Abstract
Kidney cancer is frequently metastatic on presentation at which point the disease is associated with a 95% mortality. Assessment of tumor grade on pathological examination is the most powerful means for prognostication as well as for stratification of patients into those who might respond to conventional or targeted therapy. Although there exist several grading systems in common use, all suffer from significant disparity among observers. In an attempt to objectify this process as well as to acquire grade-specific mechanistic information, we performed LC-MS/MS-based proteomics analysis on 50 clear cell kidney cancers equally distributed among normal tissues and Fuhrman grades 1–4. Initial experiments confirmed the utility of using archived formalin-fixed paraffin-embedded samples for LC-MS/MS-based proteomics analysis, and the LC-MS/MS findings were validated by extensive immunoblotting. We now show that changes among many biochemical processes and pathways are strongly grade-dependent with the glycolytic and amino acid synthetic pathways highly represented. In addition, proteins relating to acute phase and xenobiotic metabolism signaling are highly represented. Self-organized mapping of proteins with similar patterns of expression led to the creation of a heat map that will be useful in grade characterization as well as in future research relating to oncogenic mechanisms and targeted therapies for kidney cancer.
Kidney cancer (or renal cell carcinoma (RCC)1) is the seventh most common malignancy, the 10th most common cause of cancer death in men, and the ninth most common cancer in women. In 2009, an estimated 13,000 deaths (8,100 men and 4,900 women) will occur in the United States. The disease is frequently asymptomatic; a third of cases are diagnosed when the disease is already metastatic at which time it has 95% mortality (1).
Assessment of tumor grade is the most powerful available means to date of determining tumor prognosis; thus objective criteria for assessing grade are essential such that prognostication is unambiguous. In addition, grade criteria are useful in stratifying patients into those most likely to respond to conventional as well as new targeted therapies. There exist several systems for assigning tumor grade in RCC, although most pathologists utilize the Fuhrman grading system. As is evidenced by the abundance of extant grading systems (2), there appears to be a general lack of consensus and thus considerable variability in assigning tumor grades. Objective criteria for grade assignment utilizing specific protein markers will be useful in objectifying this process and thereby allowing for more accurate prognostication. Furthermore assessment of the biological basis of the differences among grades, as evidenced by diverse biochemical pathways altered in a grade-specific fashion, will lead to the development of novel diagnostic assays as well as therapeutic interventions.
Once objective grading criteria are put forth, molecular mechanisms by which tumors transition among grades can be identified and further investigated. Using this information, it might be possible to recapitulate the grade transition in vitro to discover novel mechanisms of oncogenesis or at least of transition from a relatively benign to a highly malignant phenotype. Moreover utilizing a systems biology approach to glean grade-specific network and pathway data has the capability to further the understanding of RCC oncogenesis. This approach can be used to identify novel mechanisms of tumor progression within grades and thereby can yield druggable targets.
We now show that validated grade-specific, highly sensitive proteomics analysis of RCC resulted in the identification of proteins that vary in expression in a grade-specific fashion. From these data, we identified pathways and networks that are relevant, and likely critical, to grade transitions, and we discovered markers that, either separately or in combination, are able to assist in differentiation among grades. Furthermore our analysis yielded pathways altered in RCC that can ultimately be used both to stratify patients to grade-specific treatments and to identify new therapeutic targets.
EXPERIMENTAL PROCEDURES
Materials—
After appropriate Institutional Review Board approval from University of California Davis, frozen and formalin-fixed paraffin-embedded (FFPE) clear cell renal cell carcinoma (ccRCC) tissues were obtained from the University of California Davis Medical Center in Sacramento, CA. The antibodies used in the study were mouse monoclonal anti-vimentin (VIM) (Dako), anti-SERPINH1 (Abcam), anti-phosphoglycerate kinase 1 (PGK1) (Santa Cruz Biotechnology), and anti-fructose-bisphosphate aldolase A (ALDOA) (Abnova) and rabbit polyclonal anti-ALDH1A1 (Abcam) and anti-AIFM1 (Abcam).
RCC Grading—
All the FFPE RCC samples were sectioned and counterstained with hematoxylin and eosin (H&E), and all were regraded by an experienced oncologic pathologist (A. D. B.) to standardize the grading of all specimens. The standard Fuhrman criteria of separation into four nuclear grades defined in order of increasing nuclear size, irregularity, and nucleolar prominence (3) were utilized.
Protein Extraction—
For comparison between frozen samples and FFPE tissues, protein was extracted utilizing the Liquid Tissue Protein Prep kit (Expression Pathology, Gaithersburg, MD). For protein preparation from frozen tissue, RCC grading was done based on the Fuhrman nuclear grade scale on H&E-stained slides and compared with the frozen block. Pieces of tissue were cut off the block and boiled at 95 °C for 90 min in 20 μl of Liquid Tissue buffer (Expression Pathology). Subsequently 1 μg of trypsin was added, and the samples were digested overnight at 37 °C. The samples were spun at 12,000 relative centrifugal force for 10 min, and the supernatant was transferred to a fresh tube. Protein preparation from FFPE tissues was performed with the Liquid Tissue Protein Prep kit (Expression Pathology) following the manufacturer's protocol.
For the grade-dependent proteomics experiments, the following protocol was used. Grading was done based on the FFPE sections counterstained with H&E. Unstained slides from adjacent sections were then deparaffinized and were used to collect regions of ∼0.5 cm in diameter with an assigned nuclear grade. 10-μm RCC FFPE sections were deparaffinized and hydrated through a xylene and ethanol series. The area corresponding to the adjacent H&E section was dissected using a 30-gauge needle and was transferred to 60 μl of RIPA buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.2, 2% SDS, 1% Triton X-100, 1% sodium deoxycholate, 5 mm EDTA). The sample was then heated at 100 °C for 20 min followed by a 3-h incubation at 80 °C with constant agitation to extract the protein (4). The samples were chilled on ice for 1 min, and chloroform/methanol precipitation was then performed to remove salt and detergents (5). After the addition of 40 μl of water, 400 μl of methanol was added, and after vortexing, 100 μl of chloroform was added. After vortexing, 300 μl of water was added. After a 1-min spin at 14,000 × g, the top aqueous layer was removed. Another 400 μl of methanol was added and mixed. After a 2-min spin at 14,000 × g, the supernatant was discarded, and the pellet was dried and suspended in 60 μl of 10 mm ammonium bicarbonate. Finally samples were trypsin-digested overnight with 1 μg of sequencing grade trypsin (Promega). The protein concentration of the digested frozen and FFPE RCC samples was quantified using the microBCA Protein Assay Reagent kit (Pierce), and a total of 3 μg from each sample was analyzed by LC-MS/MS.
Immunoblotting—
A section of tissue corresponding to what was used for LC-MS analysis was identified in an adjacent cut of the tissue block. These 10-μm FFPE sections were deparaffinized through a xylene and ethanol series, and 1 cm2 of a tissue with known nuclear grade was harvested using a needle. The protein was decross-linked and extracted using the QProteome FFPE protein extraction kit (Qiagen) according to the manufacturer's protocol. Immunoblotting was performed as reported previously (6). Ponceau S staining was used to verify equal protein loading.
Immunohistochemistry—
FFPE slides were deparaffinized through a xylene and ethanol series and were treated with 3% hydrogen peroxide, methanol prior to the antigen retrieval in sodium citrate buffer, pH 6. Following blocking, the slides were incubated with the primary antibodies at 4 °C overnight. After PBS washes, the slides were incubated with biotin-conjugated secondary antibodies followed by the avidin-biotin complex (ABC Elite kit, Vector Laboratories) according to the manufacturer's instructions. The avidin-biotin complex was visualized using diaminobenzidine (Vector Laboratories). The sections were counterstained with hematoxylin and were coverslipped. Photographs were taken using 20× or 10× objective lenses on an Axiovert microscope (Carl Zeiss).
MS/MS Spectrometric Analysis—
Data were acquired using a Nano-LC-2D system (Eksigent) coupled with an LTQ (linear trap quadrupole) ion trap mass spectrometer (Thermo Finnigan) with separations accomplished on an in-laboratory fabricated fritless reversed phase microcapillary column (75 × 180 mm packed with Magic C18AQ, 3-mm beads with 100-Å pores; Michrom Bioresources) and vented column configuration. Each digested sample was transferred by the Eksigent autosampler to the on-line trap column (Zorbax 300SB C18, 5 × 0.3 mm; Agilent) and desalted. Peptides were then eluted from the trap and separated by the aforementioned reversed phase microcapillary column at a flow rate of 300 nl/min and directly sprayed into the mass spectrometer. Buffer compositions used for reversed phase chromatography were as follows: buffer A, 0.1% formic acid in water; buffer B, 0.1% formic acid in 100% acetonitrile. Peptides were separated with a 48-min gradient (2–40% buffer B for 95 min, 40–80% buffer B for 12 min, and 80% buffer B for 13 min). MS/MS of the top 10 most intense ions was accumulated on the Thermo LTQ during each run. Peak lists were generated using the software Bioworks 3.3. The data were then analyzed with X!Tandem version 2008.01.01.1 and Ensembl human database version 49.36K (December 2006; 47,648 entries) based on human assembly NCBI 36 (October 2005). X!Tandem parameters were standard (fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 1.8 Da; iodoacetamide derivative of cysteine was specified as a fixed modification). Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulfone modification of methionine, tryptophan oxidation to formylkynurenine of tryptophan, and acetylation of the N terminus were specified as variable modifications. A threshold of −log(Expect scores) = 2.0 was used as a filtering criteria for X!Tandem; this is a standard value that is suitable for analysis with secondary statistical filtering such as clustering and enrichment calculation. Finally results were imported into the software Scaffold (version 2_00_02; Proteome Software Inc., Portland, OR) for protein identification validation, normalization, and comparison of spectral count or occurrences (that is, the number of all redundant peptide hits for a given protein). Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (7). Proteins were assigned by the Protein Prophet algorithm (8), and identifications were accepted if they could be established at greater than 95.0% probability and contained at least two identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The peptide false positive rate (FPR) was calculated using the Scaffold software. For each charge state, the incorrect assignments are tabulated to calculate the FPRi using the following formula: FPRi = ((Number assigned incorrect at 95% probability)/(Total number incorrect assigned)) × 100 where i is the charge state. The assignment is called correct if it is associated with a protein that has a 95% probability according to the Protein Prophet algorithm (13) and a minimum of two peptides each with a 95% probability based on the Peptide Prophet algorithm (10). The FPR is the sum of the values for each charge state.
Statistical and Pathway Analysis—
To assess accuracy of protein quantitation, results of proteomics analysis of 10 samples per grade were averaged, and a coefficient of variance was calculated for each identified protein. Significant grade-dependent differentials were filtered with a one-way analysis of variance (ANOVA) (9, 10) using thresholds of F-distribution cumulative probability of 0.05 or 0.01 (two standard filtering thresholds).
Statistically significant proteins were analyzed for molecular function, molecular process, and pathway enrichments using the Panther tools (11) and Gene Ontology. Network and metabolic pathways were analyzed using Ingenuity Pathways Analysis (IPA) (version 6; Ingenuity Systems Inc., Redwood City, CA). Statistically significant proteins were clustered using self-organizing maps (SOMs) with the software VisualGene (version 1.01.0024; Visipoint, Kuopio, Finland). SOMs are artificial neural networks based on an unsupervised algorithm (12). Protein levels of clusters were visualized in a heat map across grades using the script Pixelirator (University of California Davis Genome Center).
RESULTS
Frozen and FFPE Tissues Yield Similar Proteomics Results—
Initial experiments focused on the question of whether analysis of FFPE samples of RCC tissue yields proteomic profiles similar to those obtained from frozen samples. Because FFPE tissues are readily obtained and are considerably more abundant because of the availability of archived samples from pathology departments, such a finding would result in the ability to analyze substantially more samples. To address this issue, FFPE samples and their frozen tissue counterpart samples were both processed identically for the first part of this study.
Frozen tissues were obtained from nephrectomy specimens of three patients with confirmed (by oncologic pathologist A. D. B.) grade 2 ccRCC, and corresponding FFPE slides were obtained from the University of California Davis Pathology archives after appropriate Institutional Review Board approval. For the frozen samples, tumor grading was based on H&E slides from the corresponding frozen tumor; for FFPE tissues, H&E-stained tumor tissue obtained from the same block was scraped off of the slides as described under “Experimental Procedures.” All samples were processed with the Liquid Tissue® MS Protein Prep kit and subjected to tryptic digestion and tandem mass spectrometric analysis.
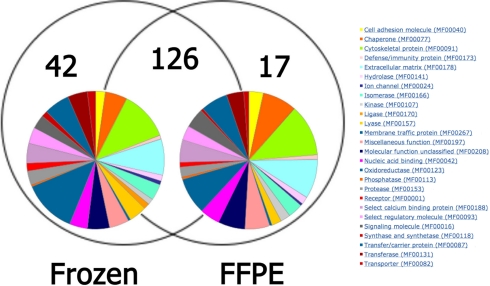
From these samples, 185 distinct proteins with two or more tryptic fragments were identified (supplemental Table 1). When proteomics analyses of FFPE and frozen samples were compared, the frozen samples yielded 168 proteins as compared with 143 for the FFPE processed samples, resulting in an overlap between the two data sets of 68% (Fig. 1). To further examine whether using FFPE samples would introduce bias, using the Panther libraries we analyzed the molecular function of the proteins identified from both tissue preparations. No significant differences in Panther molecular functions were observed between the two tissue sample preparations (Fig. 1). Oxidoreductases exhibited the most differences as expected because they are the most represented molecular function. Other functions are represented similarly in both tissue sample preparations.
Fig. 1.
Comparison of proteomics analysis from frozen and FFPE tissues. Proteins identified by a minimum of two peptides (95% confidence) per sample in either class (n = 3 samples per class) are shown by Venn diagram. The complete list of these proteins is in supplemental Table 1. The colored circles represent pie charts of Panther molecular functions constructed from proteins identified in each sample source.
Of the total 185 unique proteins identified in this experiment, only five proteins were significantly different (pairwise one-tailed t test p value <0.01) between FFPE and frozen tissue. All of these proteins, HSPA8, ANXA4, TUBB2C, YWHAB (14-3-3 β/α isoform), and TUBA1A, were found to be present at greater abundance in the FPPE samples. In addition, all of these proteins are classified by Gene Ontology as binding proteins. These experiments show that archived FFPE samples are adequate for proteomics analysis and biomarker discovery as compared with frozen tissue, and thus the readily available FFPE samples were utilized for subsequent analyses in this study.
Proteomics Analysis of ccRCC Yields Grade-specific Variations in Protein Quantities—
On the basis of the above results, additional proteomics analyses were performed on 50 FFPE tissues, which were evenly distributed among normal tissue and ccRCC of all Fuhrman grades (n = 10 per grade). Tumor tissues of confirmed grade were identified on FFPE slides in situ and removed by scraping, and proteomics analysis was performed by shotgun proteomics as described under “Experimental Procedures.” In the cases of four patients, both normal and cancer tissue was identified on the same slide; these tissues were analyzed separately.
A total of 1,470,313 spectra were analyzed, resulting in the identification of 777 proteins containing two or more peptide fragments. The rate of false positive for the filtering criteria used (minimum of two peptides at 95% confidence) is shown in supplemental Table 2 along with ages and genders of patients from whom the materials were obtained, the normalized spectral counts are shown in supplemental Table 3, the raw spectral counts are shown in supplemental Table 4, the unique peptide counts are shown in supplemental Table 5, the protein coverage is shown in supplemental Table 6, and the protein confidence is shown in supplemental Table 7. From the set of 777 identified proteins, 105 showed significant differences among the four Fuhrman grades and the normal kidney tissue using a one-way ANOVA (p value <0.01; Table I), and 180 proteins were significantly different using a slightly less stringent one-way ANOVA (p value <0.05; shown in supplemental Table 3).
Table I.
105 grade-specific proteins that were significantly different among grades (ANOVA p value <0.01)
The average of normalized spectral counts are shown for normal renal cells (N) and RCC grades 1–4 (G1–G4) cells. ENSP, Ensembl peptide; UpG1, highest level in grade 1 or 2; UpG3, highest level in grade 3 or 4.
| Symbol | Name | ENSP ID | Entrez ID | SOM cluster | ANOVA p value | Average
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | G1 | G2 | G3 | G4 | ||||||
| GATM | Glycine amidinotransferase | ENSP00000303263 | 2628 | Down | 1.3e−9 | 17 | 2.6 | 1.2 | 0.6 | 0 |
| ALDH6A1 | Methylmalonate-semialdehyde dehydrogenase | ENSP00000342564 | 4329 | Down | 2.8e−9 | 14.9 | 1.8 | 0.4 | 0.3 | 0 |
| ALDOB | Fructose-bisphosphate aldolase B | ENSP00000363988 | 229 | Down | 5.4e−8 | 15.5 | 1.8 | 1.2 | 0 | 0 |
| GPX3 | Glutathione peroxidase 3 precursor | ENSP00000373477 | 2878 | Down | 9.6e−8 | 6 | 0.7 | 0.2 | 0 | 0.1 |
| ALDH2 | Aldehyde dehydrogenase | ENSP00000261733 | 217 | Down | 1.1e−7 | 7.3 | 0.6 | 1.1 | 0 | 0.2 |
| TINAGL1 | Tubulointerstitial nephritis antigen-like precursor | ENSP00000271064 | 64129 | Down | 3.7e−7 | 3.3 | 1.3 | 0.5 | 0.1 | 0 |
| CNDP2 | Cytosolic nonspecific dipeptidase | ENSP00000325548 | 55748 | UpG1 | 8.0e−7 | 8.3 | 28.2 | 27.5 | 8.6 | 4.2 |
| SHMT1 | Serine hydroxymethyltransferase, cytosolic | ENSP00000318868 | 6470 | Down | 9.7e−7 | 3.7 | 1.3 | 1.3 | 0.1 | 0 |
| COL18A1 | Collagen, type XVIII, α-1 | ENSP00000383201 | 80781 | Down | 1.2e−6 | 7.2 | 4.3 | 2.5 | 0.7 | 1.2 |
| ASS1 | Argininosuccinate synthase | ENSP00000253004 | 445 | Down | 1.7e−6 | 8.2 | 0.9 | 0 | 0 | 0 |
| HSPG2 | Heparan sulfate proteoglycan core precursor | ENSP00000363827 | 3339 | Down | 1.9e−6 | 25.6 | 14.7 | 7.7 | 1.8 | 2.1 |
| ANPEP | Aminopeptidase N | ENSP00000300060 | 290 | Down | 2.3e−6 | 8.6 | 0.5 | 0.1 | 0.1 | 0 |
| PKM2 | Pyruvate kinase isozymes M1 and M2 | ENSP00000334983 | 5315 | Up | 2.7e−6 | 14.3 | 51.7 | 52.2 | 49.5 | 42.3 |
| ALDH4A1 | Δ1-Pyrroline-5-carboxylate dehydrogenase | ENSP00000290597 | 8659 | Down | 4.8e−6 | 9.6 | 0.4 | 0.3 | 0 | 0 |
| ACADM | Medium chain-specific acyl-CoA dehydrogenase | ENSP00000359871 | 34 | Down | 5.3e−6 | 5.3 | 0.8 | 1.1 | 0.1 | 0.2 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | ENSP00000364475 | 2203 | Down | 6.6e−6 | 5.7 | 1.3 | 1.3 | 0 | 0.6 |
| SUCLG2 | Succinyl-CoA ligase β chain | ENSP00000307432 | 8801 | Down | 1.4e−5 | 2.8 | 0.2 | 0.1 | 0.6 | 0 |
| ENO1 | α-Enolase | ENSP00000234590 | 2023 | Up | 1.7e−5 | 20.9 | 78 | 48.8 | 29.5 | 34.4 |
| ACAA2 | 3-Ketoacyl-CoA thiolase, mitochondrial | ENSP00000285093 | 10449 | Down | 2.0e−5 | 11.7 | 5.7 | 3.9 | 0.3 | 0.2 |
| CFL1 | Cofilin-1 | ENSP00000309629 | 1072 | Up | 2.0e−5 | 1.9 | 2.8 | 4.1 | 5.4 | 6.7 |
| AGMAT | Agmatinase | ENSP00000364986 | 79814 | Down | 2.9e−5 | 3.6 | 0.9 | 0.4 | 0 | 0 |
| PEBP1 | Phosphatidylethanolamine-binding protein 1 | ENSP00000261313 | 5037 | Down | 3.5e−5 | 11.9 | 9.4 | 6.8 | 6.8 | 3.7 |
| KHK | Ketohexokinase | ENSP00000260598 | 3795 | Down | 5.0e−5 | 4.2 | 0.8 | 1.2 | 0.3 | 0.1 |
| ADH1C | Alcohol dehydrogenase 1B | ENSP00000306606 | 126 | Down | 5.1e−5 | 3.8 | 0.6 | 0 | 0 | 0.1 |
| NDRG1 | Protein NDRG1 | ENSP00000319977 | 10397 | UpG1 | 9.2e−5 | 0.3 | 10.2 | 11.3 | 3.8 | 3.3 |
| PCK2 | Phosphoenolpyruvate carboxykinase | ENSP00000216780 | 5106 | Down | 9.7e−5 | 12 | 1.2 | 0.4 | 0 | 0 |
| BHMT | Betaine-homocysteine S-methyltransferase 1 | ENSP00000274353 | 635 | Down | 0.0001 | 6.4 | 4 | 4.3 | 0 | 0.1 |
| GPI | Glucose-6-phosphate isomerase | ENSP00000348877 | 2821 | UpG1 | 0.0001 | 3.9 | 15.5 | 15.4 | 15.2 | 7.6 |
| LDHA | l-Lactate dehydrogenase A chain | ENSP00000227157 | 3939 | UpG1 | 0.0002 | 3.4 | 21.3 | 20.4 | 16.5 | 13.1 |
| HRSP12 | Ribonuclease UK114 | ENSP00000254878 | 10247 | Down | 0.0002 | 3.2 | 0.5 | 0.5 | 0 | 0 |
| ANXA4 | Annexin A4 | ENSP00000347164 | 307 | UpG1 | 0.0002 | 5.5 | 20.2 | 22.4 | 13.9 | 9.6 |
| ACY1 | Aminoacylase-1 | ENSP00000232907 | 95 | Down | 0.0002 | 9.5 | 2.6 | 0.8 | 0 | 0 |
| HIBCH | 3-Hydroxyisobutyryl-CoA hydrolase | ENSP00000352706 | 26275 | Down | 0.0002 | 2 | 0.2 | 0.2 | 0 | 0.1 |
| HSD17B10 | 3-Hydroxyacyl-CoA dehydrogenase type-2 | ENSP00000168216 | 3028 | Down | 0.0003 | 1.4 | 0.3 | 0.1 | 0.2 | 0 |
| ALDH9A1 | 4-Trimethylaminobutyraldehyde dehydrogenase | ENSP00000271359 | 223 | Down | 0.0003 | 1.7 | 0.2 | 0.6 | 0.3 | 0.1 |
| COL4A2 | Collagen α | ENSP00000257309 | 1284 | Down | 0.0003 | 10.6 | 4.3 | 2.3 | 3.7 | 2.5 |
| ATP5O | ATP synthase O subunit | ENSP00000290299 | 539 | Down | 0.0003 | 3.6 | 0.3 | 0.5 | 0.4 | 0.4 |
| PFKL | 6-Phosphofructokinase, liver type | ENSP00000269848 | 5211 | UpG1 | 0.0003 | 0.6 | 5.5 | 4.1 | 1.3 | 0.2 |
| AMBP | AMBP protein precursor, α1-microglobulin | ENSP00000265132 | 259 | Down | 0.0004 | 1.5 | 0.1 | 0.2 | 0 | 0.2 |
| AIFM1 | Apoptosis-inducing factor 1 | ENSP00000287295 | 9131 | Down | 0.0005 | 5.5 | 0.6 | 0.7 | 0.6 | 0.1 |
| MME | Neprilysin | ENSP00000353679 | 4311 | Down | 0.0006 | 2.4 | 0.1 | 0.1 | 0 | 0 |
| DDC | Aromatic-l-amino-acid decarboxylase | ENSP00000350616 | 1644 | Down | 0.0006 | 4 | 0.4 | 0.6 | 0 | 0 |
| PDIA6 | Protein-disulfide isomerase A6 precursor | ENSP00000272227 | 10130 | Up | 0.0006 | 1.9 | 3.3 | 4 | 3.9 | 8.3 |
| ECHS1 | Enoyl-CoA hydratase | ENSP00000357535 | 1892 | Down | 0.0006 | 4.6 | 1 | 1.2 | 0.3 | 0.1 |
| AKR1A1 | Alcohol dehydrogenase | ENSP00000312606 | 10327 | Down | 0.0006 | 9.1 | 4.3 | 3.8 | 1.8 | 1.2 |
| COL4A1 | Collagen α-1(IV) chain precursor (arresten) | ENSP00000364979 | 1282 | Down | 0.0006 | 5.2 | 1.6 | 0 | 0.1 | 0.4 |
| ALDOA | Fructose-bisphosphate aldolase A | ENSP00000336927 | 226 | Up | 0.0006 | 2.2 | 8.8 | 10.8 | 14.1 | 12.8 |
| PGK1 | Phosphoglycerate kinase 1 | ENSP00000362413 | 5230 | Up | 0.0006 | 13.5 | 31 | 35 | 27 | 24 |
| VIM | Vimentin | ENSP00000224237 | 7431 | Up | 0.0006 | 20.8 | 45.4 | 46.4 | 47.3 | 64.5 |
| HSPA8 | Heat shock cognate 71-kDa protein | ENSP00000227378 | 3312 | Up | 0.0006 | 8.5 | 6.5 | 10.4 | 13.4 | 15.6 |
| HSPA5 | 78-kDa glucose-regulated protein precursor | ENSP00000324173 | 3309 | Up | 0.0007 | 5.2 | 5.2 | 6.7 | 9.6 | 13.5 |
| CALR | Calreticulin precursor | ENSP00000320866 | 811 | Up | 0.0007 | 0.5 | 1.1 | 1.6 | 2.4 | 5 |
| PTRF | Polymerase I and transcript release factor | ENSP00000349541 | 284119 | UpG1 | 0.0007 | 0.1 | 2.5 | 1.1 | 0.1 | 0.4 |
| ACTN4 | α-Actinin-4 | ENSP00000252699 | 81 | Down | 0.0008 | 26.9 | 17.4 | 13.3 | 13.8 | 15.8 |
| GLUDP5 | Glutamate dehydrogenase 1 | ENSP00000277865 | 2746 | Down | 0.0008 | 8.3 | 4.2 | 3.9 | 1.3 | 1.3 |
| ACAT1 | Acetyl-CoA acetyltransferase | ENSP00000265838 | 38 | Down | 0.0009 | 11.4 | 2.5 | 2.1 | 5.6 | 0.5 |
| PFKP | 6-Phosphofructokinase type C | ENSP00000370517 | 5214 | UpG1 | 0.0010 | 0.1 | 10.7 | 8.8 | 5.1 | 2.7 |
| DAK | Dihydroxyacetone kinase | ENSP00000310493 | 26007 | Down | 0.0010 | 2.2 | 0.4 | 0.2 | 0 | 0 |
| CRYZ | Quinone oxidoreductase | ENSP00000339399 | 1429 | Down | 0.0010 | 3.8 | 3.9 | 5.3 | 1.9 | 1.2 |
| PBLD | Phenazine biosynthesis-like domain-containing protein | ENSP00000308466 | 64081 | Down | 0.0011 | 1.3 | 0.2 | 0.6 | 0 | 0 |
| DDX17 | Probable ATP-dependent RNA helicase DDX17 | ENSP00000216019 | 10521 | UpG1 | 0.0011 | 0 | 0.3 | 1 | 0.4 | 0.3 |
| DARS | Aspartyl-tRNA synthetase, cytoplasmic | ENSP00000264161 | 1615 | UpG1 | 0.0012 | 0.2 | 1.2 | 0.4 | 0.2 | 0.5 |
| ATP5B | ATP synthase subunit β | ENSP00000262030 | 506 | Down | 0.0014 | 33.6 | 17.3 | 19.5 | 19.1 | 10.9 |
| HADH | Hydroxyacyl-coenzyme A dehydrogenase | ENSP00000312288 | 3033 | Down | 0.0015 | 3.4 | 0.9 | 0.8 | 0.9 | 0.1 |
| SLC9A3R1 | Ezrin-radixin-moesin-binding phosphoprotein 50 | ENSP00000262613 | 9368 | Down | 0.0016 | 3.2 | 0.4 | 0.8 | 0.3 | 0.2 |
| ATP5A1 | ATP synthase subunit α | ENSP00000282050 | 498 | Down | 0.0017 | 19.3 | 7.8 | 11.2 | 12.3 | 5.8 |
| ACSM2B | Acyl-CoA synthetase medium chain family member 2B | ENSP00000327453 | 348158 | Down | 0.0017 | 7.7 | 1.8 | 0.2 | 0.3 | 0 |
| FGG | Fibrinogen γ chain precursor | ENSP00000336829 | 2266 | Up | 0.0018 | 6.3 | 3.5 | 10.4 | 23.3 | 24.3 |
| DCXR | l-Xylulose reductase | ENSP00000303356 | 51181 | Down | 0.0021 | 2.1 | 0.4 | 0.3 | 0.2 | 0.1 |
| ALDH1A1 | Retinal dehydrogenase 1 | ENSP00000297785 | 216 | UpG1 | 0.0022 | 3 | 8.2 | 10.1 | 4.7 | 1.7 |
| CRIP2 | Cysteine-rich protein 2 | ENSP00000328521 | 1397 | UpG1 | 0.0022 | 0.5 | 1.4 | 0.6 | 0 | 0.2 |
| IDH2 | Isocitrate dehydrogenase | ENSP00000331897 | 3418 | UpG3 | 0.0024 | 5.6 | 0.7 | 1.1 | 5.6 | 1.3 |
| FGB | Fibrinogen β chain precursor | ENSP00000306099 | 2244 | Up | 0.0025 | 5.6 | 2.5 | 7.4 | 20.5 | 21.6 |
| ALDOC | Fructose-bisphosphate aldolase C | ENSP00000226253 | 230 | Up | 0.0026 | 1.3 | 5.1 | 6.3 | 6.6 | 6.6 |
| ATP1A1 | Na+- and K+-transporting ATPase subunit α-1 precursor | ENSP00000358508 | 476 | Down | 0.0027 | 9.3 | 1.8 | 1.3 | 3.3 | 0.5 |
| ABAT | 4-Aminobutyrate aminotransferase | ENSP00000268251 | 18 | Down | 0.0031 | 3.9 | 0.6 | 0 | 0 | 0 |
| VTN | Vitronectin precursor | ENSP00000226218 | 7448 | 0.0033 | 5.4 | 1.4 | 0.6 | 1.7 | 3.9 | |
| GGTLA1 | γ-Glutamyltransferase 5 precursor | ENSP00000263112 | 2687 | Down | 0.0033 | 2.8 | 0.2 | 0 | 0 | 0 |
| TF | Serotransferrin precursor | ENSP00000264998 | 7018 | Down | 0.0034 | 9.5 | 6.1 | 5.2 | 2.7 | 3.8 |
| S100A9 | Protein S100-A9 | ENSP00000295382 | 6280 | Up | 0.0034 | 2.5 | 3 | 0.7 | 6.1 | 16.4 |
| FN1 | Fibronectin precursor | ENSP00000338200 | 2335 | Up | 0.0039 | 1.6 | 7.8 | 5.9 | 18.3 | 25.6 |
| LRP2 | Low density lipoprotein receptor-related protein 2 precursor | ENSP00000263816 | 4036 | Down | 0.004 | 2.8 | 0.2 | 1.3 | 0 | 0 |
| ALDH7A1 | α-Aminoadipic semialdehyde dehydrogenase | ENSP00000297542 | 501 | Down | 0.004 | 2.5 | 0.4 | 0.8 | 0.1 | 0 |
| ADFP | Adipophilin | ENSP00000276914 | 123 | UpG1 | 0.0041 | 0 | 2.2 | 0.7 | 0.1 | 0.2 |
| SERPINH1 | Serpin H1 precursor | ENSP00000350894 | 871 | Up | 0.0042 | 0.6 | 3.9 | 3 | 4.8 | 8 |
| HSP90AB1 | Heat shock protein HSP 90-β | ENSP00000325875 | 3326 | Up | 0.0044 | 8.2 | 8.8 | 11.1 | 15.1 | 15.4 |
| LAMA5 | Laminin subunit α-5 precursor | ENSP00000252999 | 3911 | Down | 0.0044 | 5 | 3.8 | 4.2 | 0.3 | 0.4 |
| SNORA67 | Eukaryotic initiation factor 4A-I | ENSP00000293831 | 26781 | Up | 0.0053 | 0.5 | 0.7 | 0.6 | 1.1 | 3.2 |
| CRYL1 | λ-Crystallin homolog | ENSP00000298248 | 51084 | Down | 0.0058 | 2.8 | 2.5 | 1.5 | 0 | 0.1 |
| HINT1 | Histidine triad nucleotide-binding protein 1 | ENSP00000304229 | 3094 | UpG1 | 0.0058 | 0.2 | 0.3 | 1.5 | 0.4 | 0.1 |
| QDPR | Dihydropteridine reductase | ENSP00000281243 | 5860 | Down | 0.0061 | 1.4 | 0.5 | 0.7 | 0.3 | 0.1 |
| ECH1 | Δ3,5,Δ2,4-Dienoyl-CoA isomerase | ENSP00000221418 | 1891 | Down | 0.0064 | 4.1 | 1.5 | 1.9 | 0.2 | 1 |
| CORO1A | Coronin-1A | ENSP00000219150 | 11151 | Up | 0.0067 | 0.1 | 0.1 | 0.4 | 0.4 | 1.1 |
| ENO2 | γ-Enolase | ENSP00000229277 | 2026 | Up | 0.0071 | 0 | 12.6 | 6.8 | 4.8 | 6.4 |
| DPYSL2 | Dihydropyrimidinase-related protein 2 | ENSP00000309539 | 1808 | UpG1 | 0.0072 | 1.3 | 3.2 | 4.3 | 0.7 | 1.3 |
| GLYAT | Glycine N-acyltransferase | ENSP00000340200 | 10249 | Down | 0.0075 | 2 | 0.1 | 0 | 0 | 0 |
| LDHB | l-Lactate dehydrogenase B chain | ENSP00000229319 | 3945 | 0.0078 | 12 | 5.6 | 7 | 11.3 | 4.9 | |
| UGT2B7 | UDP-glucuronosyltransferase 2B7 precursor | ENSP00000304811 | 7364 | Down | 0.0079 | 1 | 0.1 | 0 | 0 | 0 |
| YWHAZ | 14-3-3 protein ζ and δ | ENSP00000309503 | 7534 | Up | 0.0081 | 2.3 | 4.7 | 3.9 | 5.7 | 6.1 |
| TUFM | Elongation factor Tu | ENSP00000322439 | 7284 | Down | 0.0084 | 3.3 | 0.3 | 1.2 | 1.9 | 0.8 |
| COX2 | Cytochrome c oxidase subunit 2 | ENSP00000354876 | 4513 | 0.0086 | 3.6 | 0.2 | 0.3 | 0.9 | 0.2 | |
| EHHADH | Peroxisomal bifunctional enzyme | ENSP00000231887 | 1962 | Down | 0.0089 | 4.4 | 1.1 | 0.3 | 0 | 0 |
| PCK1 | Phosphoenolpyruvate carboxykinase | ENSP00000319814 | 5105 | Down | 0.0090 | 2.2 | 0 | 0 | 0 | 0 |
| HADHA | Trifunctional enzyme subunit α | ENSP00000370023 | 3030 | Down | 0.0091 | 6 | 3 | 3.9 | 2.4 | 0.6 |
| AGRN | Agrin precursor | ENSP00000368678 | 375790 | UpG1 | 0.0097 | 0.3 | 1.7 | 1 | 0 | 0 |
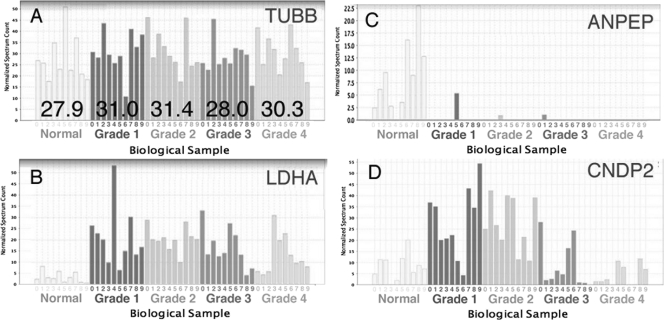
Protein levels were determined by spectral count (13, 14) after normalization. Each protein spectral count (the number of all possibly redundant peptide hits for a given protein) was normalized across the 50 samples using the total number of spectra for each sample. In addition, four proteins that had the lowest inter- and intragrade variability, MYH9, tubulin β chain (TUBB) (shown in Fig. 2A), histone H4, and peptidylprolyl isomerase A, were chosen to be used as endogenous controls. Factors using the averages in a given tumor grade of these four proteins were used to normalize spectral count when comparing samples among grades. By way of example, spectral count variability is shown for three representative proteins that have distinct patterns of expression changes within grades (Fig. 2, B–D). In these examples, analysis by one-way ANOVA (p value <0.01) revealed statistically significant changes in proteins levels across grades.
Fig. 2.
Sample variability for four representative proteins across grades. A, TUBB (ANOVA p value = 0.83), a representative of the four proteins with the lowest intra- and intergrade variability that were used as endogenous controls, is shown. The average value per grade is shown overlaid. A normalization factor was computed using the four endogenous controls and was used when comparing protein levels among grades. B–D, normalized spectrum count is shown for three representative proteins that have varying protein level grade patterns. B, aminopeptidase N (ANPEP); C, LDHA; D, CNDP2.
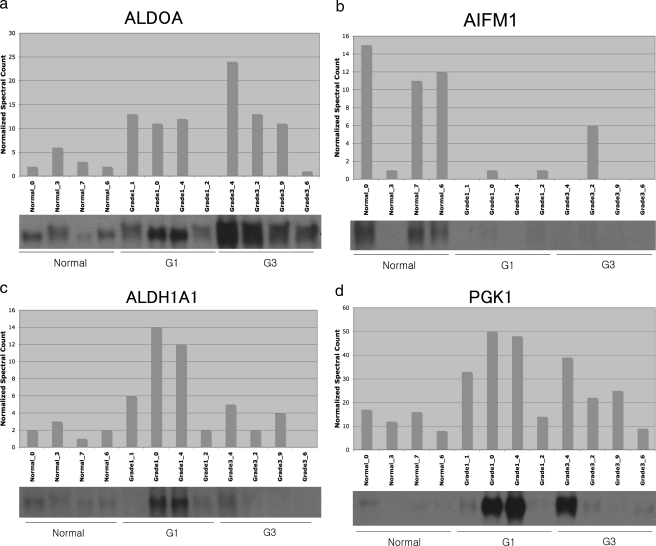
To confirm the validity of the protein identification and quantitation methods used in this study, grade-specific changes of several proteins representative of the LC-MS data were determined by immunoblotting. To handle properly the variability between individual samples among grades (for examples, see Fig. 2), we compared four samples of three different grades, from normal tissue as well as grades 1 and 3, to confirm the LC-MS data. Four representative antibodies of proteins, whose average normalized spectral counts (n = 10) were significantly altered across grades, were utilized from the 10 samples. There was a high degree of consistency between the immunoblot protein band intensities and the normalized spectral count in all cases (Fig. 3). As further validation of the veracity of the spectral count method, each of the 18 proteins that were shown to be statistically differential in our previous work based on 2D gel electrophoresis and spot optical density (15) showed consistent quantitative results in the current study. Finally our cluster analysis showed striking cluster examples (see below) that could not have been achieved if the relative quantitation was inaccurate.
Fig. 3.
Validation of proteomics MS analysis by immunoblotting. Protein was extracted from tissue samples homologous to that used for LC-MS analysis and was subjected to immunoblotting using the antibodies described under “Experimental Procedures.” For each protein, the upper section shows the normalized spectral count, and the lower section shows the corresponding sample immunoblot. a, ALDOA; b, AIFM1 (apoptosis-inducing factor 1, mitochondrial precursor); c, ALDH1A1 (aldehyde dehydrogenase family 1 member A1; retinal dehydrogenase 1); d, PGK1. G, grade.
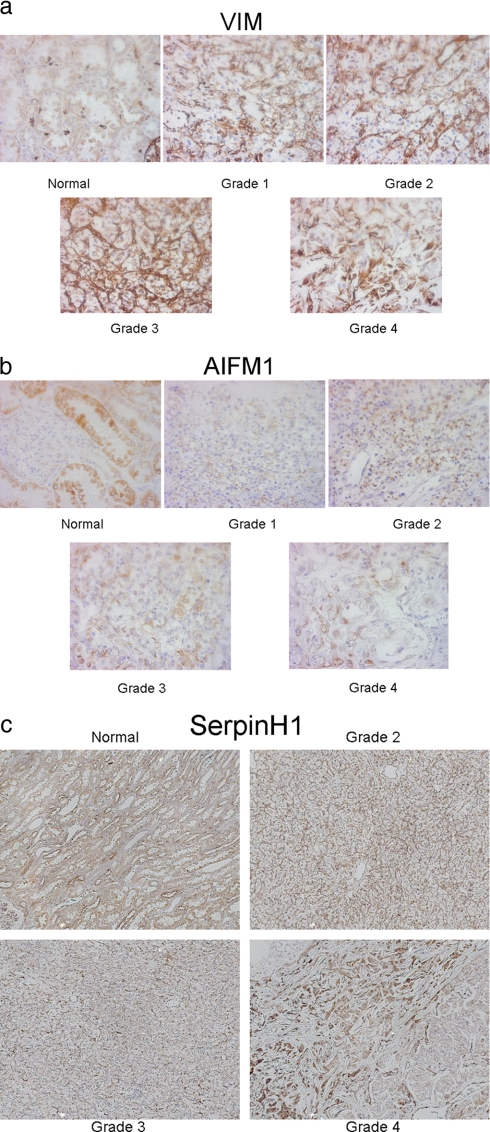
Immunohistochemistry using selected antibodies was also performed (Fig. 4), but the use of this technique was not applicable for validation due to protein compartmentalization (see Figs. 2 and 3). The MS and immunoblot protocols are based on a fairly crude sample collection where cells within the sample are mixed together, whereas the immunohistochemistry shows protein levels within specific cellular compartments.
Fig. 4.
Immunohistochemistry of representative tissue across cancer grades. Representative proteins displaying different grade-specific patterns of expression are shown analyzed by immunohistochemistry of VIM (20×) (a), AIFM1, apoptosis-inducing factor 1, mitochondrial (10×) (b), and SERPINH1, Serpin H1 precursor, HSP47, collagen-binding protein (10×) (c).
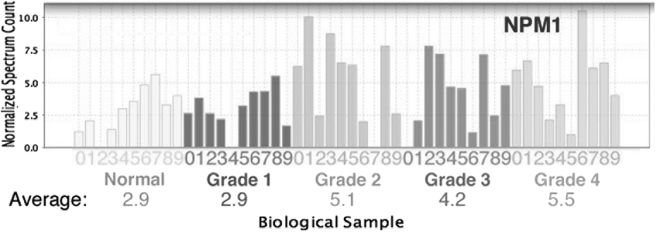
As discussed above, the Fuhrman grading system for RCC utilizes the appearance of the nucleolus for stratification (3). Thus, the level of the nucleolar protein nucleophosmin was analyzed, and statistical significance was assessed utilizing a pairwise t test between normal tissue and grade 1 samples on one hand and samples of grades 2, 3, and 4 on the second hand (Fig. 5). Consistent with criteria for classification in the Fuhrman grading system and as further confirmation of the accuracy of the grading in this study, nucleophosmin appeared to be significantly increased in grade 2, 3, and 4 tumors as compared with normal tissue and grade 1 (p value = 0.0025).
Fig. 5.
Levels of nucleophosmin are significantly increased in grades 2–4. Normalized spectrum count for the nucleolar protein nucleophosmin is analyzed by pairwise test of statistical significance (Student's t test p value = 0.0025 for normal and grade 1 versus grades 2, 3, and 4).
Proteomics Analysis Shows Biochemical Processes and Pathways Enhanced in RCC—
Comparison of the proteins in all 50 samples that are statistically differential among all grades and normal tissue (supplemental Table 2) resulted in the identification of enriched functions (Table II), molecular processes (Table III), and metabolic pathways (Table IV) with a p value <0.01 after multiple testing correction. Pie charts with the molecular functions, the biological processes, and the metabolic and signaling pathways are shown in supplemental Figs. 1–3.
Table II.
RCC grade-dependent enriched molecular functions
Significantly enriched molecular functions (p value <0.01 after Bonferroni multiple testing correction) using Panther libraries (13) are shown. Numbers of hits are the numbers of genes present in the NCBI Homo sapiens (H.s.) reference set and in the RCC significant 105-protein set.
| Molecular function | NCBI H.s. ref. | RCC 105 | p valuea |
|---|---|---|---|
| Dehydrogenase | 225 | 22 | 6.59e−22 |
| Oxidoreductase | 603 | 25 | 4.97e−17 |
| Lyase | 155 | 15 | 3.66e−15 |
| Hydratase | 27 | 7 | 4.14e−9 |
| Epimerase/racemase | 47 | 7 | 1.88e−7 |
| Isomerase | 178 | 9 | 1.49e−6 |
| Carbohydrate kinase | 27 | 5 | 1.66e−5 |
| Synthase and synthetase | 213 | 8 | 7.65e−5 |
| Aldolase | 5 | 3 | 2.07e−4 |
| Synthetase | 96 | 6 | 4.60e−4 |
| Transferase | 884 | 12 | 6.67e−3 |
| Decarboxylase | 21 | 3 | 1.46e−2 |
| Reductase | 184 | 6 | 1.72e−2 |
| Extracellular matrix | 384 | 7 | 2.75e−2 |
After Bonferroni multiple testing correction.
Table III.
RCC grade-dependent enriched biological processes
Significantly enriched biological processes (p value <0.01 after Bonferroni multiple testing correction) using Panther libraries (13) are shown. Numbers of hits are the numbers of genes present in the NCBI H. sapiens (H.s.) reference set and in the RCC significant 105-protein set.
| Biological process | NCBI H.s. ref. | RCC 105 | p valuea |
|---|---|---|---|
| Carbohydrate metabolism | 592 | 28 | 1.04e−20 |
| Glycolysis | 46 | 12 | 2.07e−16 |
| Fatty acid β-oxidation | 27 | 7 | 5.09e−9 |
| Amino acid metabolism | 230 | 11 | 8.59e−8 |
| Fatty acid metabolism | 187 | 10 | 7.71e−7 |
| Amino acid activation | 38 | 5 | 7.97e−5 |
| Lipid, fatty acid, and steroid metabolism | 770 | 14 | 8.18e−5 |
| Coenzyme and prosthetic group metabolism | 174 | 7 | 2.28e−4 |
| Other metabolism | 559 | 11 | 5.08e−4 |
| Coenzyme metabolism | 61 | 5 | 7.90e−4 |
| Other carbon metabolism | 82 | 5 | 3.25e−3 |
| Gluconeogenesis | 17 | 3 | 7.08e−3 |
| Vitamin biosynthesis | 21 | 3 | 1.80e−2 |
| Extracellular matrix protein-mediated signaling | 62 | 4 | 2.50e−2 |
After Bonferroni multiple testing correction.
Table IV.
RCC grade-dependent enriched IPA pathways
Significantly enriched pathways (p value <0.01 after Benjamini-Hochberg false discovery rate multiple testing correction) using IPA software and databases are shown. Numbers of hits are the numbers of genes present in the IPA reference database and in the RCC significant 105-protein set.
| Pathway | IPA ref. | RCC 105 | p valuea |
|---|---|---|---|
| Glycolysis/gluconeogenesis | 141 | 20 | 2.51e−19 |
| Propanoate metabolism | 126 | 17 | 1.00e−17 |
| Valine, leucine, and isoleucine degradation | 107 | 17 | 3.16e−17 |
| Butanoate metabolism | 129 | 15 | 2.00e−15 |
| β-Alanine metabolism | 99 | 14 | 2.00e−14 |
| Lysine degradation | 144 | 13 | 2.51e−12 |
| Pyruvate metabolism | 145 | 13 | 5.01e−12 |
| Fatty acid metabolism | 189 | 15 | 7.94e−11 |
| Tryptophan metabolism | 239 | 13 | 1.74e−8 |
| Fructose and mannose metabolism | 139 | 8 | 1.86e−6 |
| Arginine and proline metabolism | 178 | 9 | 4.90e−6 |
| Pentose phosphate pathway | 89 | 7 | 4.90e−6 |
| Ascorbate and aldarate metabolism | 82 | 5 | 3.24e−5 |
| Urea cycle and metabolism of amino groups | 80 | 6 | 3.98e−5 |
| Histidine metabolism | 129 | 7 | 7.94e−5 |
| Inositol metabolism | 25 | 4 | 9.12e−5 |
| Glycerolipid metabolism | 144 | 7 | 1.05e−4 |
| Citrate cycle | 59 | 4 | 3.47e−3 |
| Glutamate metabolism | 78 | 4 | 7.94e−3 |
After Benjamini-Hochberg false discovery rate multiple testing correction.
We note the abundance of dehydrogenases, such as ALDH4A1, ALDH1A1, ALDH6A1, ALDH9A1, and ALDH7A1, in the 105 RCC grade-dependent proteins corresponding to a significant enrichment of dehydrogenases in RCC biology (p value = 6.6e−22). These proteins, among others, are known to be involved in xenobiotic metabolism signaling, which itself exhibits an enrichment p value of 1.15e−4. Xenobiotic metabolism is associated with apoptosis and tumorigenesis and thus may be playing a critical role in RCC oncogenesis and response to therapy (see “Discussion”). Another set of identified proteins (TF, AMBP, fibronectin (FN1), FGB, FGG, and FGA) is part of the acute phase response signaling pathway, which is also enriched in this data set (p value = 1.33e−5). TF and AMBP have decreased levels in higher RCC grades, and FN1 has increased levels in higher RCC grades. FN1 has been associated with cell migration and could be involved in the metastatic process. Fibrin, the proteolytic product of fibrinogen, has been associated with positive regulation of cell proliferation, and an increased expression of this protein has been shown in various malignancies (see “Discussion”).
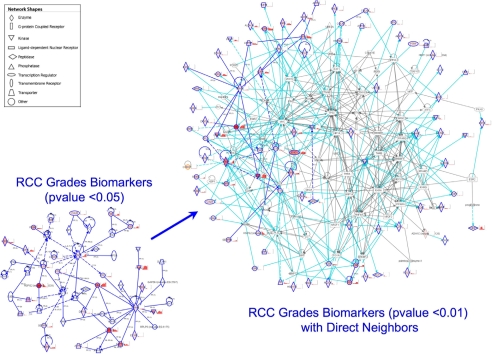
From the 180 proteins with ANOVA p value <0.05 (supplemental Table 3), 51 form a network using the IPA software (Fig. 6, lower left corner). Using the 27 from the list of 105 proteins with one-way ANOVA p value <0.01, we queried the IPA knowledge database to display 64 direct first-order neighbors in the IPA network. These 64 proteins have been associated to 85 of the 105 RCC grade one-way ANOVA p value <0.01 proteins. Among them, the network shows that p53, Myc, and HIF-1α are major hubs (Fig. 6, right). These proteins have known roles in kidney cancer oncogenesis and progression (see “Discussion”), and their presence in this ccRCC network diagram confirms the relevance of the 105-protein data set and their association to kidney cancer.
Fig. 6.
Network analysis of proteins showing significant grade-specific changes. Proteins showing significant grade-specific changes (p < 0.05) are assembled into a network showing direct (solid line) and indirect (dashed line) interactions (left) shown in dark blue. Colored proteins (red hues) are part of the 105 significantly grade-dependent (p < 0.01) proteins and have accompanying grade-specific histograms (normal to grade 4 from left to right). The network was grown (right) to include direct first-order neighbors of the significant proteins (p < 0.01). Connections between a member of the 105 significantly (p < 0.01) grade-dependent proteins and a direct first-order neighbor are shown in cyan. Connections between direct first-order neighbors are shown in gray.
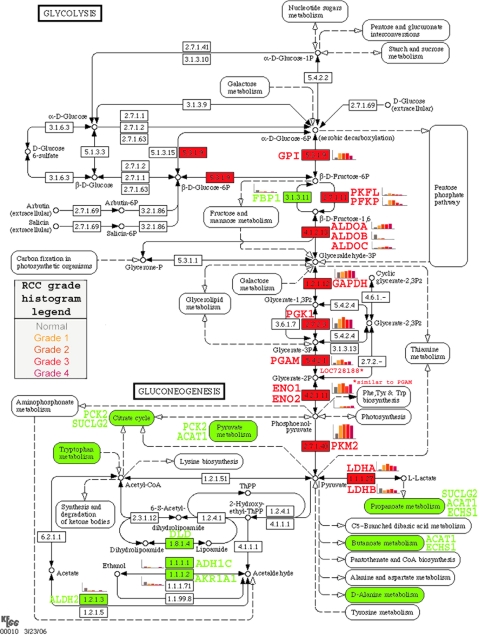
20 of the proteins significantly different among grades lie in the glycolysis pathway (Tables II and III). The highly significant processes and pathways altered in RCC that were found in this data set are consistent with what was found in previously published work using two-dimensional gel electrophoresis on frozen RCC samples from our (15) and other (16) laboratories. The changes in glycolysis pathway proteins observed in the present study converge on pyruvate (Fig. 7) demonstrating the importance of aerobic glycometabolism (the Warburg effect (16, 17)) in RCC. Furthermore we show (in red in Fig. 7) that cancer cells increase many of the proteins involved in glycolysis but also may use other related proteins or isoforms. For example EC 2.7.1.11 (6-phosphofructokinase) shows an increase of both PFKL and PFKP, whereas EC 3.1.3.11 (fructose bisphosphatase; FBP1), which governs the reverse reaction, is down-regulated. Similarly EC 4.2.1.11 (phosphopyruvate hydratase) shows an increase of both ENO1 and ENO2. For EC 4.1.2.13 (fructose-bisphosphate aldolase), there is an increase of ALDOA and ALDOC, whereas ALDOB is reduced, and for the anaerobic section of the glycolysis, EC 1.1.1.27 (l-lactate dehydrogenase), the LDHA level increases significantly, whereas LDHB decreases.
Fig. 7.
The glycolysis pathway is altered in a grade-specific manner. As representative of grade-specific proteomics data, elements of the glycolysis pathway are shown as a function of tumor grade. Colors indicate significantly (p < 0.05) higher (red) or lower (green) in RCC versus normal. Histograms of grade-specific changes are shown adjacent to significantly altered proteins. PGAM, phosphoglycerate mutase; GPI, glucose-phosphate isomerase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; P, phosphate.
Self-organizing Mapping Shows Grade-specific Trends in Protein Levels—
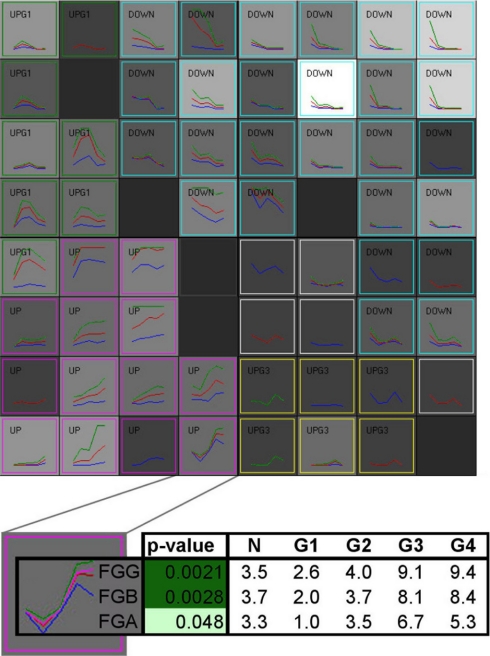
It can be seen from the previous data that there exist a variety of patterns of protein levels as a function of tumor grade such that some proteins are most highly expressed in low grades, others are most highly expressed in high grades, and still others are most highly expressed in intermediate grades. Clustering using a 64-node self-organizing map algorithm as a function of the modulation of the protein level across grades (Fig. 8) revealed that the three chains of fibrinogen, which make up the fully assembled protein (18), form together a cluster node (Fig. 8, inset). Indeed FGA, FGB, and FGG are co-localized on chromosome 4 tightly in a 50-kb region, and they have been shown to be co-regulated by the transcription factor STAT3 (19). Similarly the α and β chain of ATP synthase clustered together, whereas the three other chains at a ratio of 1:3 of these two are present in neighboring cluster nodes with slightly different level patterns. These findings are consistent with the quaternary structure and regulation of these two proteins: this further demonstrates the validity of our methods and results.
Fig. 8.
Cluster analysis of proteins showing significant grade-specific changes. 181 proteins with an RCC grade ANOVA p value <0.05 were clustered using a 64-node SOM algorithm on the basis of the similarity between protein levels across grades. The density of each node is shown by the gray intensity (black = 0; white = 10). In each node, minimum (blue), maximum (green), and average (red) across normal kidney tissue and four RCC grade are plotted. The nodes were then grouped based on their trend: up, pink; down, cyan; UpG1, highest level in grade 1 or 2, green; UpG3, highest level in grade 3, yellow). The bottom panel shows a typical cluster magnified. It is made of three proteins: the three chains (α, β, and γ) of fibrinogen, the precursor of fibrin. The table shows the underlying spectral count data for these three protein chains. G, grade.
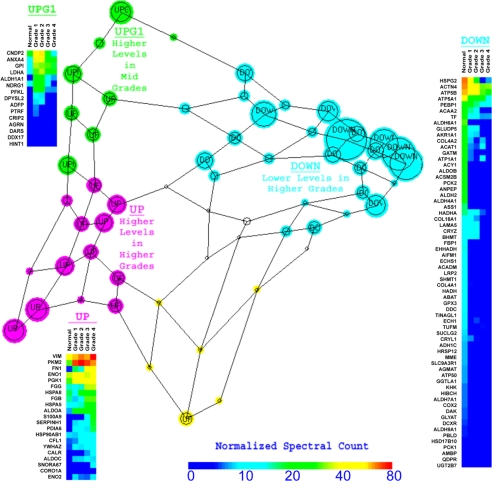
From the cluster data using 180 proteins that were significantly different among grades (p value <0.05), we generated a Sammon graphic, representing the self-organizing map clustering with distance and placement of the nodes based on the similarity of the RCC grade protein level modulation (Fig. 9). This unsupervised neural network learning algorithm representation shows varying node sizes representative of the quantity of proteins that display the given pattern with the length of lines that link the nodes proportional to the degree of relatedness between groups (12). Protein levels from specific regions of similar patterns of expression corresponding to regions of the Sammon diagram are displayed in a heat map format (Fig. 9, insets). These heat maps have utility for diagnosis using immunohistochemistry of grade-specific protein abundance to confirm grade diagnosis in cases where pathological classification is ambiguous.
Fig. 9.
Sammon map of grade-specific protein SOM clusters. Using the patterns of levels of the proteins that were significantly different among grades, the SOM nodes shown in Fig. 8 were plotted into a Sammon map. Each node is represented by a circle with a size proportional to the density of the cluster node. The distance and relative position of each node represent the relative similitude of the protein level pattern across grade between cluster nodes. For each group of cluster nodes (up, pink; down, cyan; UpG1, highest level in grade 1 or 2, green) a heat map of the normalized spectral count in normal tissue and in the four RCC grades is plotted.
DISCUSSION
Kidney cancer is often discovered incidentally, a scenario that frequently occurs when the disease is already metastatic at which point 5-year survival is a dismal 5% (1). Given the staggering resistance of this disease to conventional treatment, the discovery of new druggable therapeutic targets has the potential to improve these statistics. This is already evidenced by the clinical translation of knowledge of kinase pathways activated in the disease, resulting in trials of the kinase inhibitors sorafenib and sunitinib, which have vastly improved survival of metastatic RCC. Along these lines, elucidation of additional (if not all) biochemical pathways that are altered in RCC has the likelihood of leading to new interventions; this is a major strength of proteomics and was the goal of this study.
The use of archived FFPE tissues for proteomics analysis, as has been done in this study for the first time to our knowledge in RCC, has several advantages over the use of frozen tissue. Tumor tissue can be readily and visually identified and separated from normal tissue on unstained slides where it can by removed with a scalpel and used directly for shotgun proteomics; and tissues that are in the possession of a pathology department, either paraffin blocks or previously cut slides, can be utilized for such analysis, greatly expanding the range and variety of tissues available for proteomics analysis. Ours is the first report on this technique for kidney cancer, and we show here that it is entirely consistent with the use of “gold standard” frozen tissue blocks for similar analysis.
RCC can be pathologically classified into several distinct subtypes, for example clear cell, papillary, and chromophobe RCC (1). To preserve the homogeneity of the samples, in this study we examined only ccRCC. Several observations from this study relate to the accuracy of the Fuhrman grading system (3), the most commonly used of the several grading systems currently in use (2). The finding that the nucleolar protein nucleophosmin is increased in grades 2–4 as compared with control and grade 1 (Fig. 5) is consistent with the dictates of the Fuhrman system in which nucleoli appear in grades 2 and higher. In addition, data in this study (heat maps in Fig. 9) suggest that the four existing Fuhrman grades can, on a purely biochemical basis, be distilled down to two because protein alterations appear more similar within grades 1 and 2 and within grades 3 and 4. This separation, as it relates to actuarial survival, has been previously noted: one report showed no statistical difference in outcome of patients with grades 1 and 2 (20), whereas another showed that actuarial survival segregated with grades 1–2 and 3–4 (21). There have been several other studies showing a similar trend of prognosis as a function of the two-tiered grading system (for a review, see Ref. 22), supporting our proteomics data and our approach to discover a set of biomarkers for prognosis.
There exist several techniques for non-target driven proteomics analysis that are especially suited for cancer. The use of two-dimensional gel electrophoresis and mass spectrometry has proved useful in this regard, but they require large amounts of protein, are quite labor-intensive, and require manual removal of spots on the gel that introduces operator error. Another technique, known as PROTEOMEX, utilizes sera from both healthy donors as well as RCC patients and relies on immunologic methods of protein identification; this technique relies on the presence of a robust immunological response of the RCC patient. Finally shotgun proteomics, the technique utilized in this study, relies on liquid chromatography and tandem mass spectrometry to identify trypsin-digested proteins and subsequent computer analysis to reconstruct proteins contained within the sample. The advantage of this technique is that it can identify low abundance proteins and membrane proteins that are poorly represented in 2D gels, and its adaptation for use in FFPE samples is relatively straightforward. There exist several genomics and proteomics studies in kidney cancer, including one from our laboratory (15). Other groups have examined RCC proteomics using the PROTEOMEX (23) and two-dimensional gel electrophoresis/mass spectrometry techniques (16). Although many of the proteins discovered by these groups are consistent with the findings reported here, neither of these groups utilized “shotgun” proteomics, which has yielded a greater number of identified proteins, and neither examined RCC proteomics as a function of tumor grade. Our results clearly demonstrate that comparing cancer cells with normal without taking into account cancer grades will likely not work to identify usable biomarkers.
As for the quantitation method used in this study, it is important to remember that spectral counting is a discrete process, whereas immunoblotting is a continuous one. Further, we counted peptides that may originate from the same protein, whereas immunoblotting is proportional to protein quantity. This can result in an accentuation of the differences when comparing LC-MS results with immunoblots, whereas it is not a factor when comparing samples processed with the same technology. Every effort was made to specifically extract tumor tissue in each slide, but the tissue analyzed by LC-MS and immunoblotting was necessarily a mixture of cell types. This is in contrast to the immunohistochemistry that shows discrete cell types and cellular compartments. In the future, we intend to refine our sampling, cancer grading, and cell collection (using for example laser capture microdissection) in an effort to reduce the observed variability between samples. Nevertheless the sampling size (n = 10 per grade) and the pathway and clustering analyses have allowed us to present meaningful results as evidenced by the data in this study and prior published work (15). We therefore conclude that spectral counting is a reliable quantitative proteomics method for the study of kidney cancer.
The uniqueness and importance of the current study relates to a fine tuning of the proteomics analysis to not simply discover pathways that are altered in RCC in general but to examine proteomic variations as a function of tumor grade and, by extension, of prognosis. It is important to emphasize that the 105 proteins identified as significantly altered in RCC (Table I) were identified by one-way analysis of variance across normal tissues and the four tumor grades (p value <0.01); more data of statistical significance can be mined by examining pairwise t testing comparing specific grades as was utilized in the nucleophosmin data (Fig. 5). Reassuringly, data in the current study show a surprising degree of concordance with our earlier study (15) despite the use of different separation techniques (two-dimensional gel electrophoresis versus liquid chromatography) and the use of different quantitation methods despite having used substantially fewer samples in the earlier study. The current study has greater “resolution” and utility as compared with other proteomics studies of RCC that are blind to grade, and for this reason data obtained here will help answer such questions as 1) what biochemical events account for progression of RCC within grade?, 2) what are objective criteria that distinguish among grades?, and 3) can grade changes be recapitulated by alteration of specific protein expression? The first two issues have been addressed in this study, and the third is actively being pursued in our laboratory.
Previous proteomics studies in our laboratory showed that several glycolytic enzymes were up-regulated as compared with the normal renal tissues (15); the current study corroborated these findings and demonstrated grade-dependent changes in individual pathway proteins (Fig. 7). PGK1 is a hypoxia-inducible gene under the regulation of p53, and change in its level may be indicative of the increased glycolytic activity that has been observed in cancer cells for decades (16, 17). Alternatively this increase in PGK1 level may indicate the hypoxic response via activated p53, a prevalent phenomenon in cancer tissues, with the consequent up-regulation in glycolysis. Interestingly the level of this protein was increased in grades 1 and 2 more than in higher grades, possibly suggesting that it is a relatively early event in cancer progression. Myc, which was found to be a hub in the network scheme (Fig. 6) and targets nucleophosmin (Fig. 6 and Ref. 24), can also activate glycolytic genes (25). Pyruvate kinase is a critical enzyme in the present study. Its fetal isoform, pyruvate kinase isoform M2 (PKM2), was found to be markedly increased in all RCC grades, consistent with a recent finding from another laboratory (26) suggesting that embryonic PKM2 is up-regulated in cancer tissues and may be advantageous for tumor cell growth. This study demonstrates which grades have higher expression of glycolysis pathway proteins, lending support to the hypothesis that the Warburg effect may be more important at discrete points of cancer progression. It also appears that the protein isoforms used in the glycolysis enzymatic reactions may be different in cancer cells than in normal tissue (Fig. 7).
Grade-dependent changes in ccRCC are also associated with alterations in protein expression within the intrinsic apoptosis pathway, a key mechanism by which tumor cells attempt to evade therapy. Apoptosis-inducing factor (AIFM1) is a mitochondrial membrane-associated protein that was originally identified by its translocation from permeabilized mitochondria to the nucleus to cause DNA fragmentation. Data presented here indicate significant reduction in AIFM1 in cancer tissues with the magnitude of the decrease being inversely proportional to grade. These data provide mechanistic insight into how RCC is able by evading the extrinsic apoptotic pathway, thereby conferring the growth advantage on cancer versus non-cancerous cells and contributing to the high degree of chemotherapy resistance seen in RCC (1).
Our finding of a high degree of significance (p value = 6.6e−22) in changes of proteins of the dehydrogenase class that are important in xenobiotic metabolism is of considerable interest in light of research on environmental influences on oncogenesis (27). There are various reports of dietary factors that may increase the risk of kidney cancer, and it is conceivable that the observed increase in xenobiotic metabolism is a marker for this process. In addition, the xenobiotic pathway may be involved in the response of the tumor to chemotherapeutic agents. Changes in proteins comprising the acute phase response are also highly significant (p = 1.33e−5). Although the acute phase response is seen in infectious and cardiovascular diseases, its relevance in cancer, specifically RCC, has also been demonstrated (28, 29). This acute phase response may underlie the use of immune modulating agents in classical therapies of RCC, but whether this finding indicates that novel anti-inflammatory therapies may also be useful in treatment of this disease remains to be investigated.
Biomarkers can be segregated into diagnostic and predictive (prognostic) markers. The heat maps generated (Fig. 9) have utility for confirming grade assignment when pathological classification is ambiguous. Although several candidate markers are promising, there are currently none in regular clinical use. Proteomics analysis has the potential to yield a profile of altered proteins that will lead to a prediction of tumor behavior. Although the current study was not designed to yield these data, subsequent analyses in our laboratory using similar techniques and prospective patient data are currently being undertaken; these data will lead to a better understanding of the source of variability between patients and subsequently to prognostic biomarkers from proteomics data.
In summary, we performed extensive proteomics analysis as a function of RCC tumor grade and identified proteins and pathways altered in specific grades. Future work in this field will correlate outcome data with proteomics analyses and lead to specific prognostic biomarkers for this disease.
Supplementary Material
Acknowledgments
We acknowledge the University of California Davis Genome Center Proteomics Core for running the samples on its MS proteomics platform and H. Xu and A. Kozik from the University of California Davis Genome Center for providing the Pixelirator script.
Footnotes
Published, MCP Papers in Press, January 21, 2009, DOI 10.1074/mcp.M800252-MCP200
The abbreviations used are: RCC, renal cell carcinoma; FFPE, formalin-fixed paraffin-embedded; ccRCC, clear cell renal cell carcinoma; VIM, vimentin; ALDO, fructose-bisphosphate aldolase; H&E, hematoxylin and eosin; FPR, false positive rate; ANOVA, analysis of variance; IPA, Ingenuity Pathways Analysis; SOM, self-organizing map; TUBB, tubulin β chain; 2D, two-dimensional; TF, transferrin; FN, fibronectin; AMBP, α1-microglobulin/bikunin precursor; FG, fibrinogen; PFK, 6-phosphofructokinase; LDH, l-lactate dehydrogenase; PGK1, phosphoglycerate kinase 1; NCBI, National Center for Biotechnology Information.
This work was supported, in whole or in part, by National Institutes of Health Grant 5UO1CA86402 from the Early Detection Research Network, NCI. This work was also supported by the Research Service of the United States Department of Veterans Affairs.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Weiss, R. H., and Lin, P.-Y. ( 2006) Kidney cancer: identification of novel targets for therapy. Kidney Int. 69, 224–232 [DOI] [PubMed] [Google Scholar]
- 2.Novara, G., Martignoni, G., Artibani, W., and Ficarra, V. ( 2007) Grading systems in renal cell carcinoma. J. Urol. 177, 430–436 [DOI] [PubMed] [Google Scholar]
- 3.Fuhrman, S. A., Lasky, L. C., and Limas, C. ( 1982) Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 6, 655–663 [DOI] [PubMed] [Google Scholar]
- 4.Hwang, S. I., Thumar, J., Lundgren, D. H., Rezaul, K., Mayya, V., Wu, L., Eng, J., Wright, M. E., and Han, D. K. ( 2007) Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene 26, 65–76 [DOI] [PubMed] [Google Scholar]
- 5.Wessel, D., and Flugge, U. I. ( 1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 6.Weiss, R. H., Joo, A., and Randour, C. ( 2000) p21Waf1/Cip1 is an assembly factor required for PDGF-induced vascular smooth muscle cell proliferation. J. Biol. Chem. 275, 10285–10290 [DOI] [PubMed] [Google Scholar]
- 7.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 8.Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 9.Fisher, R. ( 1921) Studies in crop variation: an examination of the yield of dressed grain. J. Agric. Sci. 11, 107–135 [Google Scholar]
- 10.Stoline, M. R. ( 1981) The status of multiple comparisons: simultaneous estimation of all pairwise comparisons in one-way ANOVA designs. Am. Stat. 35, 134–141 [Google Scholar]
- 11.Thomas, P. D., Campbell, M. J., Kejariwal, A., Mi, H., Karlak, B., Daverman, R., Diemer, K., Muruganujan, A., and Narechania, A. ( 2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohonen, T. ( 1995) Self-Organizing Maps, Springer, Berlin
- 13.Hendrickson, E. L., Xia, Q., Wang, T., Leigh, J. A., and Hackett, M. ( 2006) Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst 131, 1335–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A., and Ahn, N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 15.Perroud, B., Lee, J., Valkova, N., Dhirapong, A., Lin, P. Y., Fiehn, O., Kultz, D., and Weiss, R. H. ( 2006) Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol. Cancer 5, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unwin, R. D., Craven, R. A., Harnden, P., Hanrahan, S., Totty, N., Knowles, M., Eardley, I., Selby, P. J., and Banks, R. E. ( 2003) Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 3, 1620–1632 [DOI] [PubMed] [Google Scholar]
- 17.Warburg, O. ( 1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 18.Yu, S., Sher, B., Kudryk, B., and Redman, C. M. ( 1984) Fibrinogen precursors. Order of assembly of fibrinogen chains. J. Biol. Chem. 259, 10574–10581 [PubMed] [Google Scholar]
- 19.Zhang, Z., Fuentes, N. L., and Fuller, G. M. ( 1995) Characterization of the IL-6 responsive elements in the gamma fibrinogen gene promoter. J. Biol. Chem. 270, 24287–24291 [DOI] [PubMed] [Google Scholar]
- 20.Grignon, D. J., Ayala, A. G., el Naggar, A., Wishnow, K. I., Ro, J. Y., Swanson, D. A., McLemore, D., Giacco, G. G., and Guinee, V. F. ( 1989) Renal cell carcinoma. A clinicopathologic and DNA flow cytometric analysis of 103 cases. Cancer 64, 2133–2140 [DOI] [PubMed] [Google Scholar]
- 21.Gelb, A. B., Shibuya, R. B., Weiss, L. M., and Medeiros, L. J. ( 1993) Stage I renal cell carcinoma. A clinicopathologic study of 82 cases. Am. J. Surg. Pathol. 17, 275–286 [DOI] [PubMed] [Google Scholar]
- 22.Goldstein, N. S. ( 1999) Grading of renal cell carcinoma. Urol. Clin. North Am. 26, 637–642, vii [DOI] [PubMed] [Google Scholar]
- 23.Kellner, R., Lichtenfels, R., Atkins, D., Bukur, J., Ackermann, A., Beck, J., Brenner, W., Melchior, S., and Seliger, B. ( 2002) Targeting of tumor associated antigens in renal cell carcinoma using proteome-based analysis and their clinical significance. Proteomics 2, 1743–1751 [DOI] [PubMed] [Google Scholar]
- 24.Zeller, K. I., Haggerty, T. J., Barrett, J. F., Guo, Q., Wonsey, D. R., and Dang, C. V. ( 2001) Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J. Biol. Chem. 276, 48285–48291 [DOI] [PubMed] [Google Scholar]
- 25.Kim, J. W., and Dang, C. V. ( 2006) Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66, 8927–8930 [DOI] [PubMed] [Google Scholar]
- 26.Christofk, H. R., Vander Heiden, M. G., Wu, N., Asara, J. M., and Cantley, L. C. ( 2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 [DOI] [PubMed] [Google Scholar]
- 27.Yang, C. S. ( 2006) Dietary factors may modify cancer risk by altering xenobiotic metabolism and many other mechanisms. J. Nutr. 136, 2685S–2686S [DOI] [PubMed] [Google Scholar]
- 28.Pejovic, M., Djordjevic, V., Ignjatovic, I., Stamenic, T., and Stefanovic, V. ( 1997) Serum levels of some acute phase proteins in kidney and urinary tract urothelial cancers. Int. Urol. Nephrol. 29, 427–432 [DOI] [PubMed] [Google Scholar]
- 29.Jabs, W. J., Busse, M., Kruger, S., Jocham, D., Steinhoff, J., and Doehn, C. ( 2005) Expression of C-reactive protein by renal cell carcinomas and unaffected surrounding renal tissue. Kidney Int. 68, 2103–2110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.