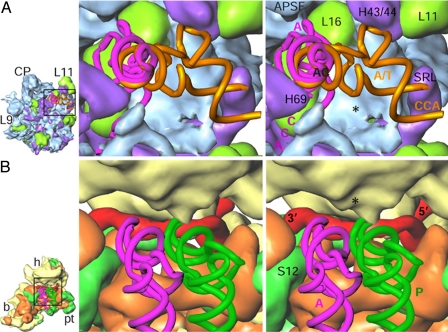
Stereo representation of topology of the mRNA and tRNA paths on the Lmr. (A) The tRNA path from A/T state to A site on the LSU. (B) The mRNA and tRNA paths encompassing A and P sites on the SSU. Positions of the A/T state (A/T, orange) and the A-site (A, pink) tRNAs are adopted from ref. 33, and the position of P-site tRNA (P, green) is adopted from ref. 29. The Lmr-specific protein densities (blue and yellow, respectively, in A and B) are marked by *. Landmarks: AC and CCA, anticodon and acceptor ends, respectively, of tRNAs. All other landmarks are as in previous figures.
Several rRNA segments that are involved in binding of tRNAs at all 3 sites, namely the A, P, and E (exit) sites in eubacterial ribosomes (22, 23, 29), are absent in the Lmr. Although most missing rRNA components of the A and P sites (SSU rRNA helices h31 and h34 and LSU rRNA helix H89) are replaced by proteins in the Lmr, those of the E site are not compensated. Furthermore, the E site lacks most of the binding rRNA components in both SSU (h23, modified and shifted helices h24, h29, and h42, and h43) and LSU (H11, H68, H77, and H88) of the eubacterial ribosome, suggesting that the E site is either very weak or absent altogether in the Lmr.
H89 and H91 form much of the lining of the corridor with which an incoming aminoacyl-tRNA interacts during its movement from initial binding A/T state (33) to the A site in the eubacterial ribosome (34). Although both of these helices are severely truncated in Lmr, their missing segments are partially replaced by Lmr-specific proteins (Fig. 5A). A dramatic change in chemical environment at the initial tRNA binding stage would have profound impact on how an aminoacyl-tRNA is accommodated during each elongation cycle in the Lmr. Similarly, the passage of tRNA from the A site to P site is dominated by Lmr-specific proteins (Figs. 5B and 6A). Thus, the protein–mRNA and protein–tRNA interactions are much more extensive in the Lmr than in any other ribosome for which a structure has been determined.
Fig. 6.
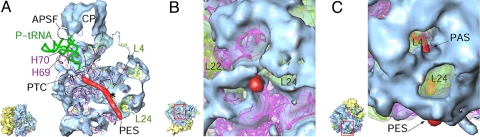
Topology of the polypeptide-exit tunnel. (A) Cut-away view of the LSU (white surfaces correspond to the cutting plane) to reveal the tunnel. (B) The PES, as seen from the bottom of the LSU. (C) The LSU is shown from the side opposite to that in A to reveal the PAS. A model of an α-helical polypeptide chain (red) is used to delineate the tunnel (35). * in A points to location of PAS that lies behind the cutting plane. All other landmarks are as in previous figures. The orientations of the Lmr, with the area boxed in red in the thumbnail enlarged, are shown in the corresponding panels to the left.
The Polypeptide-Exit Tunnel.
Consistent with the absence of eubacterial rRNA segments within domains I and III of the 12S Lm-rRNA, domains that occupy the bottom portion of the nascent polypeptide exit tunnel in bacterial LSU (35), the Lmr structure in this area is quite different from its eubacterial counterpart. The tunnel in the Lmr LSU has 2 openings on the solvent side, one corresponding to the conventional polypeptide-exit site (PES) and the second at ≈25 Å away but before the PES (Fig. 6A). We refer to the second exit as the polypeptide-accessible site (PAS). The solvent-side openings of both PES and PAS are predominantly encircled by Lmr-specific proteins (Fig. 6 B and C). The PAS has not been observed in any of the cytoplasmic or the chloroplast ribosomes and appears to be characteristic of mitochondrial ribosomes (20) necessary for the cotranslational insertion of polypeptides in the inner mitochondrial membrane.
Conclusions
We find several features that are unique to organellar (mitochondrial and chloroplast) ribosomes, such as the presence of a protein-rich base of the SSU body and the exit of the polypeptide tunnel (20, 24). Both of these regions could be involved in attachment of organellar ribosomes to the membrane. Furthermore, although the mitochondrial ribosomes differ dramatically in structure between mammalian (20) and protistan (this study) organisms, they evidently have some common characteristic features, such as a protein-rich mRNA path, a P-site finger, a tunnel in the SSU body, the PAS, and bridge B9.
We find that, despite its strikingly small complement of rRNA, the Lmr is similar to its eubacterial counterpart in overall size and morphology. The Lmr proteins that have bacterial homologues retain segments crucial for their incorporation in the Lmr, suggesting that all of the components that are required to build a functional translational machine are present in this highly-divergent form of the ribosome. Our results also suggest that the maintenance of a certain minimum size and the retention of key architectural elements have underpinned a notably-conserved basic functioning of the ribosome, despite the vicissitudes of the structure's long evolution. Nevertheless, several functionally-relevant regions in the Lmr have relative positions that differ from those seen in a eubacterial ribosome. From these characteristics, we predict that translational elongation factors in the Leishmania mitochondrion are also quite distinctive, if they are to complement the Lmr structure. Distinct topological differences, as compared with the eubacterial and eukaryotic ribosomes, in the mRNA and tRNA paths and PES in the Lmr suggest that the process of protein synthesis on the Lmr is mechanistically very different. Further structural and functional characterization of the protein components present at these key sites will be critical in establishing detailed mechanisms for the recruitment of mRNAs to the Lmr, movement of the tRNAs, and insertion of the nascent chains into the inner mitochondrial membrane.
Materials and Methods
Isolation of the Lmr.
The kinetoplast-mitochondrial fraction from the L. tarentolae was isolated by flotation in Percoll gradients as described (16, 36). Mitochondria were lysed with dodecyl maltoside, and 50S mitochondrial monosomes were purified on 2 successive sucrose gradients (16).
Cryo-EM and 3D Image Reconstruction.
Cryo-EM grids were prepared according to standard procedures (37). Data were collected on a Philips FEI Tecnai F20 field emission gun electron microscope, equipped with low-dose kit and an Oxford cryo-transfer holder, at a magnification of ×50,760. A total of 267 micrographs were scanned on a Zeiss flatbed scanner with a step size of 14 μm, corresponding to 2.76 Å on the object scale. The projection-matching procedure within the SPIDER software (37) was used to obtain the 3D map. Because the 50S Lmr displays greater overall morphological similarity to the eubacterial 70S ribosome than to the mammalian 55S mitoribosome (see ref. 17), we initially used an 11.5-Å resolution Escherichia coli 70S ribosome map (18) as the reference volume, to obtain a low-resolution map of the Lmr from ≈5,000 cryo-EM images. The Lmr 3D map so obtained was low-pass-filtered and then used as reference for the alignment of the larger dataset. Initially, 185,413 images, sorted into 30 groups according to defocus value (ranging from 1.6 to 4.5 μm under focus), were picked. Because of the problem of inherent heterogeneity of the Lmr (see SI Text), we had to eliminate a large proportion of images to improve the resolution. From the pool of manually screened 185,413 images, only 62,911 were retained, after elimination of the images that yielded low values for the cross-correlation coefficient with the 2D projection images generated from the initial model. Cross-correlation-based elimination was done in several steps (10–15% removed in each step) and was iterated until significant improvement in the map resolution was achieved. After removal of images from overrepresented groups within 83 equi-spaced views of the ribosome, another 9,436 images were eliminated. A total of 53,475 images were thus included in the final 3D reconstruction. The resolution of the final CTF-corrected 3D map, estimated by using the Fourier shell correlation with a cutoff value of 0.5 (see ref. 18), was 14.1 Å [or 9.8 Å by the 3σ criterion (38)].
Interpretation of the Cryo-EM Map.
Conserved regions in published X-ray structures of eubacterial ribosomes (13, 14, 22, 23) and homology models of the mammalian mitoribosome (25, 26) were used for molecular interpretation of the majority of the Lmr map. Furthermore, we applied energy minimization-based analysis, using program Mfold (27), to model structures of Lm-rRNA segments that were not available from X-ray or previously-built models. Among the top 5 secondary structures predicted by Mfold for each rRNA segment, the one that best explained the cryo-EM density was incorporated into our model. The cross-correlation coefficient (CCC) values between the fitted atomic coordinates and the corresponding cryo-EM density maps were determined after conversion of the fitted coordinates into the density map, through computation of averaged densities within volume elements scale-matched to those of the cryo-EM map (i.e., a pixel size of 2.76 Å, and after filtration of the atomic structures to the resolution of the cryo-EM density map). The CCC values between the filtered atomic coordinates and cryo-EM densities were in the 0.69 to 0.77 range. O (39) was used for docking of atomic coordinates and visualization was performed with SPIDER, IRIS Explorer (Numerical Algorithms Group), and Ribbons (40).
Supplementary Material
Acknowledgments.
We thank Melissa Breedlove and Giridhar K. Venkata for assistance with image processing and Linda Spremulli for discussions in early stages of this study. This work was supported in part by National Institutes of Health Grant GM61576 (to R.K.A.) and Human Frontier Science Program Grant RGY232003 (to R.K.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM map of the 50S Lmr has been deposited in the EM database, http://emdep.rutgers.edu (accession code EMD-5113). Fitted rRNA and protein components of the Lmr SSU and LSU have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3IY8 and 3IY9, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901631106/DCSupplemental.
References
- 1.Vickerman K, Preston TM. In: Biology of the Kinetoplastida. Lumsden WHR, Evans DA, editors. London: Academic; 1976. pp. 35–130. [Google Scholar]
- 2.Lukeš J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 3.Stuart KD, Schnaufer A, Ernst NL, Panigrahi A. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim Biophys Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horváth A, Neboháčová M, Lukeš J, Maslov DA. Unusual polypeptide synthesis in the kinetoplast-mitochondria from Leishmania tarentolae. Identification of individual de novo translation products. J Biol Chem. 2002;277:7222–7230. doi: 10.1074/jbc.M109715200. [DOI] [PubMed] [Google Scholar]
- 6.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat GJ, Souza AE, Feagin JE, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 1992;52:231–240. doi: 10.1016/0166-6851(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 8.Militello KT, Read LK. Coordination of kRNA editing and polyadenylation in Trypanosoma brucei mitochondria: Complete editing is not required for long poly(A) tract addition. Nucleic Acids Res. 1999;27:1377–1385. doi: 10.1093/nar/27.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feagin JE, Shaw JM, Simpson L, Stuart K. Creation of AUG initiation codons by addition of uridines within cytochrome b transcripts of kinetoplastids. Proc Natl Acad Sci USA. 1988;85:539–543. doi: 10.1073/pnas.85.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw J, Feagin JE, Stuart K, Simpson L. Editing of mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988;53:401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- 11.de la Cruz V, Lake JA, Simpson AM, Simpson L. A minimal ribosomal RNA: Sequence and secondary structure of the 9S kinetoplast ribosomal RNA from Leishmania tarentolae. Proc Natl Acad Sci USA. 1985;82:1401–1405. doi: 10.1073/pnas.82.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz V, Simpson A, Lake J, Simpson L. Primary sequence and partial secondary structure of the 12S kinetoplast (mitochondrial) ribosomal RNA from Leishmania tarentolae: Conservation of peptidyl-transferase structural elements. Nucleic Acids Res. 1985;13:2337–2356. doi: 10.1093/nar/13.7.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 14.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4-Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 15.Smith TF, Lee J, Gutell RR, Hartman H. The origin and evolution of the ribosome. Biol Direct. 2008;3:16. doi: 10.1186/1745-6150-3-16. http://www.biology-direct.com/content/3/1/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maslov DA, et al. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol Biochem Parasitol. 2006;148:69–78. doi: 10.1016/j.molbiopara.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Maslov DA, et al. Proteomics and electron microscopic characterization of the unusual mitochondrial ribosome-related 45S complex in Leishmania tarentolae. Mol Biochem Parasitol. 2007;152:203–212. doi: 10.1016/j.molbiopara.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabashvili IS, et al. Solution structure of the E. coli 70S ribosome at 11.5-Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 19.Spahn CMT, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae: tRNA–ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 20.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 21.Spahn CMT, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5-Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 23.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 24.Sharma MR, et al. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA. 2007;104:19315–19320. doi: 10.1073/pnas.0709856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mears JA, et al. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol. 2002;321:215–234. doi: 10.1016/s0022-2836(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 26.Mears JA, et al. A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zíková A, et al. Trypanosoma brucei mitochondrial ribosomes: Affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yusupov MM, et al. Crystal structure of the ribosome at 5.5-Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 30.Agrawal RK, et al. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol. 2000;150:447–459. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal RK, et al. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 33.Valle M, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 34.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 36.Braly P, Simpson L, Kretzer F. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J Protozool. 1974;21:782–790. doi: 10.1111/j.1550-7408.1974.tb03752.x. [DOI] [PubMed] [Google Scholar]
- 37.Frank J, Penczek P, Agrawal RK, Grassucci RA, Heagle AB. Three-dimensional cryoelectron microscopy of ribosomes. Methods Enzymol. 2000;317:276–291. doi: 10.1016/s0076-6879(00)17020-x. [DOI] [PubMed] [Google Scholar]
- 38.Orlova EV, et al. Structure of keyhole limpet hemocyanin type 1 (KHL1) at 15-Å resolution by electron cryomicroscopy and angular reconstruction. J Mol Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:103–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.