Abstract
Idiopathic pulmonary fibrosis is a lethal parenchymal lung disease characterized by denudation of the lung epithelium, fibroblast proliferation, and collagen deposition. Cellular changes underlying disease progression involve injury to alveolar epithelial cells, epithelial to mesenchymal transition, proliferation of α-smooth muscle actin (α-SMA)–expressing myofibroblasts and of fibroblasts resulting in enhanced deposition of extracellular matrix proteins. Hepatocyte growth factor (HGF) inhibits progression of bleomycin-induced pulmonary fibrosis in mice. The mechanism underlying the inhibitory effect of HGF was investigated in an in vitro model. We show that HGF markedly antagonizes basal and transforming growth factor (TGF)-β–induced expression of myofibroblast markers such as α-SMA, collagen type 1, and fibronectin in rat alveolar epithelial cells. HGF also inhibited TGF-β–induced α-SMA expression in primary murine alveolar epithelial cells. Since TGF-β is known to regulate α-SMA expression, the effect of HGF on components of TGF-β signaling was investigated. HGF induced expression of Smad7, an inhibitor of TGF-β signaling, in a mitogen-activated protein kinase–dependent manner. HGF also induced the nuclear export of Smad7 and Smad ubiquitin regulatory factor 1 (Smurf1) to the cytoplasm. HGF-dependent decrease in α-SMA was abolished with specific siRNAs targeted to Smad7. Thus, induction of Smad7 by HGF serves to limit acquisition of the myofibroblast phenotype in alveolar epithelial cells.
Keywords: EMT, HGF, Smad7
CLINICAL RELEVANCE
This study shows the ability of hepatocyte growth factor (HGF) to up-regulate Smad7 expression and function in epithelial cells, which antagonizes generation of the fibroblast phenotype. Thus, development of small molecules that can activate the HGF receptor to achieve the same results would be beneficial in the treatment of pulmonary fibrosis.
Idiopathic pulmonary fibrosis (IPF) is a devastating progressive disease of the lung characterized by diffuse fibroblastic foci in the parenchyma of patients. These fibrotic foci contain fibroblasts and α-smooth muscle actin (α-SMA)–expressing myofibroblasts responsible for enhanced deposition of extracellular matrix proteins. The source of these fibrotic features is not fully understood.
Recent evidence suggests that IPF may originate from repeated, minor injuries to alveolar epithelial cells leading to formation of the fibroblastic foci (1–4). Continuous epithelial cell insults lead to apoptosis and increased cytokine and growth factor secretion. Transforming growth factor (TGF)-β, an important growth factor mediating pro-fibrotic signals, is thought to be required for the generation of fibrotic foci (3–5). TGF-β is important for fibrosis in both the liver and kidney (6, 7), and in the lung (5, 8–11). There is an increasing body of evidence underscoring the importance of TGF-β in the induction of epithelial to mesenchymal transition (EMT), typically characterized by loss of cell adhesion, de novo expression of α-SMA and other fibrogenic mediators (12, 13). Treatment of rat or human alveolar epithelial cells with TGF-β results in a significant increase in the expression of myofibroblast marker α-SMA and reduction in epithelial cell markers (4, 14, 15). TGF-β–dependent induction of myofibroblasts from lung epithelial cells is considered to be an important step toward fibroblastic foci formation in IPF (3, 4, 16).
TGF-β transmits its signal through type I and II serine/threonine kinase receptors and phosphorylates downstream targets Smad2 and Smad3. Subsequently, Smad2 and Smad3 interact with Smad4, translocate to the nucleus, and activate TGF-β–responsive genes (17). Various regulators of Smad signaling have been identified that abrogate TGF-β responses. For example, the transcriptional corepressors SnoN, TGIF, and Ski bind activated Smads in the nucleus and prevent TGF-β–mediated transcription (18). In addition, Smad7, through interactions with the activated TGF-β receptor at the cell membrane, prevents the interactions of Smad 2/3 with the receptor (19). These negative regulators of Smad signaling keep TGF-β effects in check and may be important targets for modulating TGF-β–driven myofibroblast formation in lung fibrosis. In vivo, transient expression of Smad7 inhibited development of bleomycin-induced pulmonary fibrosis (20).
Hepatocyte growth factor (HGF) is a potent mediator of cellular proliferation, migration, survival, and tissue regeneration. First identified in the liver, HGF and its receptor, c-met, have now been found in lung, heart, kidney, and brain (21). A number of studies suggest that HGF plays a protective role in bleomycin-induced pulmonary fibrosis in mice. HGF expression plasmid or recombinant protein administration reduces bleomycin-induced collagen deposition as assessed histologically and by hydroxyproline content in the lung (22–25). HGF has been shown to induce MMP-9–dependent apoptosis of fibroblastic MRC5 cells (26). In contrast, HGF induces proliferation of alveolar epithelial cells and reduction in apoptosis, indicating protective functions of HGF on epithelial cells (22–25). However, the mechanisms responsible for HGF-dependent protection of alveolar epithelial cells and inhibition of pulmonary fibrosis are poorly understood.
In this study, we examined the effect of HGF on the development of a myofibroblast phenotype using an epithelial cell culture system previously used to demonstrate TGF-β1–dependent change of alveolar epithelial cells to myofibroblasts (15). We have found that HGF markedly antagonizes TGF-β receptor–dependent expression of not only the characteristic myofibroblast marker α-SMA, but also other pro-fibrotic mediators such as collagen type 1 and fibronectin in rat alveolar epithelial cells. HGF also antagonized TGF-β–induced α-SMA expression in primary murine alveolar epithelial cells. In addition, HGF induced expression of the negative regulator of TGF-β signaling, Smad7, in a mitogen-activated protein kinase (MAPK)-dependent manner, which reduced α-SMA expression in the cells. We also show that HGF induces the export of Smad7 and Smurf1 proteins from the nucleus to the cytoplasm, where TGF-β signaling can be antagonized. Therefore, induction of Smad7 by HGF serves to inhibit lung alveolar epithelial cells from assuming a myofibroblast phenotype.
MATERIALS AND METHODS
Cell Culture Treatments
Rat lung epithelial (RLE)-6TN cells were obtained from American Type Culture Collection (Manassas, VA) and grown in 1:1 mixture of Dulbecco's modified Eagle's medium and nutrient mixture F-12 Ham (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini Bioproducts, West Sacramento, CA), 40 mmol/L HEPES (Gibco/Invitrogen), 100 U/ml penicillin G, and 100 μg/ml streptomycin (Gibco/Invitrogen). For preparation of protein extracts for Western blotting, RLE-6TN cells were incubated in medium alone or in medium containing 2.5 ng/ml TGF-β1 (R&D Systems, Minneapolis, MN) ± 10, 50, or 100 ng/ml HGF (R&D Systems) for 24 hours for α-SMA protein and β-actin expression measurements by Western blotting. For phosphoprotein assessment, RLE cells were serum starved for 24 hours, followed by 100 ng/ml HGF treatment for 5 or 25 minutes. To inhibit specific cell signaling pathways, cells were pretreated with either a specific inhibitor of p38 (20 μM SB203580; Calbiochem, San Diego, CA), Akt inhibitor (10 μM; Calbiochem) or a specific inhibitor of MEK (50 μM PD98059; Calbiochem) for 30 minutes followed by treatment with 2.5 ng/ml TGF-β1 alone or in combination with 100 ng/ml HGF.
Primary Cell Isolation
Isolation of alveolar type 2 epithelial cells was performed using a previously described protocol with some modifications (27). Briefly, lungs from 6- to 8-week-old C57BL6 male mice were perfused with PBS containing 20 U/ml dispase (Gibco/Invitrogen), 0.1 mg/ml collagenase/dispase (Roche, Branchburgh, NJ), and 0.1 mg/ml DNase II (Sigma Aldrich, St. Louis, MO). Lungs were then digested at 37°C for 30 minutes, followed by mincing and sequentially passing through 100-μm and 40-μm sieves (BD Biosciences, San Diego, CA). Cells were washed and red blood cells were lysed using PharmLyze (BD Biosciences). CD45+ and CD32+ cells were magnetically depleted using biotinylated antibodies specific for CD45 and CD16/32 (BD Biosciences), respectively, followed by incubation with anti-biotin magnetic beads (Miltenyi Biotech, Auburn, CA). The epithelial cell–enriched fraction was collected and resuspended in bronchial epithelial growth media serum-free media with growth supplements (Clontech, Mountain View, CA). Cells were plated on collagen-coated plastic chambered slides. A quantity of 100 μg/ml cis hydroxy proline (Sigma Aldrich) was added during plating to inhibit fibroblast proliferation.
Immunocytochemistry
RLE cells were plated on glass chamber slides at 20% confluence. After 24 hours, cells were fixed with 2% paraformaldehyde in PBS for 10 minutes at room temperature. Nonspecific binding was blocked with 10% goat serum in 5% BSA/PBS. The antibodies used to examine expression of specific proteins were: mouse monoclonal anti–α-SMA (1:100 dilution; Dako, Carpinteria, CA), rabbit polyclonal anti-zonula occludens-1 (ZO-1) (2 μg/ml; Zymed, San Francisco, CA), mouse monoclonal anti-fibronectin (5G7, Abcam), rabbit polyclonal anti-human Collagen I (AB675P; Chemicon, Billerica, MA) or rabbit polyclonal E-cadherin (24E10; Cell Signaling, Danvers, MA). Antibody incubation was performed for 1 hour at room temperature followed by incubation with secondary conjugated Alexa 555 and Alexa 488 antibodies (Molecular Probes/Invitrogen). Nuclei were stained with a 1:1,000 dilution of DRAQ5 (Alexis Biochemical, Lausen, Switzerland). For Smad7 subcellular localization experiments, RLE cells were plated at 20% confluence. After 24 hours, cells were treated with 50 mM MEK inhibitor for 30 minutes and HGF for 3 hours, then fixed in 2% paraformaldehyde as described above. Cells were sequentially immunostained with rabbit anti-Smad7 (1:100 dilution; Santa Cruz Biotechnology) overnight at 4°C and then with mouse anti–E-cadherin (1:50 dilution; BD Transduction Labs, Franklin Lakes, NJ) for 1 hour. Appropriate secondary conjugated Alexa 488 and Alexa 555 antibodies were added for 1 hour, followed by DRAQ5 nuclear stain. For Smurf1 immunostaining, cells were incubated with 1:75 dilution of anti-goat Smurf1 antibody (Novus Biologicals, Littleton, CO). Confocal images were captured using an Olympus Fluoview 1000 Confocal system with an Olympus IX81 inverted microscope (Olympus, Center Valley, PA). Acquired images were analyzed with MetaMorph 7.1 software (Molecular Devices, Sunnyvale, CA). To calculate intensity levels of nuclear Smad7 and Smurf1. Nuclear regions were defined by DRAQ5 nuclear stain.
Nuclear, Cytoplasmic, and Total Protein Extract Preparation and Western Blotting
RLE nuclear and cytoplasmic extracts were prepared as previously described (28). RLE total cell extracts were prepared in SDS lysis buffer (0.0625 M Tris pH 6.8, 1.5% SDS, 10% glycerol) warmed to 95°C. Thirty-microgram protein samples were separated using 4 to 15% gradient polyacrylamide gels, transferred to PVDF membranes, and probed with the following antibodies: mouse monoclonal anti–α-SMA (1:2,000), rabbit anti-Smad7 (1:750); and the following antibodies purchased from Cell Signaling Technology: rabbit anti-phospho Smad2 (pSmad2) (Ser465/467) (1:1,000), rabbit anti-phospho Smad3 (pSmad3) (1:1,000), rabbit anti-Smad2/3 (1:1,000), rabbit anti-pSmad3 (Ser433/435) (1:1,000), rabbit anti–phospho-p44/42 MAPK (Thr202/Tyr204) (1:1,000), rabbit anti–total p44/42 MAPK (1:1,000), rabbit anti–phospho-JNK (1:1,000), rabbit anti–total JNK (1:1000), rabbit anti-Akt (Thr 308) (1:1,000), rabbit anti-Akt (Ser 473) (1:1000), rabbit anti–total Akt (1:1000), rabbit anti–phospho-p38 MAP Kinase (Thr180/Tyr182) (1:1,000), rabbit anti-p38 MAP kinase (1:1,000), rabbit anti–β-actin (1:1,000). All blots were exposed for optimal lengths of time for visualization.
Smad7 Knockdown Using siRNA
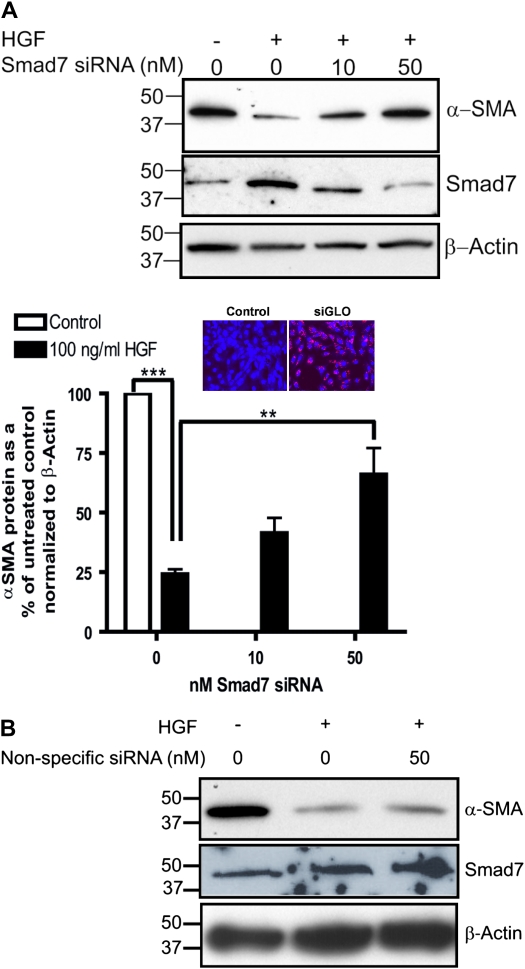
RLE cells were left untransfected, or transfected with 10 or 50 nM On-target plus Smartpool Smad7 siRNA or 50 nM nonspecific siRNA (Dharmacon, Chicago, IL) with siPORT NeoFX (Ambion) according to manufacturer's recommendations. Twenty-four hours after transfection, fresh medium containing 100 ng/ml HGF was added to the cells, followed by 24 hours of incubation. Cells were scraped and Smad7, α-SMA, and β-actin proteins were assessed by Western blotting. As a control for transfection efficiency, fluorescently tagged siRNA (siGLO; Dharmacon) was introduced into cells using the above protocol.
Statistical Analysis
All experiments were done independently three times unless otherwise indicated. Data are presented as means ± SE. Data were analyzed using Student's unpaired t test or Mann Whitney t test. In each case, P ≤ 0.05 was considered significant.
RESULTS
RLE-6TN Cells Express Basal ZO-1, E-Cadherin, Aquaporin-5, and α-SMA
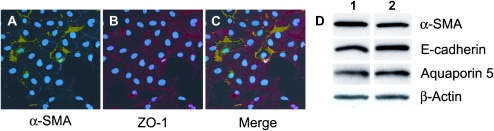
To characterize the cellular properties of the rat alveolar cell line, RLE-6TN, we performed immunostaining for the epithelial marker, ZO-1 (15, 29) and the myofibroblast marker, α-SMA. All cells expressed ZO-1 with a fraction of the cells also showing basal co-expression of α-SMA (Figures 1A–1C). In areas where cells exhibited cobblestone morphology, ZO-1 was plasma membrane–associated. A pattern of intracellular, fibril-associated α-SMA expression noted in some cells was consistent with several other reports (15, 30, 31). In addition, as shown by immunoblotting in Figure 1D, RLE cells express the epithelial cell markers, E-cadherin and aquaporin-5. Cells in lane 1 were plated at a low density and in lane 2 at a higher density, with no clear difference in protein expression suggesting that cell–cell contact did not influence myofibroblast or epithelial cell marker expression. Cumulatively, these results suggested that RLE-6TN cells possess characteristics of both epithelial cells and myofibroblasts. It should be noted that given the uniform expression of ZO-1 in all cells, it was clear that some cells spontaneously assumed a fibroblast phenotype. However, this property was not observed in other epithelial cell lines from human or rodent origin that are routinely maintained in our laboratory. This feature of RLE-6TN cells, however, prompted us to investigate whether the observed inhibitory effects of HGF on bleomycin-induced pulmonary fibrosis (20–24) could be studied at a mechanistic level using these cells as a model system.
Figure 1.
The alveolar epithelial cells rat lung epithelial (RLE)-6TN express basal myofibroblast marker α-smooth muscle actin (α-SMA) and epithelial cell markers E-cadherin, aquaporin-5, and ZO-1. RLE cells were plated on glass slides for 24 hours, fixed in 2% paraformaldehyde, and immunostained for α-SMA (green) and ZO-1 (red). Nuclei were stained with DRAQ5 (blue). A–C are representative images captured at a ×40 magnification. Note the fibrous strands present across these cells. A number of α-SMA and ZO-1 double-positive cells are evident along with cells expressing ZO-1 alone. (D) RLE cells were analyzed by Western blotting methods for epithelial cell markers E-cadherin and aquaporin-5, and the myofibroblast marker α-SMA. β-actin was assessed for equal protein loading. Cells were plated at a low density for samples in lane 1 and high density for samples in lane 2 harvested after 24 hours in culture. Results shown are representative of two independent experiments.
HGF Prevents Expression of TGF-β–Induced Fibrogenic Markers in Type 2 Pneumocytes and RLE Cells
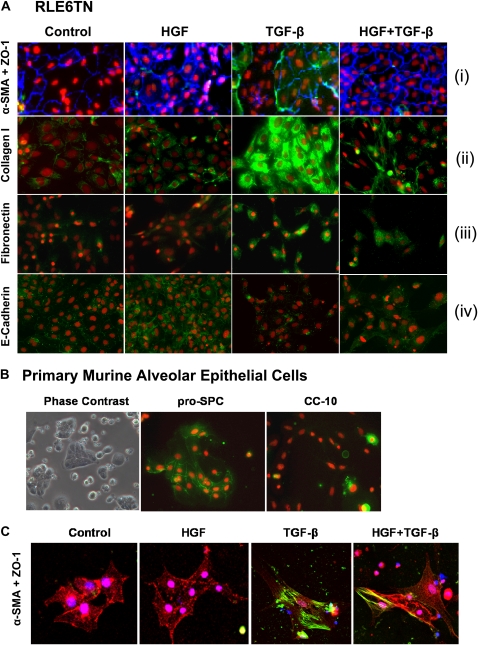
Studies in renal, hepatic, and lung fibrosis indicate that HGF may completely antagonize the effects of TGF-β and may contribute to the inhibition of epithelial to mesenchymal transition (7, 22–25, 32). We therefore sought to determine whether HGF may up-regulate the expression of epithelial proteins, and down-regulate fibrogenic proteins induced by TGF-β, in cultures of RLE cells. The expression of epithelial markers ZO-1 and E-cadherin was appreciably induced by HGF, but reduced by TGF-β. Simultaneous exposure to HGF and TGF-β did not inhibit the expression of epithelial markers (Figure 2A, panels i and iv). Cellular expression of pro-fibrotic markers such as α-SMA, collagen type 1, and fibronectin in response to HGF and TGF-β, alone or in combination, was also assessed in RLE cells. The results of our experiments indicate that the expression of these proteins was up-regulated by TGF-β but not by HGF, and HGF treatment along with TGF-β abrogated the effect of TGF-β (Figure 2A, panels i–iii).
Figure 2.
Treatment of primary pneumocytes and RLE cells with hepatocyte growth factor (HGF) results in inhibition of transforming growth factor (TGF)-β–mediated EMT. (A) RLE cells treated with or without HGF ± TGF-β were fixed and immunofluorescently stained for (i) α-SMA (green) and ZO-1 (blue); (ii) collagen type I (green); (iii) fibronectin (green); or (iv) E-cadherin (green). Nuclei were stained with DRAQ5 (red). (B) Primary murine alveolar epithelial cells in culture were observed by phase contrast (left panel); or immunofluorescence microscopy for pro–surfactant protein C (green, middle panel) or CC-10 (green, right panel) expression. Nuclei were stained with DRAQ5 (red). (C) Primary murine alveolar epithelial cells were cultured as shown and were fixed and stained for ZO1 (red), α-SMA (green), and nuclei (blue). Results shown represent at least two independent experiments. All images were digitally captured at ×40 magnification.
We also investigated whether the effects of HGF could be observed in primary murine alveolar type II cells. Murine primary pneumocyte cultures were generated using a previously established protocol and used to study the ability of HGF to prevent EMT (Figure 2B, left panel). Robust expression of the alveolar type 2 epithelial marker pro–surfactant protein C (Figure 2B, middle panel), and minimal expression of the Clara cell–specific airway epithelial cell marker CC-10 (Figure 2B, right panel) indicated an enriched population of alveolar epithelial cells. Cells cultured in the presence of HGF demonstrated retention of epithelial morphology with intact cell–cell contacts and strong expression of ZO-1. Treatment of cells with TGF-β, however, caused a change in morphology to fibroblast-like spindle-shaped cells with lower expression of ZO-1 and up-regulation of α-SMA. Upon exposure of the cells to HGF and TGF-β in combination, we observed a marked induction of ZO-1 and reduction of α-SMA (Figure 2C). These results suggest that HGF inhibits TGF-β signaling and fibroblastic transition in lung epithelial cells.
Interference with TGF-β Type I Receptor Signaling Down-Regulates the Expression of α-SMA in RLE Cells
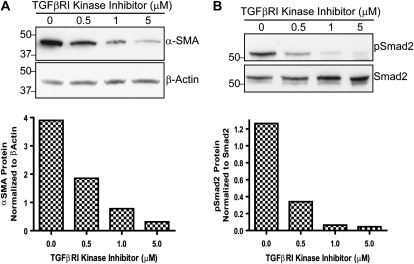
We next explored the basis for basal α-SMA protein expression in RLE cells. As noted in many studies, we chose α-SMA as a marker for the myofibroblast phenotype which has semblance to both fibroblasts and smooth muscle cells (33). We hypothesized that activation of the TGF-β receptor was responsible for the basal expression of α-SMA. Therefore, we treated cells with a TGF-β type I receptor kinase inhibitor, SB-431542, to block TGF-β signaling. This inhibitor has been shown to specifically block signaling downstream of the TGF-β type I receptor (activin receptor-like kinase 5 [ALK5]). It also inhibits the activin type I receptor ALK4 and the nodal type I receptor ALK7, which are very highly related to ALK5 in their kinase domains (34). Basal α-SMA protein decreased in response to increasing concentrations of the inhibitor (0.5–5μM) (Figure 3A). As expected, the inhibitor also blocked Smad2 phosphorylation (Figure 3B). This suggests that activation of the TGF-β type I receptor kinase or a related receptor kinase promotes α-SMA protein expression and a myofibroblast phenotype in RLE cells.
Figure 3.
The TGF-β type I receptor kinase inhibitor, SB-431542, significantly reduces α-SMA protein expression in RLE-6TN cells. Western blot analysis of RLE cells incubated without or with 0.5, 1, or 5 μM inhibitor for 24 hours. (A) Blots were probed for α-SMA and β-actin protein expression. The blots were analyzed by densitometry and α-SMA expression was normalized to β-actin. (B) Blots were re-probed with pSmad2 and total Smad2. Blots were analyzed by densitometry and levels of pSmad2 were normalized to total Smad2. The Western blots are representative of at least three independent experiments.
HGF Significantly Decreases α-SMA Protein Expression in RLE Cells
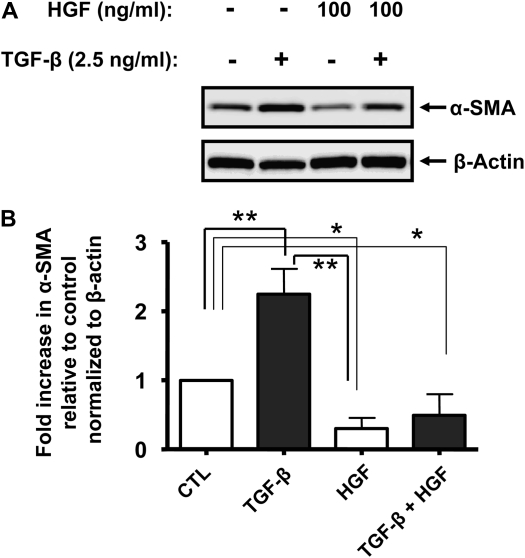
Through the activation of its tyrosine kinase receptor, HGF modulates cell motility, morphogenesis, survival, and proliferation. These effects may be important for the protective action of HGF in models of pulmonary fibrosis in vivo, as demonstrated in multiple studies (22–25). We examined whether HGF could alter levels of α-SMA protein in RLE cells. Cells were exposed to increasing concentrations (10–100 ng/ml) of HGF for 24 hours in the presence or absence of TGF-β, lysed, and assayed for α-SMA protein expression by immunoblotting. While lower concentrations of HGF did decrease basal and TGF-β–induced α-SMA (data not shown), a concentration of 100 ng/ml significantly inhibited expression of the protein (Figures 4A and 4B).
Figure 4.
HGF decreases basal and TGF-β–induced α-SMA protein in RLE cells. (A) RLE cells were incubated without or with 100 ng/ml HGF alone or in combination with 2.5 ng/ml TGF-β. After 24 hours, cells were lysed and samples containing equal amounts of total protein were resolved by SDS-PAGE. Expression of α-SMA and β-actin proteins was assessed by immunoblotting using specific monoclonal antibodies. (B) Relative expression of α-SMA was evaluated by densitometric analysis by calculating α-SMA to β-actin ratios for each sample. Statistical significance was established using Mann-Whitney t test. Results shown are representative of at least three independent experiments. *P < 0.05; **P < 0.01.
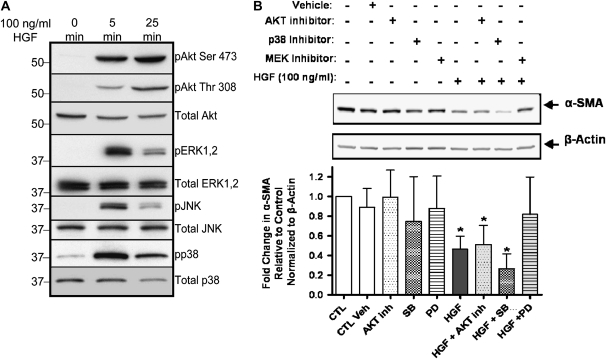
Inhibition of α-SMA Expression by HGF Is ERK1/2 Dependent
To elucidate the molecular mechanisms responsible for HGF-dependent inhibition of α-SMA, we first assayed the activation status of a variety of signaling molecules after HGF treatment. Exposure of RLE cells to HGF for 5 minutes significantly activated Akt (Ser 473 and Thr 308) and the MAPK family members ERK1/2, JNK, and p38 (Figure 5A). Akt activation continued through 25 minutes of HGF treatment, while ERK1/2, JNK, and p38 phosphorylation diminished slightly. Interestingly, we observed basal activation of p38 in the absence of HGF stimulation, suggesting that other endogenous factors, such as TGF-β, may be responsible for the basal levels of phospho-p38 (35, 36). We next examined the effect of blocking HGF-induced signaling molecules on α-SMA protein levels. RLE cells were exposed to optimal concentrations of pharmacologic inhibitors widely used to block specific signaling pathways. Either a MEK inhibitor (37, 38), an Akt inhibitor (39), or a p38 inhibitor (40, 41) was used for 30 minutes, followed by HGF treatment for 24 hours. The Akt inhibitor did not alter the HGF-mediated suppression of α-SMA protein. However, treatment of RLE cells with the MEK inhibitor, PD98059, significantly antagonized the HGF-mediated decrease in α-SMA (Figure 4B). Interestingly, exposure to the p38 inhibitor before addition of HGF further diminished the expression of α-SMA protein, tempting us to speculate that TGF-βR–dependent α-SMA protein production in these cells may potentially involve p38 activation (Figure 5B). Overall, these results showed that HGF reduces α-SMA protein in an ERK-dependent manner.
Figure 5.
HGF-mediated reduction of α-SMA proceeds through the MEK-ERK1/2 signaling pathway. (A) Stimulation of RLE cells with 100 ng/ml HGF for 5 and 25 minutes results in activation of Akt, ERK1/2, JNK, and p38 as shown by western blotting with phospho-specific antibodies. Total Akt, ERK1/2, JNK, and p38 protein levels were also assessed as controls for protein loading. (B) RLE cells were pre-treated for 30 minutes without or with vehicle (DMSO) or specific pharmacologic inhibitors for the kinases AKT (AKT inhibitor, 10 μM), p38 (SB203580, 20 μM),or MEK (PD98059, 50 μM). Cells were then treated with HGF (100 ng/ml) for 24 hours. Analysis of α-SMA expression was performed using immunoblotting and densitometric analysis of α-SMA/β-actin ratios. Results shown are representative of at least three independent experiments. *P < 0.05 compared with control (CTL) using Mann Whitney t test.
HGF Induces Smad7 Expression and Inhibits TGF-β Signaling
In two models of chronic renal disease, administration of HGF expression plasmids resulted in reduction in renal injury (42, 43). HGF has been reported to intercept Smad2/3 translocation to the nucleus in intestinal fibroblasts (44), up-regulate Smad transcriptional corepressor TGIF in mesangial cells (45), and increase expression of the transcriptional corepressor SnoN in tubular epithelial cells (46). To date, no report exists on effects of HGF on myofibroblast generation from lung epithelial cells.
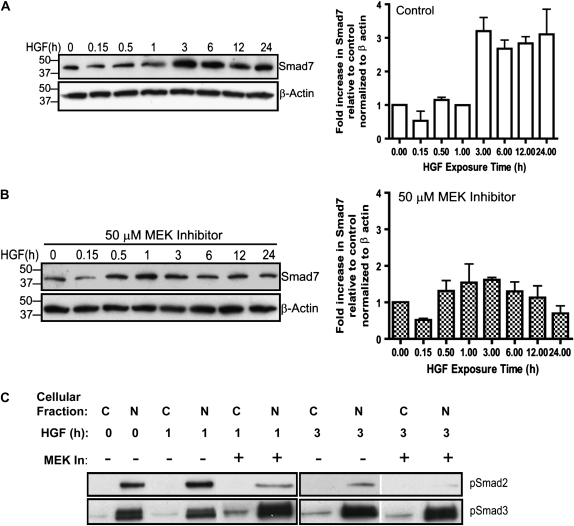
To explore whether HGF intersects the TGF-β signaling pathway in rat lung epithelial cells, we assessed Smad7 protein expression in the cells. RLE cells were treated with HGF, total protein was extracted, and Western blots were probed for Smad7 protein. Smad7 levels began to increase at 3 hours after HGF stimulation, and remained elevated through 24 hours (Figure 6A).
Figure 6.
HGF treatment increases Smad7 protein in RLE cells. (A) RLE cells were treated with HGF for 0 to 24 hours. Western blotting demonstrates Smad7 protein increases in a time-dependent manner after HGF treatment with protein levels peaking at 3 hours and remaining elevated through 24 hours. The blot was analyzed by densitometry and data are shown as percentage of control levels (0 h) as a ratio to β-actin. Standard error was calculated from the average of three independent experiments. (B) Thirty minutes of pretreatment with 50 μM PD98059 antagonized the HGF-dependent increase in Smad7 protein. Each blot was analyzed by densitometry and data are shown as percentage of control levels (0 h time-point) as a ratio to β-actin. Standard error was calculated from the average of three independent experiments. pSmad2 and pSmad3 levels were determined in cells treated with 50 μM PD98059 for 30 minutes and HGF for 1 or 3 hours. Nuclear (N) and cytoplasmic (C) extracts were collected and Western blots generated. pSmad2 levels were increased in response to HGF treatment with the MEK inhibitor antagonizing the activation. The MEK inhibitor had no effect on pSmad3 levels.
To determine if HGF-dependent Smad7 induction involved MEK-ERK activation, RLE cells were pretreated with the MEK inhibitor for 30 minutes before HGF stimulation. The MEK inhibitor essentially blocked the HGF-dependent increase in Smad7 protein without any effect on basal Smad7 levels (Figure 6B). Given that the MEK inhibitor appreciably blocked the HGF-dependent induction of Smad7, we also wanted to investigate the effects of the inhibitor on pSmad2 and pSmad3 levels. In Figure 6B, we show that pSmad2 and pSmad3 profiles were different in response to HGF and the MEK inhibitor. There was a small increase in pSmad2 in response to HGF, peaking at 1 hour after stimulation, which was inhibited by the MEK inhibitor. pSmad3 levels peaked at 3 hours after treatment with HGF and were refractory to treatment with the MEK inhibitor. Thus, the decrease observed for pSmad2 was not due to unequal loading and the pSmad3 level was useful as a loading control. Since all three proteins—Smad2, Smad3, and Smad4—have been shown to be required for Smad7 transcription (47), inhibition of pSmad2 by the MEK inhibitor is in agreement with the reduced levels of Smad7 observed in Figure 6B.
In contrast to SnoN protein up-regulation by HGF in kidney epithelial cells (7), no effect on SnoN protein in rat alveolar epithelial cells was observed after HGF treatment (data not shown).
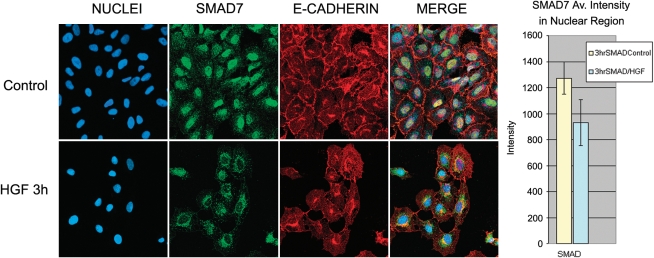
HGF Induces Nuclear Export of Smad7 and Smurf1
Smad7-dependent inhibition of TGF-β signaling occurs at the TGF-β receptor. Therefore, to assess cellular localization of Smad7 in the presence or absence of HGF, we performed immunocytochemistry in RLE cells with antibodies to Smad7 and a cell membrane–associated protein, E-cadherin. The majority of Smad7 was found to be localized to the nucleus in untreated RLE cells, as has been previously noted in many cell types (48). However, after 3 hours of HGF treatment, nuclear Smad7 decreased and cytoplasmic Smad7 increased (Figure 7). Quantitative analysis of 100 cells averaged across five fields per treatment revealed a significant shift of Smad7 out of the nuclear region after treatment with HGF. It should be noted that the average values shown in a bar graph format include cells that appeared to have lost all nuclear Smad7 and others that still retained an appreciable amount. These results show that HGF treatment promotes export of Smad7 from the nucleus, which is required for its inhibitory functions.
Figure 7.
HGF induces export of Smad7 from the nucleus to the cytoplasm. RLE cells were plated on glass slides. After 24 hours, cells were treated with 100 ng/ml HGF for 3 hours, fixed with 2% paraformaldehyde for 10 minutes at room temperature, and immunostained for Smad7 (green) and E-cadherin (red). Nuclei (blue) were stained with DRAQ5. Representative images were captured using ×40 magnification. HGF-treated cells exhibit predominantly cytoplasmic Smad7 staining. Average intensity of the nuclear region was determined by using the nuclear stain to define the areas of interest and then performing intensity analysis with MetaMorph software. Intensity was calculated on a per-cell basis, averaged, and presented for control and HGF-treated cells. Five fields containing a total of 100 cells were analyzed from images captured using a ×40 objective.
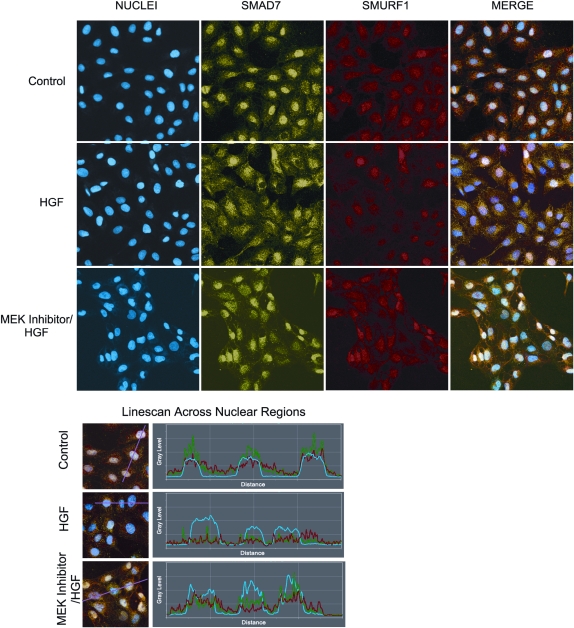
Smad ubiquitin regulatory factor 1 (Smurf1) exports Smad7 from the nucleus to the cytoplasmic region, where it can interact with the type I TGF-β receptor, leading to inhibition of TGF-β signaling (49, 50). Because we observed a change in the distribution of Smad7, we asked whether Smurf1 also displayed a change in localization after HGF exposure. RLE cells were treated with the MEK inhibitor for 30 minutes and HGF for 3 hours, fixed, and observed with confocal immunofluorescent microscopy. Intensity analysis of over 300 cells was conducted in MetaMorph using DRAQ5 labeling of the nuclei to define the nuclear region. In untreated cells, both Smad7 and Smurf1 were predominantly in the nucleus (Figure 8). After 3 hours of exposure to HGF, analysis of nuclear intensity of both Smad7 and Smurf1 was similarly decreased. However, pretreatment with the MEK inhibitor blocked the HGF-dependent nuclear export of Smad7 and Smurf. Exposure to HGF and the inhibitor resulted in maximum nuclear expression of both Smad7 and Smurf1. Linescan profiles (Figure 8) through the center of the nuclei underscore the results of the image analysis. Nuclear linescan regions are identified by the blue line, Smad7 by the green line, and Smurf1 by the red line. In the control cells, Smad7 and Smurf1 expression is predominantly in the nuclear region. After treatment with HGF, the nuclear expression of Smad7 and Smurf1 drops dramatically. After treatment with the MEK inhibitor and HGF, the majority of expression remains in the nuclear region. These results suggest that HGF regulates the Smad7-Smurf1 cellular localization in an ERK-dependent fashion.
Figure 8.
HGF-dependent translocation of Smad7/Smurf1. RLE cells were plated on glass microscope slides and treated with 50 μM PD98059 (MEK inhibitor), 100 ng/ml HGF, or the combination for 3 hours. Cells were fixed in 2% paraformaldehyde and immunostained with antibodies against Smad7 (green) and Smurf1 (red). Nuclei (blue) were stained with DRAQ5. Nuclear intensities of Smad7 and Smurf1 proteins were determined as described in the legend to Figure 6. Linescan profiles of representative cells in MetaMorph correspond to results of image analysis and show a dramatic reduction in the nuclear presence of both Smad7 and Smurf1 in response to HGF which was antagonized by the MEK inhibitor.
Knockdown of Smad7 Relieves HGF-Mediated Inhibition of α-SMA Protein Levels
To establish that the HGF effect was dependent on Smad7 up-regulation, we examined the effect of Smad7-specific siRNA on α-SMA protein expression. Cells were either left untransfected or transfected with 10 or 50 nM Smad7 siRNA, treated with 100 ng/ml HGF for 24 hours, lysed, and protein collected for assessment of α-SMA, Smad7, and β-actin protein. As a control for the transfection, we used fluorescently tagged siRNA to label transfected cells in a separate experiment and observed a near 100% transfection efficiency of the protocol that was used in all other experiments (Figure 9A). Cells devoid of Smad7 siRNA and exposed to 100 ng/ml HGF for 24 hours demonstrated the characteristic decrease in α-SMA protein compared with untreated cells (Figure 9A). Increasing concentrations of Smad7 siRNA antagonized the HGF-dependent decrease in α-SMA protein (Figure 8A). Transfection of 50 nM Smad7 siRNA restored the level of α-SMA protein to 60% of the levels in untreated controls (Figure 9A). Transfection of nonspecific siRNA had no effect on Smad7 or α-SMA protein levels (Figure 9B). An overall 60% inhibition of Smad7 expression appreciably blocked the HGF-mediated inhibition of α-SMA protein levels. These results suggested that HGF-dependent down-modulation of α-SMA is regulated, in part, by induction of Smad7 and inhibition of Smad signaling.
Figure 9.
Smad7 siRNA antagonizes the HGF-dependent decrease in α-SMA protein in RLE cells. (A) RLE cells were untransfected or transfected with 10 or 50 nM Smad7-specific siRNA or (B) 50 nM nonspecific siRNA followed by treatment with 100 ng/ml HGF for 24 hours. Cell extracts were collected and Western blotting was performed for detection of α-SMA, Smad7, and β-actin protein. Each blot was analyzed by densitometry, and averages and standard errors were determined from three independent experiments. The level of α-SMA protein was normalized to β-actin and the untreated control and presented as a percentage of the untreated control. **P < 0.05; ***P < 0.01. Inset: RLE cells transfected with fluorescently-tagged siRNA (siGLO) or mock transfected with delivery reagent only (Control) exhibited approximately 100% transfection efficiency.
DISCUSSION
In this study, we have shown that the rat alveolar epithelial cell line RLE-6TN has the ability to change to a phenotype that co-expresses both epithelial cell–specific proteins such as ZO-1 and the myofibroblast-specific protein α-SMA. The RLE-6TN cells model the characteristic change in lung tissue in patients with IPF in whom α-SMA and epithelial markers are co-expressed in many cells in the parenchyma, suggesting the presence of cells in transition from epithelial to a myofibroblast phenotype (15). We also observe a population of cells that exclusively expresses ZO-1 without α-SMA. This model system has allowed us to study the influence of HGF, previously shown to inhibit experimental fibrosis in mice (22–25), on myofibroblastic transition of cells. Here, we show that HGF inhibits this transition via upregulation of Smad7 expression and its export to the cytoplasmic compartment in the cell.
While many studies have noted the inhibitory effects of HGF on bleomycin-induced collagen deposition and lung fibrosis, little is known regarding the underlying mechanism(s). Figures 2 and 4 clearly demonstrate that HGF reduces TGF-β–induced α-SMA protein in RLE cells, which was also observed in primary murine alveolar epithelial cells. Others have shown that EMT involves deposition of collagen, fibronectin, and loss of cell–cell contacts (51). We report that HGF abrogates the expression of TGF-β–induced collagen type 1 and fibronectin while up-regulating cell adhesion proteins such as E-cadherin and ZO-1, thereby potentially reversing EMT. HGF binds to the c-met receptor inducing auto-phosphorylation of tyrosine residues in the cytoplasmic domain and in turn transmits downstream signals (7). Different molecular mechanisms have been identified for antagonism of TGF-β signaling by HGF. In kidney fibroblasts, translocation of phosphorylated Smad2/3 is blocked by HGF (52). Furthermore, in renal tubular epithelial cells, SnoN, a transcriptional corepressor of TGF-β–responsive genes, is increased upon HGF treatment (46). However, we have observed that HGF does not modulate SnoN protein levels in RLE cells (data not shown). Thus, HGF-mediated up-regulation of Smad7 expression, as also alteration of its subcellular localization, adds to the repertoire of mechanisms used by HGF to influence signaling downstream of the TGF-β receptor.
Interestingly, the activation of p38 has been shown to enhance expression of both Smad7 and α-SMA (53). In our investigations of the participation of MAPK pathways in HGF-mediated decrease in α-SMA levels, it was evident that ERK and p38 MAPK have different roles in mediating HGF effects. While ERK was required for inhibitory effects of HGF, HGF-induced p38 promoted α-SMA levels. Therefore, we were unable to reverse the HGF response with blocking of the MEK-ERK pathway because HGF was still able to increase α-SMA levels through the activation of p38. Significant attenuation of α-SMA protein levels was observed only when the p38 inhibitor was included with HGF.
On one hand, the receptor activated Smads, Smad2 and Smad3, are constitutively expressed and activated by phosphorylation upon activation of the TGF-β receptor. On the other hand, the inhibitory Smad7, typically repressed, gets transcriptionally induced to provide negative feedback to Smads 2/3 during this process. Since new protein synthesis is involved in Smad7 up-regulation, it is not surprising that we begin to observe a detectable increase in Smad7 protein, not immediately but at 3 hours after HGF treatment, staying elevated up to 24 hours (Figure 6A). Since Smad2 and Smad3 have been shown to play a role in Smad7 transcription (47, 50), we assessed their protein levels in the nucleus under the influence of HGF. Interestingly, nuclear pSmad2 peaked at 1 hour after HGF stimulation, while pSmad3 peaked at 3 hours. Thereafter, pSmad3 levels began to increase and remained elevated through 24 hours, which was not the case for pSmad2 (data not shown). It therefore seems likely that pSmad3 induction may occur via a mechanism secondary to HGF stimulation. In a previous study, EGF was shown to promote Smad3 phosphorylation (54). Given that binding of activated Smad3 to the Smad7 promoter has been documented (55), we believe that HGF-dependent activation of Smad3 plays a role in sustaining Smad7 levels. Activation of Smad2, however, also plays a role in Smad7 transcription (47). We observed a dramatic reduction in nuclear pSmad2 (but not pSmad3) and cytosolic Smad7 levels in response to HGF in the presence of the MEK inhibitor (Figure 6B). Collectively, it seems likely that activation of both Smad2 and Smad3 is required to initiate Smad7 transcription, but that pSmad3 may be required to sustain it. It will be interesting to determine whether Smad2 plays a recruiting role in initiation of Smad7 transcription.
Previous reports suggest that the subcellular localization of Smad7 is important for its inhibitory function (56). For example, in COS1 cells, the majority of ectopically expressed Smad7 was identified in the nucleus, but upon transfection with TGF-βRI, Smad7 was exported to the cytoplasm (48). Once in the cytoplasm, Smad7 is transported to the plasma membrane, where it inhibits Smad2/3 activation (50). We show that not only does HGF increase Smad7 protein, but that it also promotes its export to the cytoplasm, which is important to exert its inhibitory effects (48). Smurf1 and Smurf2, two E3 ubiquitin ligases have been shown to complex with nuclear Smad7, promoting its export to the cytoplasm, leading to inhibition of TGF-β signaling by degradation of the type I TGF-β receptor (49, 50, 57). Interestingly, the nuclear export of the Smad7/Smurf complex was found to be independent of TGF-β stimulation but was dependent on the Smad7 inducer IFN-γ (57). Our experimental observation that HGF up-regulates Smad7 and promotes the nuclear export of Smad7/Smurf1 is consistent with previous reports indicating that the relative abundance of Smad7 and Smurf2 (which is highly homologous to Smurf1) dictates the subcellular localization of Smad7. The complexity of TGF-β signaling, which is well documented, frequently involves subcellular compartmentalization of downstream proteins. The temporal and spatial distribution of signaling proteins downstream of the TGF-β receptors in response to HGF in lung or other epithelial cell types needs to be studied in more detail.
How does MEK-ERK signaling contribute to HGF-induced increase in Smad7 levels? Numerous reports indicate the involvement of ERK in the inhibition of TGF-β signaling (53, 58). ERK has been shown to specifically promote Smad1/Smurf1 nuclear export downstream of BMP-7 signaling (59). In line with these findings, we show that MEK inhibition not only blocks Smad7 protein induction, but that it also blocks the nuclear export of Smad7/Smurf1. Future studies will investigate the role of ERK activation in Smad7/Smurf1 association and the transcriptional machinery responsible for HGF-dependent up-regulation of Smad7 in lung epithelial cells.
The mechanism by which elevated Smad7 inhibits α-SMA is unclear. As discussed above, Smad7 inhibits TGF-β signaling through competitively binding to type I receptors and inhibits activation of Smad2/3 (60Smad7 has been shown to inhibit activation of integrin linked kinase (ILK) previously reported to promote TGF-β–induced EMT and α-SMA expression in renal fibrosis (61). The latter study also reported inhibition of ILK by HGF, thereby contributing to the inhibitory effect of Smad7 on α-SMA expression. In previous reports, increased Smad7 protein levels in response to IFN-γ (62), or the cytokines TNF-α and IL-1 and lipopolysaccharide (63), was shown to cause a substantial decrease in Smad2/3 activation. The relative phosphorylation status of Smad2 and 3 may contribute to preferential inhibition of α-SMA gene expression without compromising Smad7 gene expression. While pSmad2 may not play a role in sustaining Smad7 gene expression, it may be essential for α-SMA gene expression downstream of the TGF-β receptor activation. It is also possible that Smad7 uses alternate mechanisms to regulate α-SMA gene expression.
Recently, it has been reported that Smad7 overexpression leads to Smurf2-dependent degradation of β-catenin in mice, resulting in abnormal hair follicle growth (64). The level of Smad7 overexpression required to affect hair follicle growth was not sufficient to block Smad2/3 phosphorylation and transcription of Smad-inducible genes. However, even at the lower Smad7 expression level, it interacted with β-catenin, leading to Smurf2-dependent ubiquitination and degradation of β-catenin, causing abnormal hair follicle formation (64). This suggests alternative roles of Smad7 in regulating cellular functions that may be important for the Smad7-dependent effects on α-SMA protein levels. It is also possible that Smad7 may antagonize other transcription factors that mediate α-SMA gene expression in the lung epithelium. Other transcriptional corepressors of Smad signaling, such as c-Myc, Evi-1, and TGIF, may be involved in HGF-dependent down-regulation of α-SMA (60), which was not explored here. Collectively, our studies highlight a role for Smad7 in preventing acquisition of a myofibroblast phenotype in lung epithelial cells in response to HGF, which involves, at least in part, activation of the MEK-ERK pathway.
Acknowledgments
The authors thank Dr. Robert Hendricks for the use of the confocal microscope.
This work was supported by National Institutes of Health grants RO1 HL69810, P50 HL 70807, P50 84932 (to P.R.), TG HL007563 (to S.D.S.), and EY08098 (R.H.), the Core Grant for Vision Research.
Originally Published in Press as DOI: 10.1165/rcmb.2008-0217OC on November 6, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Selman M. Idiopathic pulmonary fibrosis challenges for the future. Chest 2001;120:8–10. [DOI] [PubMed] [Google Scholar]
- 2.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc 2006;3:364–372. [DOI] [PubMed] [Google Scholar]
- 3.Thannickal VJ, Toews GB, White ES, Lynch JP III, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med 2004;55:395–417. [DOI] [PubMed] [Google Scholar]
- 4.Gharaee-Kermani M, Gyetko MR, Hu B, Phan SH. New insights into the pathogenesis and treatment of idiopathic pulmonary fibrosis: a potential role for stem cells in the lung parenchyma and implications for therapy. Pharm Res 2007;24:819–841. [DOI] [PubMed] [Google Scholar]
- 5.Khalil N, Bereznay O, Sporn M, Greenberg AH. Macrophage production of transforming growth factor beta and fibroblast collagen synthesis in chronic pulmonary inflammation. J Exp Med 1989;170:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest 2002;110:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y. Hepatocyte growth factor in kidney fibrosis: therapeutic potential and mechanisms of action. Am J Physiol Renal Physiol 2004;287:F7–F16. [DOI] [PubMed] [Google Scholar]
- 8.Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med 1994;331:1286–1292. [DOI] [PubMed] [Google Scholar]
- 9.Broekelmann TJ, Limper AH, Colby TV, McDonald JA. Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc Natl Acad Sci USA 1991;88:6642–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westergren-Thorsson G, Hernnas J, Sarnstrand B, Oldberg A, Heinegard D, Malmstrom A. Altered expression of small proteoglycans, collagen, and transforming growth factor-beta 1 in developing bleomycin-induced pulmonary fibrosis in rats. J Clin Invest 1993;92:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Flanders KC, Phan SH. Cellular localization of transforming growth factor-beta expression in bleomycin-induced pulmonary fibrosis. Am J Pathol 1995;147:352–361. [PMC free article] [PubMed] [Google Scholar]
- 12.Willis BC, Borok Z. TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol 2007;293:L525–L534. [DOI] [PubMed] [Google Scholar]
- 13.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 2005;24:5764–5774. [DOI] [PubMed] [Google Scholar]
- 14.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir Res 2005;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalil N, Greenberg AH. The role of TGF-beta in pulmonary fibrosis. Ciba Found Symp 1991;157:194–207. (discussion 207–111). [DOI] [PubMed] [Google Scholar]
- 17.Massague J. How cells read tgf-beta signals. Nat Rev Mol Cell Biol 2000;1:169–178. [DOI] [PubMed] [Google Scholar]
- 18.Luo K. Ski and snon: negative regulators of TGF-beta signaling. Curr Opin Genet Dev 2004;14:65–70. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA Jr, Wrana JL, et al. The mad-related protein smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 1997;89:1165–1173. [DOI] [PubMed] [Google Scholar]
- 20.Nakao A, Fujii M, Matsumura R, Kumano K, Saito Y, Miyazono K, Iwamoto I. Transient gene transfer and expression of smad7 prevents bleomycin-induced lung fibrosis in mice. J Clin Invest 1999;104:5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurle RA, Davies G, Parr C, Mason MD, Jenkins SA, Kynaston HG, Jiang WG. Hepatocyte growth factor/scatter factor and prostate cancer: a review. Histol Histopathol 2005;20:1339–1349. [DOI] [PubMed] [Google Scholar]
- 22.Yaekashiwa M, Nakayama S, Ohnuma K, Sakai T, Abe T, Satoh K, Matsumoto K, Nakamura T, Takahashi T, Nukiwa T. Simultaneous or delayed administration of hepatocyte growth factor equally represses the fibrotic changes in murine lung injury induced by bleomycin: a morphologic study. Am J Respir Crit Care Med 1997;156:1937–1944. [DOI] [PubMed] [Google Scholar]
- 23.Umeda Y, Marui T, Matsuno Y, Shirahashi K, Iwata H, Takagi H, Matsumoto K, Nakamura T, Kosugi A, Mori Y, et al. Skeletal muscle targeting in vivo electroporation-mediated HGF gene therapy of bleomycin-induced pulmonary fibrosis in mice. Lab Invest 2004;84:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dohi M, Hasegawa T, Yamamoto K, Marshall BC. Hepatocyte growth factor attenuates collagen accumulation in a murine model of pulmonary fibrosis. Am J Respir Crit Care Med 2000;162:2302–2307. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe M, Ebina M, Orson FM, Nakamura A, Kubota K, Koinuma D, Akiyama K, Maemondo M, Okouchi S, Tahara M, et al. Hepatocyte growth factor gene transfer to alveolar septa for effective suppression of lung fibrosis. Mol Ther 2005;12:58–67. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno S, Matsumoto K, Li MY, Nakamura T. Hgf reduces advancing lung fibrosis in mice: a potential role for MMP-dependent myofibroblast apoptosis. FASEB J 2005;19:580–582. [DOI] [PubMed] [Google Scholar]
- 27.He M, Kubo H, Ishizawa K, Hegab AE, Yamamoto Y, Yamamoto H, Yamaya M. The role of the receptor for advanced glycation end-products in lung fibrosis. Am J Physiol Lung Cell Mol Physiol 2007;293:L1427–L1436. [DOI] [PubMed] [Google Scholar]
- 28.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor gata-3 is differentially expressed in murine th1 and th2 cells and controls th2-specific expression of the interleukin-5 gene. J Biol Chem 1997;272:21597–21603. [DOI] [PubMed] [Google Scholar]
- 29.Hermanns MI, Unger RE, Kehe K, Peters K, Kirkpatrick CJ. Lung epithelial cell lines in coculture with human pulmonary microvascular endothelial cells: development of an alveolo-capillary barrier in vitro. Lab Invest 2004;84:736–752. [DOI] [PubMed] [Google Scholar]
- 30.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol 2002;3:349–363. [DOI] [PubMed] [Google Scholar]
- 31.Yao HW, Xie QM, Chen JQ, Deng YM, Tang HF. TGF-beta1 induces alveolar epithelial to mesenchymal transition in vitro. Life Sci 2004;76:29–37. [DOI] [PubMed] [Google Scholar]
- 32.Inagaki Y, Higashi K, Kushida M, Hong YY, Nakao S, Higashiyama R, Moro T, Itoh J, Mikami T, Kimura T, et al. Hepatocyte growth factor suppresses profibrogenic signal transduction via nuclear export of smad3 with galectin-7. Gastroenterology 2008;134:1180–1190. [DOI] [PubMed] [Google Scholar]
- 33.Phan SH. The myofibroblast in pulmonary fibrosis. Chest 2002;122:286S–289S. [DOI] [PubMed] [Google Scholar]
- 34.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. Sb-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (alk) receptors alk4, alk5, and alk7. Mol Pharmacol 2002;62:65–74. [DOI] [PubMed] [Google Scholar]
- 35.Moustakas A, Heldin CH. Non-smad TGF-beta signals. J Cell Sci 2005;118:3573–3584. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Capelo A. Dual role for TGF-beta1 in apoptosis. Cytokine Growth Factor Rev 2005;16:15–34. [DOI] [PubMed] [Google Scholar]
- 37.Hayashida T, Decaestecker M, Schnaper HW. Cross-talk between ERK MAP kinase and smad signaling pathways enhances TGF-beta-dependent responses in human mesangial cells. FASEB J 2003;17:1576–1578. [DOI] [PubMed] [Google Scholar]
- 38.Lee PJ, Zhang X, Shan P, Ma B, Lee CG, Homer RJ, Zhu Z, Rincon M, Mossman BT, Elias JA. Erk1/2 mitogen-activated protein kinase selectively mediates IL-13-induced lung inflammation and remodeling in vivo. J Clin Invest 2006;116:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y, Qiao L, Wang S, Rong SB, Meuillet EJ, Berggren M, Gallegos A, Powis G, Kozikowski AP. 3-(hydroxymethyl)-bearing phosphatidylinositol ether lipid analogues and carbonate surrogates block pi3-k, akt, and cancer cell growth. J Med Chem 2000;43:3045–3051. [DOI] [PubMed] [Google Scholar]
- 40.Tsukada S, Westwick JK, Ikejima K, Sato N, Rippe RA. Smad and p38 MAPK signaling pathways independently regulate alpha1(i) collagen gene expression in unstimulated and transforming growth factor-beta-stimulated hepatic stellate cells. J Biol Chem 2005;280:10055–10064. [DOI] [PubMed] [Google Scholar]
- 41.Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Smad7 is required for TGF-beta-induced activation of the small GTPase cdc42. J Cell Sci 2004;117:1835–1847. [DOI] [PubMed] [Google Scholar]
- 42.Azuma H, Takahara S, Matsumoto K, Ichimaru N, Wang JD, Moriyama T, Waaga AM, Kitamura M, Otsuki Y, Okuyama A, et al. Hepatocyte growth factor prevents the development of chronic allograft nephropathy in rats. J Am Soc Nephrol 2001;12:1280–1292. [DOI] [PubMed] [Google Scholar]
- 43.Mizuno S, Kurosawa T, Matsumoto K, Mizuno-Horikawa Y, Okamoto M, Nakamura T. Hepatocyte growth factor prevents renal fibrosis and dysfunction in a mouse model of chronic renal disease. J Clin Invest 1998;101:1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Liu Y. Blockage of tubular epithelial to myofibroblast transition by hepatocyte growth factor prevents renal interstitial fibrosis. J Am Soc Nephrol 2002;13:96–107. [DOI] [PubMed] [Google Scholar]
- 45.Dai C, Liu Y. Hepatocyte growth factor antagonizes the profibrotic action of TGF-beta1 in mesangial cells by stabilizing smad transcriptional corepressor TGIF. J Am Soc Nephrol 2004;15:1402–1412. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Dai C, Liu Y. A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition. J Am Soc Nephrol 2005;16:68–78. [DOI] [PubMed] [Google Scholar]
- 47.Stopa M, Anhuf D, Terstegen L, Gatsios P, Gressner AM, Dooley S. Participation of smad2, smad3, and smad4 in transforming growth factor beta (TGF-beta)-induced activation of smad7: the TGF-beta response element of the promoter requires functional smad binding element and e-box sequences for transcriptional regulation. J Biol Chem 2000;275:29308–29317. [DOI] [PubMed] [Google Scholar]
- 48.Itoh S, Landstrom M, Hermansson A, Itoh F, Heldin CH, Heldin NE, ten Dijke P. Transforming growth factor beta1 induces nuclear export of inhibitory smad7. J Biol Chem 1998;273:29195–29201. [DOI] [PubMed] [Google Scholar]
- 49.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through smad7 and induces receptor degradation. J Biol Chem 2001;276:12477–12480. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. Smurf1 regulates the inhibitory activity of smad7 by targeting smad7 to the plasma membrane. J Biol Chem 2002;277:39919–39925. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y. Epithelial to mesenchymal transition in renal fibrogenesis: Pathologic significance, molecular mechanism, and therapeutic intervention. J Am Soc Nephrol 2004;15:1–12. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Dai C, Liu Y. Hepatocyte growth factor suppresses renal interstitial myofibroblast activation and intercepts smad signal transduction. Am J Pathol 2003;163:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dowdy SC, Mariani A, Janknecht R. Her2/neu- and tak1-mediated up-regulation of the transforming growth factor beta inhibitor smad7 via the ets protein er81. J Biol Chem 2003;278:44377–44384. [DOI] [PubMed] [Google Scholar]
- 54.Matsuura I, Wang G, He D, Liu F. Identification and characterization of ERK MAP kinase phosphorylation sites in smad3. Biochemistry (Mosc) 2005;44:12546–12553. [DOI] [PubMed] [Google Scholar]
- 55.Cutroneo KR, Phan SH. TGF-beta1-induced smad 3 binding to the smad 7 gene: knockout of smad 7 gene transcription by sense phosphorothioate oligos, autoregulation, and effect on TGF-beta1 secretion: bleomycin acts through TGF-beta1. J Cell Biochem 2003;89:474–483. [DOI] [PubMed] [Google Scholar]
- 56.Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. The n domain of smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J Cell Biol 2001;155:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to smurf2 to form an e3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 2000;6:1365–1375. [DOI] [PubMed] [Google Scholar]
- 58.Yang YC, Piek E, Zavadil J, Liang D, Xie D, Heyer J, Pavlidis P, Kucherlapati R, Roberts AB, Bottinger EP. Hierarchical model of gene regulation by transforming growth factor beta. Proc Natl Acad Sci USA 2003;100:10269–10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Balancing bmp signaling through integrated inputs into the smad1 linker. Mol Cell 2007;25:441–454. [DOI] [PubMed] [Google Scholar]
- 60.Feng XH, Derynck R. Specificity and versatility in TGF-beta signaling through smads. Annu Rev Cell Dev Biol 2005;21:659–693. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Yang J, Dai C, Wu C, Liu Y. Role for integrin-linked kinase in mediating tubular epithelial to mesenchymal transition and renal interstitial fibrogenesis. J Clin Invest 2003;112:503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-beta/smad signalling by the interferon-gamma/stat pathway. Nature 1999;397:710–713. [DOI] [PubMed] [Google Scholar]
- 63.Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Bottinger EP. A mechanism of suppression of TGF-beta/smad signaling by NF-kappa b/rela. Genes Dev 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- 64.Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, et al. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell 2006;11:301–312. [DOI] [PubMed] [Google Scholar]