Abstract
Multidrug transporters provide a survival strategy for living organisms. As expected given their central role in survival, these transporters are ubiquitous, and in many genomes, several genes coding for putative transporters have been identified. However, in an organism such as Escherichia coli mutations in genes coding for transporters other than the major AcrAB-TolC multidrug efflux transporter have only a marginal effect on phenotype. Thus, whether the physiological role of the transporters identified is indeed drug export has been questioned. We show here that the minor effect of single mutations is due to the overlapping functionality of several transporters. This was revealed by generating multiple chromosomal deletion mutations in genes coding for transporters that share the same substrate and testing their effect on the resistance phenotype. In addition, complementation studies imply that AcrAB-TolC confers robust resistance provided that single-component transporters in the plasma membrane are functional. This finding supports the contention that hydrophobic drugs are removed in a 2-stage process: AcrAB-TolC removes substrates from the periplasmic space, while single-component transporters remove them from the cell. The overlapping specificities of the transporters ensure coverage of a wide range of xenobiotics and provide robustness in the response to environmental stress. This strategy also confers evolvability to the organism by reducing constraints on change and allowing the accumulation of nonlethal variation.
Keywords: AcrAB, drug resistance, EmrE, MdfA, multidrug transporters
Living organisms are constantly assailed by a host of harmful chemicals from the environment. Because of the diversity of these “xenobiotics,” cellular survival mechanisms must deal with an immense variety of molecules. Polyspecific drug transporters provide one means of doing so. These transporters recognize a wide range of dissimilar substrates that may differ in structure, size, or electrical charge and actively remove them from cells. As such, they provide an essential survival strategy for the organism. However, given that the substrates of these polyspecific transporters include many antibiotics as well as antifungal and anticancer drugs, they are associated with the phenomenon of multidrug resistance (MDR), which poses serious problems in the treatment of cancers and infectious diseases; consequently, some of them are known as multidrug transporters (MDTs) (1–3).
As expected given their central role in survival, these transporters are ubiquitous, and several genes coding for putative MDTs have been identified in many genomes. Genomic analysis has found that most bacteria have a large number of intrinsic potential drug exporter genes. Several different approaches have been taken to explore the role of these numerous putative MDTs. The ability of each putative MDT in a single organism (Escherichia coli) to confer resistance to a series of toxic compounds has been systematically questioned. Sulavik et al. (4) generated strains with null mutations in several efflux genes and tested their susceptibility to 35 compounds, including antibiotics, detergents, antiseptics, and dyes. Nishino and Yamaguchi (5) thoroughly tested the ability of each putative MDT in E. coli to confer resistance when expressed in a multiple-copy plasmid, either with its own promoter or in an expression vector. They identified 20 genes that confer resistance to at least 1 of a series of 26 antimicrobial agents.
Both of these studies confirmed, as suggested by many before, that AcrAB-TolC provides the major intrinsic resistance of these cells. In the study of Sulavik et al., null AcrAB strains became more susceptible to 29 of the 35 compounds tested (4); in the study of Nishino and Yamaguchi, expression of plasmidic AcrAB conferred significant resistance to 19 of the 24 compounds tested (5). Because the role of AcrAB-TolC is so central, the questions are raised as to why the organism needs all of the other drug exporter genes and whether their physiological role is indeed drug export or transport of specific physiological compounds, with the ability to expel drugs being only a fortuitous side effect (6–9). The phenotype of single null mutations of transporters other than AcrAB-TolC would suggest a minor role in resistance to the compounds tested, because their sensitivity to ethidium and acriflavine is not dramatically modified compared with that in the wild-type cells. Moreover, multiple null mutations in genes from the same families do not necessarily have a synergistic effect (4, 10).
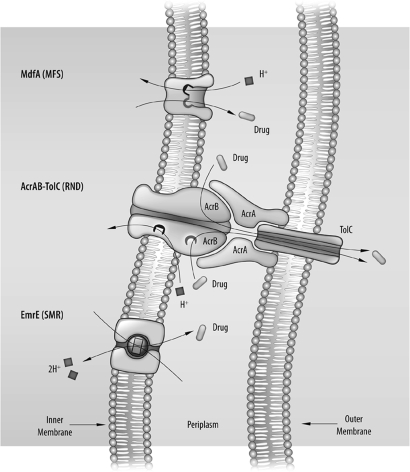
A possible explanation for the minor effect of null mutations in so many transporters is that this is due to backup compensation in which transporters with overlapping functionality cover for the loss of other transporters. To identify potential functional interactions of the transporters, we tested the effect of multiple deletion mutations on the phenotype of resistance to a single toxicant at a time. Here we chose to study the resistance to ethidium and to acriflavine, because other transporters besides AcrAB-TolC have been shown to confer resistance to either one (5). In addition, 2 of the aforementioned transporters have been thoroughly characterized. MdfA, a major facilitator superfamily (MFS) transporter that functions as a single 12 TM polypeptide, and EmrE, a small multidrug resistance (SMR) family transporter that functions as a homodimer, remove toxic compounds, including acriflavine and ethidium, from the cytoplasm in exchange for protons (11–17).
Using the approach described herein, we found that a double-null mutation in genes coding for transporters (emrE and mdfA) that share some of the same substrates (acriflavine and ethidium) has a strong effect on the resistance phenotype, and that cells exhibit sensitivity to these substrates almost as high as that seen in the ΔacrB strain. Remarkably, in strains in which the genes coding for MdfA and EmrE were both inactivated, the AcrAB-TolC complex provided only feeble resistance to ethidium and acriflavine. We propose that this is the result of a 2-stage functional organization in which MFS and SMR transporters in the plasma membrane transport the toxic compounds from the cytoplasm away from their targets and AcrAB-TolC removes these compounds from the periplasmic space out of the cell.
Results
AcrAB-TolC Is the Major Intrinsic MDT, but Other MDTs Play Important Roles as well.
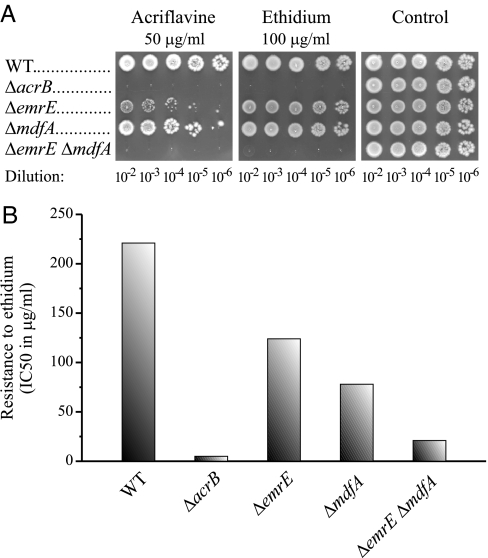
AcrAB-TolC from E. coli provides the major intrinsic resistance of these cells. Genetic inactivation of AcrB results in cells that are highly sensitive to a wide variety of compounds (3–5). We tested the effect of inactivation of genes coding for 2 other E. coli MDTs: EmrE, an SMR transporter and MdfA, an MFS transporter. As expected based on previous reports (4, 18), the single knockouts exhibited a distinct, but relatively weak, effect on the resistance to ethidium and acriflavine compared with the AcrB knockout. In the experiments reported here, the ability of cells to grow under various conditions was assessed in solid media containing ethidium (100 μg/mL) or acriflavine (50 μg/mL) in which 5 μL of logarithmic dilutions of an overnight culture were spotted (Fig. 1A). All of the cells tested grew well in the medium without toxicants at all of the dilutions tested (Fig. 1A, control). In the presence of either acriflavine or ethidium at the above concentrations, cells from a ΔacrB strain did not grow at any of the dilutions. In contrast, the growth of ΔemrE and ΔmdfA cells was only marginally impaired in the ethidium plate and only slightly more affected in the presence of acriflavine. Because EmrE and MdfA share some substrates, we tested whether they functionally overlap by generating a double-null mutation. When both genes were inactivated in the same strain, the resulting cells became practically as sensitive as the AcrB knockout (Fig. 1A).
Fig. 1.
The effect of MDT null mutations on the intrinsic resistance to acriflavine and ethidium. (A) Serial dilutions of overnight cultures were spotted (5 μL) on plates with or without toxic compounds (control). Growth was analyzed after overnight incubation at 37 °C. (B) Growth in liquid media was performed as described in Materials and Methods with increasing concentrations of ethidium. IC50 values were calculated from fits of the data obtained by Origin 8 software (the lowest R2 value is 0.93).
To quantify these results, we also tested growth in liquid media, and determined the IC50 for growth of the various strains (Fig. 1B). Whereas growth of BW25113 cells with intact MDTs (WT) was inhibited by 50% at 221 μg/mL of ethidium, the IC50 for ethidium for the other strains was 5 μg/mL for ΔacrB, 124 μg/mL for ΔemrE, 78 μg/mL for ΔmdfA, and 21 μg/mL for ΔemrE ΔmdfA. These results strongly support the observations in solid media and imply an additive effect of the double mutation. It should be noted that BW25113 cells exhibited a much higher intrinsic resistance to ethidium and acriflavine than any of the other commonly used E. coli strains (Fig. S1). Thus, E. coli DH5α, BL21 (DE3), and JM109 did not grow in plates containing either 50 μg/mL of acriflavine or 100 μg/mL of ethidium, whereas BW25113 cells grew even at concentrations as high as 150 μg/mL of acriflavine and 400 μg/mL of ethidium. E. coli TA15 displayed an intermediate phenotype (Fig. S1).
The results demonstrate that EmrE and MdfA are functional in the cell and when both of them are inactivated by deletion of the appropriate genes, the cell sensitivity to ethidium and acriflavine is almost as high as that in the ΔacrB strain.
AcrAB-TolC, the Major Intrinsic MDT, Confers only Marginal Resistance to Substrates of EmrE and MdfA in the Double-Null Mutant.
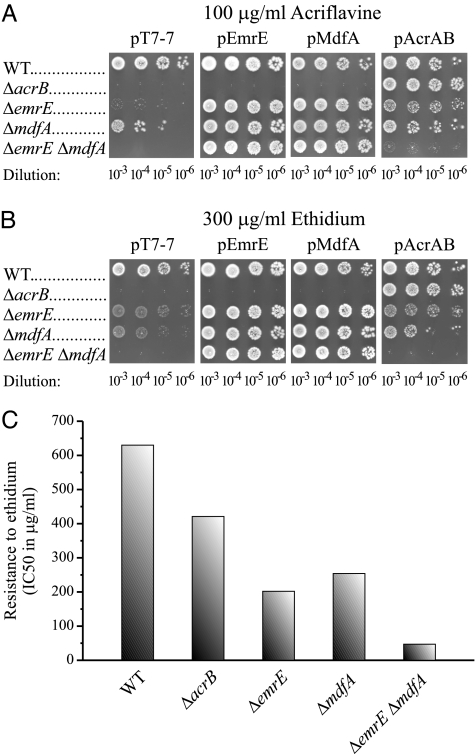
The phenotype of the double-null mutant indicates overlapping functionality between EmrE and MdfA. We further analyzed the interaction of the transporters coded by these genes with each other and with AcrAB-TolC by testing for functional complementation. We tested the ability of plasmidic genes to complement for the genomic mutations in solid medium (Fig. 2 A and B) and found that plasmidic emrE and mdfA by themselves complemented the defective phenotype of the single- and double-null mutations of the single-component transporters but not that of the ΔacrB strain. In contrast, plasmidic AcrAB closely complemented the phenotype of the ΔacrB strain, partially complemented the phenotype of the single null mutations, but could not restore full resistance to the double-null mutation ΔemrE ΔmdfA (Fig. 2 A and B).
Fig. 2.
Plasmidic expression complements the defective phenotype only for exporters of the same type. (A and B) Cells harboring pT7–7, pEmrE, pMdfA, or pAcrA together with pAcrB (pAcrAB in short) were tested for resistance on solid media with acriflavine (A) or ethidium (B). Growth was analyzed after overnight incubation at 37 °C. (C) The indicated strains carrying plasmidic AcrAB were grown as illustrated in Fig. 1B, and the calculated IC50 values are shown.
We further explored the notion that AcrAB-TolC provides only very weak resistance in the strain devoid of the 2 single-component transporters by determining the IC50 values for ethidium in the 5 strains expressing plasmidic AcrAB (Fig. 2C). IC50 values without plasmidic AcrAB are shown in Fig. 1B, and the effect of the plasmidic expression of acrAB is illustrated in Fig. 2C. In wild-type and ΔacrB cells, very robust resistance was conferred, with IC50 values of 630 and 421 μg/mL, respectively. Resistance also was significant (albeit lower) in the single-null mutants, with IC50 values of 202 μg/mL for ΔemrE and 254 μg/mL for ΔmdfA. Strikingly, however, only very marginal resistance was conferred to the double-null mutant, ΔemrE ΔmdfA, with an IC50 value of 47 μg/mL.
We conclude that for the compounds tested in the present study, efficient intrinsic resistance mediated by AcrAB-TolC requires the activity of single-component transporters.
In ΔemrE ΔmdfA Cells, AcrAB-TolC Confers full Resistance to Substrates That Are Not Shared with EmrE or MdfA.
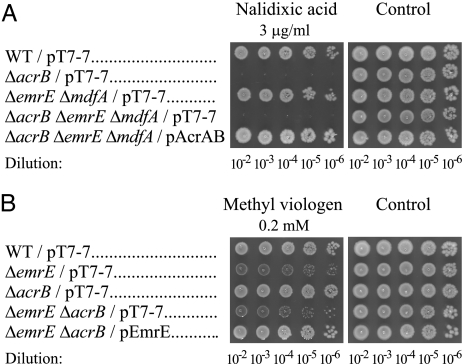
The finding of no cross-complementation of phenotype between the 2 types of transporters (i.e., AcrAB-TolC and the single-component transporters EmrE and MdfA) implies that in the process of removing ethidium from the cell to the medium, the AcrAB-TolC complex and the single-component transporters EmrE and MdfA function in a sequential manner. To support this conclusion, we ruled out the possibility that the lack of complementation may be due to a general effect on the membrane composition, permeability, or expression levels of the transporters. We did this through direct evaluation of the function of each transporter by assessing its ability to confer resistance to substrates that are not shared with the other transporters. The results of this direct test of transporter function are summarized in Fig. 3.
Fig. 3.
Plasmidic EmrE and AcrAB are fully functional in all null strains. Cells harboring the indicated plasmids were spotted on plates with and without nalidixic acid (A) or with and without methyl viologen (B). Growth was analyzed after overnight incubation at 37 °C.
Nalidixic acid is a substrate of AcrAB-TolC that is not recognized by either EmrE or MdfA (19, 20). This contention was confirmed by the fact that the null single (not shown) and double ΔemrE and ΔmdfA cells grew in the presence of 3 μg/mL of nalidixic acid as well as in its absence (Fig. 3A). The ΔacrB cells and a triple-null mutant of acrB, emrE, and mdfA did not grow at this concentration of nalidixic acid. This phenotype was fully complemented by plasmidic AcrAB in all of the foregoing strains, but not by either plasmidic emrE or mdfA (Fig. 3A; only complementation of the triple-null mutant by pAcrAB is shown). These results confirm that AcrAB-TolC is fully functional also in cells devoid of any EmrE or MdfA. They also imply that AcrAB-TolC may be handling nalidixic acid by itself or in interaction with MDTs other than MdfA or EmrE.
We performed the converse experiment to demonstrate that the inability of plasmidic emrE to restore ethidium and acriflavine resistance to the ΔacrB cells is not due to its impaired expression in this strain or to some unknown change in the membrane composition or permeability. We did this by analyzing the resistance of the various strains to methyl viologen, a substrate of EmrE (20) but not of MdfA (19) or AcrAB-TolC (5) (Fig. 3B). Null acrB (Fig. 3B) or mdfA (not shown) strains grew normally in solid media in the presence of 0.2 mM methyl viologen, whereas the growth of ΔemrE or ΔemrE ΔacrB strains was impaired (Fig. 3B). Plasmidic EmrE complemented this defect in both strains, supporting the conclusion that the lack of complementation for ethidium and acriflavine phenotypes is not due to some nonspecific effect.
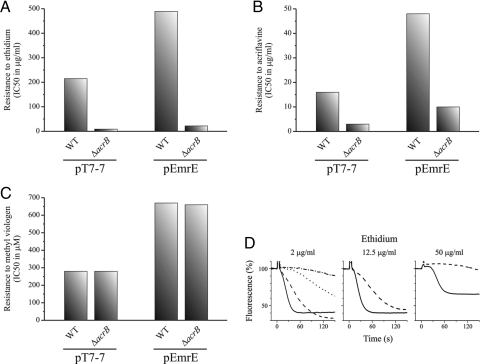
We quantified the increased resistance to methyl viologen by assessing growth in liquid media at various concentrations. The growth of wild-type and ΔacrB cells was inhibited by 50% at a similar methyl viologen concentration (280 μM) (Fig. 4C). This finding confirms the contention that methyl viologen is not a substrate of AcrAB-TolC. Plasmidic emrE increased the resistance to methyl viologen to a practically identical level in both strains (Fig. 4C). We conclude that the removal of methyl viologen from the cytoplasm to the periplasm and then out of the cell is independent of AcrAB-TolC, and that EmrE is functional in the ΔacrB mutant as well.
Fig. 4.
Plasmidic EmrE in ΔacrB cells confers full resistance to methyl viologen but only partial to ethidium and acriflavine. (A–C) WT and ΔacrB cells harboring pT7–7 or pEmrE were grown in liquid media with increasing concentrations of ethidium (A), acriflavine (B), or methyl viologen (C). IC50 values were calculated from fits of the data by Origin 8 software (the lowest R2 value is 0.95). (D) Ethidium efflux activity of EmrE. WT cells harboring pT7–7 (dotted line) or pEmrE (solid lines) and ΔacrB cells harboring pT7–7 (dashed-dotted line) or pEmrE (dashed lines) were assayed for ethidium efflux at the indicated concentrations. Glucose (0.36%) was added to initiate the active efflux of ethidium.
Partial Resistance to Ethidium Is Conferred by EmrE in the ΔacrB Strain.
In apparent contrast to the lack of functional complementation by EmrE in the ΔacrB mutant described here, Nishino and Yamaguchi (5) showed that plasmidic emrE confers additional resistance to ethidium over the intrinsic resistance in the KAM3 strain. The KAM3 strain is a derivative of E. coli K12 with impaired AcrAB-TolC activity (21). As described above, BW25113 and K12 derivatives display very different intrinsic resistance, which may be one reason for the differing effectiveness of EmrE in the 2 strains. In addition, it is possible that the levels of expression of the plasmidic EmrE differ because of differences in strain, promoter, or copy number. To support our findings, we conducted a more detailed analysis of the activity of plasmidic emrE in the BW25113 ΔacrB strain by growth in liquid media in the presence of multiple concentrations of ethidium and acriflavine. Our results confirm that plasmidic emrE confers only weak resistance to ethidium and acriflavine to the ΔacrB strain, at levels much lower than those detectable in wild-type strains (Fig. 4 A and B); whereas the IC50 values in the wild-type strains were 489 μg/mL for ethidium and 48 μg/mL for acriflavine, they were only 22 and 10 μg/mL, respectively, in the ΔacrB strain.
In ΔacrB Cells, Ethidium Is Not Removed Efficiently from the Periplasmic Space.
Although the fold increase in resistance conferred by EmrE is similar in both strains, the amount of ethidium that EmrE needs to transport to confer the resistance to 489 μg/mL or to 22 μg/mL is more than 1 order of magnitude greater. We postulate that in the ΔacrB strain, leakage into the cells of compounds such as ethidium is higher, because these compounds are not efficiently removed from the periplasm, and thus their concentration in this compartment is higher than in wild-type cells. To further elucidate this issue, we directly measured transport of ethidium in whole cells (Fig. 4D). In these studies, cells were starved in the presence of ethidium by incubation in the absence of an energy source and in the presence of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP). After the uncoupler was removed, transport was started by the addition of glucose. Although ethidium was present in both the medium and the cells, the fluorescence observed originated almost exclusively from the intracellular ethidium bound to nucleic acid. After the addition of glucose to wild-type cells transformed with plasmidic emrE (—), a rapid decrease in fluorescence occurred, which represents the removal of ethidium from the cell against its concentration gradient (Fig. 4D, Left Panel; 2 μg/mL of ethidium). This decrease was much slower in cells without plasmidic emrE (….) and was completely prevented when the cells were resuspended in a medium with CCCP (not shown). The rate of extrusion was much lower in the ΔacrB cells (-·-·-), as expected, but was significantly accelerated by plasmidic emrE (—-), and it reached very similar equilibrium to that in wild-type cells (albeit more slowly, after about 2 min). This indicates that EmrE is capable of removing ethidium from the cytoplasm against its concentration gradient also in the absence (albeit somewhat more slowly) of AcrAB. However, when the ethidium load was increased, the differences became greater (Fig. 4D, Center Panel; 12.5 μg/mL of ethidium) up to the point at which the activity of EmrE is undetectable in the ΔacrB cells (Fig. 4D, Right Panel; 50 μg/mL of ethidium). In the wild-type cells, the increase in concentrations caused changes in the apparent kinetic behavior, with an increasing lag that became more apparent at the highest ethidium concentration. Because the fluorescence is due mostly to the ethidium bound to the nucleic acid in the cell and this binding is concentration-dependent, we presume the lag is a function of the time required to remove ethidium below the equilibrium concentration. We conclude that in the ΔacrB cells, EmrE cannot cope with the increased leaks at the maximum concentrations tested, cannot efficiently decrease the intracellular concentration of ethidium, and thus confers only very feeble resistance.
Discussion
AcrAB-TolC is the major intrinsic MDT in E. coli and confers resistance to a wide range of toxic compounds (22–25). In addition, 19 other genes that code for proteins that can confer resistance to at least 1 of the 26 drugs tested have been identified (5). A high level of redundancy seems to exist for some drugs; thus, in E. coli 7 different MDTs confer resistance to ethidium and 6 different MDTs confer resistance to acriflavine (5). The phenotype of single-null mutations of emrE and mdfA suggests a minor role for this type of transporter in intrinsic resistance, because their sensitivity to ethidium and acriflavine is not dramatically modified compared with that in wild-type cells. Moreover, multiple null mutations in genes from the same families do not necessarily have a synergistic effect (4, 10). Resistance did not differ between the multiple null mutations in all of the genes coding for proteins from the SMR family and the single null emrE (Fig. S2). Thus, any redundancy in function that may provide backup is not necessarily from proteins of the same family.
To evaluate the possible functional overlap between exporters, we performed a simple genetic analysis of multiple null mutations in 3 of these genes that have at least 2 different drugs in common: EmrE, MdfA, and AcrAB, which have been reported to confer resistance to both ethidium and acriflavine. When genes of proteins catalyzing the same reaction were removed, a very obvious functional interaction was revealed. The higher sensitivity of the double-null mutation indicates that both proteins are functional in the intact cell, and they provide quite a good backup for one another. The finding that the null mutations in emrE and mdfA have an additive effect on the resistance suggests that they function at the same stage in the process of drug removal, that is, in the cell membrane, removing the toxic compounds from the cytoplasm to the periplasm. We propose that the additive effect is due simply to the decreased rates of drug removal that bring about an increase in the drug concentration in the cytoplasm above toxic levels. If this is a simple rate effect, then null mutations of emrE combined with another transporter could have a similar effect. Indeed, null mutations of emrE and yjiO (coding for another transporter that confers resistance to acriflavine) display a level of resistance similar to that of the ΔemrE ΔmdfA cells (not shown). In other words, when the rate of removal is lower than the leak into the cell, the compound accumulates to nonpermissive intracellular concentrations. A third null mutation (ΔemrE, ΔyjiO, and ΔmdfA) displayed very little additional effect (data not shown), implying that lowering the rate of removal from the cytoplasm even further below a threshold level (relative to the leak rate of the compound) has only a marginal effect on the intracellular concentration.
The findings presented here provide evidence supporting a significant role of the single-component transporters in the intrinsic resistance of E. coli. They also support the conclusion that each of the MDTs tested here (EmrE, MdfA, and YjiO) is functional in wild-type cells and removes toxicants at significant rates. In addition, and most strikingly, AcrAB-TolC provides only very weak resistance to ethidium and acriflavine in the absence of transporters in the plasma membrane that remove the foregoing compounds from the cytoplasm.
The effects of simultaneous expression of several efflux pumps on antibiotic resistance were previously investigated in E. coli and Pseudomonas aeruginosa to test the existence of functional cooperation among various MDTs. Simultaneous expression of a single-component efflux pump that exports antibiotics into the periplasm and a multicomponent efflux pump that accomplishes efflux directly into the external medium has been reported to have a synergistic effect on drug resistance (10, 26). No synergistic effects were detected in strains with multiple deletions from the same family, either MFS or SMR (4), or in a mdfA and norE double-null strain (10).
It has been suggested that transporters such as the AcrAB-TolC multidrug efflux transporter are capable of capturing their substrates in the periplasmic space rather than in the membrane or from the cytoplasm (3, 27, 28). This claim is supported by the high-resolution structures of AcrB, in which access pathways from the periplasm but not from the cytoplasm have been identified (29, 30).
The results described here and elsewhere (10, 26) provide strong evidence that AcrAB-TolC removes drugs only from the periplasm (Fig. 5). For the drugs tested, AcrAB-TolC was seen to be almost completely dependent on the activity of other MDTs. To investigate whether AcrAB-TolC is dependent on other MDTs for all of the drugs it transports, a careful and detailed assessment of overlapping specificities of other MDTs in E. coli is needed. On the other hand, dependence on AcrAB-TolC is not necessarily universal. Methyl viologen is a substrate of EmrE but not of AcrAB-TolC, and plasmidic expression of emrE in an acrB null strain confers full resistance as good as that in wild-type cells. The possibility that one of the other 3-component transporters may function to remove methyl viologen from the periplasm cannot be ruled out; however, we favor the possibility that for hydrophilic compounds such as methyl viologen, the diffusion from the periplasm to the outside is faster than the leak back to the cell through the cell membrane, and thus further mechanisms for its active removal from the cell are not needed. Indeed, the findings from our experiments on ethidium transport support the model proposed here (Fig. 5) and in previous work (26) that imply that if drug removal from the periplasmic space is not fast enough, the drug will quickly leak back into the cytoplasm, and the single-component transporter cannot cope with the load.
Fig. 5.
Model of functional interaction among AcrAB-TolC, EmrE and MdfA in E. coli. The single-component transporters, EmrE and MdfA, remove the drug (acriflavine or ethidium) from the cytoplasm and into the periplasm, where AcrAB-TolC captures it and extrudes it to the medium.
The interactions between the MDTs demonstrated here and in previous studies (10, 26) imply the development of a very robust survival mechanism provided by overlapping specificities of several MDTs. Along with the transporters, the cell response includes such components as drug-degrading enzymes, and this response provides the cell with a survival strategy to cope with resistance determinants present in the environment. This redundancy of versatile protein elements reduces the interdependence of components and confers robustness and flexibility in the response to harmful chemicals from the environment. Because of the diversity of these “xenobiotics,” cellular survival mechanisms must deal with a wide variety of molecules. MDTs with partially overlapping specificities and with functional interactions such as described here provide one means for doing so. They also confer evolvability on the organism by reducing constraints on change and allowing the accumulation of nonlethal variation. Evolvability may have been generally selected in the course of selection for robust, flexible processes suitable for life in hostile environments.
Materials and Methods
E. coli Strains and Growth Conditions.
E. coli BW25113 (31) and its isogenic deletion mutants (32) were used throughout this work; the mutants used are listed in Table S1. E. coli DH5α (Invitrogen) was used as host for cloning procedures, and UTL2mdfa::kan (18) was used as donor strain for P1 transductions. E. coli JM109 (33), BL21(DE3) (Novagen), and TA15 (34) were used for comparison of resistance phenotypes.
Cells were grown at 37 °C with shaking in LB or medium A with or without kanamycin (50 μg/mL), ampicillin (100 μg/mL), and chloramphenicol (34 μg/mL).
Plasmids.
The plasmids used in this work are listed in Table S1. pT7–7 (35) was used as the control plasmid. pEmrE (pT7-7-EmrE) (20) and pAcrA contain emrE and acrA, respectively, under the control of phage T7 RNA polymerase promoter. pMdfA is pT7–5-MdfA (19), which allows the expression of mdfA from its native promoter. pACYC is a derivative of pACYC184 (36). pAcrB is composed of AcrB with the MycHis tag in pACYC. The identity of the constructs was verified by sequencing.
Strain Construction.
The multiple-deletion mutants were constructed in E. coli BW25113 essentially as described previously (31). Plasmid pCP20 (37) was used to eliminate the kanamycin-resistance gene from the single-deletion mutants (32). The corresponding second and third mutations were introduced by P1 transduction with P1 prepared from appropriate E. coli BW25113 donors, except for the ΔmdfA::kan mutation introduced from UTL2mdfa::kan (18). Similarly, the ΔmdfA::kan mutation was transferred to BW25113 to generate the single-deletion mutant BW25113 ΔmdfA. The correct configuration of all deletion mutants was confirmed by PCR.
Resistance to Toxic Compounds.
For testing resistance on solid medium, 5-μL logarithmic dilutions of overnight cultures were spotted on LB agar plates with 30 mM Bis-Tris propane (pH 7) and the corresponding toxic compounds. Growth was analyzed after overnight incubation at 37 °C. For resistance in liquid medium, overnight cultures were diluted 100-fold into LB medium, and after 1 h at 37 °C, the cultures were diluted to OD600 of 0.01 in LB medium containing 30 mM Bis-Tris propane (pH 7) and the indicated concentrations of toxic compounds. After 6 h of growth at 37 °C, absorption at 600 nm was measured to estimate cell density. Growth (3 mL) was performed in 1.5 × 15-cm glass tubes (Fig. 4) or in 1.2-mL storage plates (ABgene), 300 μL per well, covered with gas permeable adhesive seal (Figs. 1 and 2). Data analysis was performed using Origin 8.0 software (OriginLab, Northampton, MA). The lowest R2 value in the fits for all of the experiments was 0.93.
Transport of Ethidium in Whole Cells.
Transport was assayed essentially as described previously (20). Cells grown to mid-exponential phase were harvested by centrifugation and resuspended in medium A without glucose to an OD600 of 0.5. Ethidium at the indicated concentrations and CCCP (40 μM) were added, and after 1 h at 37 °C, the cells were collected by centrifugation and resuspended in CCCP-free medium without glucose and with the original concentrations of ethidium. The reaction was initiated by the addition of 0.36% glucose. Fluorescence was measured at 30 °C with a PerkinElmer fluorometer (LS 50 B luminescence spectrometer) using FL WinLab software with an excitation wavelength at 525 nm and emission at 585 nm.
Supplementary Material
Acknowledgments.
We thank Prof. E. Bibi, Weizmann Institute of Science, for providing the UTL2mdfa::kan strain and a plasmid carrying the mdfA gene, and Sonia Steiner-Mordoch for constructing the plasmid-bearing AcrB. This work was supported by National Institutes of Health Grant NS16708 and Israel Science Foundation Grant 11/08. S.S. is Mathilda Marks–Kennedy Professor of Biochemistry at the Hebrew University of Jerusalem.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902400106/DCSupplemental.
References
- 1.Gottesman MM, Ling V. The molecular basis of multidrug resistance in cancer: The early years of P-glycoprotein research. FEBS Lett. 2006;580:998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 4.Sulavik MC, et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother. 2001;45:1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neyfakh AA. Mystery of multidrug transporters: The answer can be simple. Mol Microbiol. 2002;44:1123–1130. doi: 10.1046/j.1365-2958.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 7.Neyfakh A. Natural functions of bacterial multidrug transporters. Trends Microbiol. 1997;5:309–313. doi: 10.1016/S0966-842X(97)01064-0. [DOI] [PubMed] [Google Scholar]
- 8.Saier MH, Jr, et al. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 9.Krulwich TA, Lewinson O, Padan E, Bibi E. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat Rev. 2005;3:566–572. doi: 10.1038/nrmicro1181. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Clayton SR, Zechiedrich EL. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J Antimicrob Chemother. 2003;51:545–556. doi: 10.1093/jac/dkg126. [DOI] [PubMed] [Google Scholar]
- 11.Sigal N, Cohen-Karni D, Siemion S, Bibi E. MdfA from Escherichia coli, a model protein for studying secondary multidrug transport. J Mol Microbiol Biotechnol. 2006;11:308–317. doi: 10.1159/000095633. [DOI] [PubMed] [Google Scholar]
- 12.Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim Biophys Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Schuldiner S, et al. Small is mighty: EmrE, a multidrug transporter as an experimental paradigm. News Physiol Sci. 2001;16:130–134. doi: 10.1152/physiologyonline.2001.16.3.130. [DOI] [PubMed] [Google Scholar]
- 14.Paulsen I, et al. The SMR family: A novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol. 1996;19:1167–1175. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 15.Bay DC, Rommens KL, Turner RJ. Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim Biophys Acta. 2008;1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Schuldiner S. When biochemistry meets structural biology: The cautionary tale of EmrE. Trends BiochemSci. 2007;32:252–258. doi: 10.1016/j.tibs.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophy Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Edgar R, Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 1999;18:822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 21.Morita Y, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D, Cook DN, Hearst JE, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura H. Genetic determination of resistance to acriflavine, phenethyl alcohol, and sodium dodecyl sulfate in Escherichia coli. J Bacteriol. 1968;96:987–996. doi: 10.1128/jb.96.4.987-996.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zgurskaya HI, Nikaido H. Multidrug resistance mechanisms: Drug efflux across two membranes. Mol Microbiol. 2000;37:219–225. doi: 10.1046/j.1365-2958.2000.01926.x. [DOI] [PubMed] [Google Scholar]
- 25.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three's company: Component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struct Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Lee A, et al. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ. Waltzing transporters and “the dance macabre” between humans and bacteria. Nat Rev Drug Discov. 2007;6:56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- 29.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 30.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg EB, et al. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabor S, Richardson C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.