Abstract
When DNA-binding proteins search for their specific binding site on a DNA molecule they alternate between linear 1-dimensional diffusion along the DNA molecule, mediated by nonspecific binding, and 3-dimensional volume excursion events between successive dissociation from and rebinding to DNA. If the DNA molecule is kept in a straight configuration, for instance, by optical tweezers, these 3-dimensional excursions may be divided into long volume excursions and short hops along the DNA. These short hops correspond to immediate rebindings after dissociation such that a rebinding event to the DNA occurs at a site that is close to the site of the preceding dissociation. When the DNA molecule is allowed to coil up, immediate rebinding may also lead to so-called intersegmental jumps, i.e., immediate rebindings to a DNA segment that is far away from the unbinding site when measured in the chemical distance along the DNA, but close by in the embedding 3-dimensional space. This effect is made possible by DNA looping. The significance of intersegmental jumps was recently demonstrated in a single DNA optical tweezers setup. Here we present a theoretical approach in which we explicitly take the effect of DNA coiling into account. By including the spatial correlations of the short hops we demonstrate how the facilitated diffusion model can be extended to account for intersegmental jumping at varying DNA densities. It is also shown that our approach provides a quantitative interpretation of the experimentally measured enhancement of the target location by DNA-binding proteins.
Keywords: gene regulation, search processes, random processes, intersegmental jumps, single-molecule biophysics
At any given instant of time a biological cell only uses part of its genes for production of other molecules (RNA, proteins). During the development of the cell, or the organism the cell belongs to, different genes may be turned on. A classical example is the lactose metabolism of bacteria: in absence of lactose the Lac repressor is specifically bound at the lacZ gene and prevents unnecessary production of the lactose enzyme used to digest the milk sugar; when only lactose and no glucose is present the lacZ gene is activated and lactose-digesting enzymes are produced. Equally famed is the λ switch in Escherichia coli bacteria that are infected by bacteriophage λ. There, the λ switch decides between the dormant lysogenic state or the state of lysis that leads to the production of new phages and ultimate death of the E.coli host cell. Molecularly, the regulation of a gene relies on the presence of certain DNA-binding proteins, so-called transcription factors, that bind to a specific binding site close to the starting sequence of the related gene. The transcription factor then promotes or inhibits binding of RNA polymerase and thus regulates the transcription of this gene. This strategy allows eukaryotic and prokaryotic cells as well as viruses to respond to different internal or external signals (1, 2).
The binding of the protein to its specific binding site (cognate site, “target”) is necessarily preceded by a search process of the protein for this target site, this search being of a purely stochastic nature. Yet, some active binding proteins can locate their target orders of magnitude faster (3) than estimated for pure 3-dimensional diffusion limited search according to the celebrated Smoluchowski result
for the search rate kon. Here, the target is assumed spherical and of radius b, whereas D3d denotes the 3-dimensional diffusion coefficient D3d (4). An explanation for this discrepancy is based on the linear topology of the DNA chain in which the target site is embedded. In fact, in their seminal work Berg and von Hippel analyzed the search by DNA-binding proteins and demonstrated that the combination of different search mechanisms explains the observed binding rates. According to the Berg–von Hippel-facilitated diffusion model (5) DNA-binding proteins combine 3-dimensional diffusion in the bulk and 1-dimensional diffusion along the DNA chain (6–9). This linear diffusion is mediated by nonspecific binding, the protein's binding affinity to noncognate DNA made possible by the heteropolymeric nature of DNA (10–12). An additional search mechanism already proposed in the works of Berg and von Hippel are intersegmental transfers: some proteins with two DNA-binding sites can intermittently bind nonspecifically to 2 DNA-segments that are far apart along the DNA contour but are brought in close contact in the embedding (real) space by DNA looping. Such proteins thereby transfer across a point of close contact without actually leaving the DNA (5).
Facilitated diffusion has recently received renewed interest, following a variety of new conceptual viewpoints of gene regulation (13–18). Fresh impetus has also been given by the possibility to monitor the target search by DNA-binding proteins on the level of single DNA molecules. Various aspects of the search mechanisms of DNA-binding proteins have been studied either by fluorescence methods or indirectly by probing the response to external manipulation by optical tweezers (6–8, 19, 20). In particular, it was demonstrated that the local DNA density indeed significantly influences the target search rate (20). This observation is consistent with the Berg–von Hippel idea of intersegmental transfers mentioned above. However, in contrast to the Lac repressor protein employed in the original work by Berg and von Hippel, in the optical tweezers assay reported in ref. 20 the target search was performed by EcoRV restriction enzymes that only have one DNA-binding site. The idea of intersegmental transfers therefore has to be extended, such that the protein dissociates from one DNA segment, shortly diffuses three-dimensionally, and then immediately rebinds to another DNA segment that is close by because of DNA looping. We refer to this extended mechanism as intersegmental jumps. In what follows we develop a theoretical framework to include intersegmental jumps. This extension of the facilitated diffusion model allows us to study the influence of the local DNA density on the rate of search of DNA-binding proteins for their specific binding site. Our generalized model therefore accounts for different degrees of DNA coiling. We show that this systematic extension of the Berg–von Hippel model is quantitative and consistent with the results from our experimental study in ref. 20.
Model
Previous approaches to facilitated protein diffusion consider bulk exchange for a straight cylindrical chain (21); they invoke scaling properties in different chain configurations (22) without quantitative comparison of the search rates between these; or they reformulate the problem with uncorrelated bulk excursions (23–26). Here, we relate a multiparticle picture to single protein jumps along the DNA contour allowing us to consider DNA as a fluctuating chain.
In our description of the target search process, we use the density per length n(x,t) of proteins on the DNA as the relevant dynamical quantity (x is the distance along the DNA contour). We include 1-dimensional diffusion along the DNA with diffusivity D1d, protein dissociation with rate koffns and (re)adsorption after diffusion through the bulk fluid with diffusion constant D3d. The dynamics of n(x,t) is thus governed by
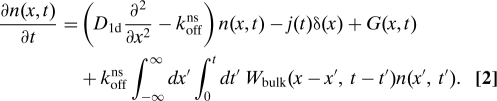 |
Here, j(t) is the flux into the target site located at x = 0, Wbulk(x − x′,t − t′) is the joint probability that a protein returns to the point x at time t after leaving the DNA at x′, t′ for a bulk excursion. G(x,t) is the flux onto the DNA of proteins that have not previously been bound to the DNA (virgin flux). j(t) is determined by imposing the boundary condition n(x = 0,t) = 0 at the target. To consider the transfer kernel Wbulk as homogeneous in space and time we made 2 assumptions that we found to be valid for the system studied in ref. 20: First, that the DNA can be treated as being infinitely long when considering the search rate. In practice this requires that the DNA should be longer than the effective sliding length lsleff introduced in Eq. 14. Note that even in bacteria the DNA length measures several millimeters, whereas DNA's persistence length is of the order of 50 nm. The second assumption is that the coiled DNA conformation fluctuates sufficiently quickly such that subsequent excursion distances x − x′ can be treated as independent.
To proceed, we Laplace and Fourier transform Eq. 2:
where we denote the Fourier and Laplace transform of a function by explicit dependence on the transformed variable. Thus n(q,u) = L[n(q,t)], n(q,t) = F[n(x,t)] and n0(x) = n(x,t = 0). The Fourier–Laplace transform of W(x,t) is here defined through
We rewrite these equations in the form
where the density
is the solution of the diffusion problem in absence of the target, i.e., when only nonspecific binding between proteins and the DNA occurs. In position space at x = 0 one obtains j(u) = nnsx(x = 0,u)/W(x = 0,u).
In this study we focus on the long time behavior of j(t) for which the density nnsx(x,t) will reach an equilibrium value neqns. Since, by Tauberian theorems, large t correspond to small u we study the u → 0 behavior of j(u), with nns(x,u) ∼ neqns/u. For W0(x = 0,u) we assume that the limit W(x = 0,u = 0) (time average) is finite and nonzero; this is true when the distance distribution of bulk excursions is sufficiently long tailed, as fulfilled for the transfer kernel Wbulk encountered below. In practice, being interested in the first arrival at the target, we assume that the protein concentration is sufficiently dilute such that this arrival happens after reaching neqns (“rapid equilibrium”) but still with many searching proteins. Thus j(u) ∼ k1dneqns/u, and therefore at long times the stationary current jstat ∼ k1dneqns into the target is reached. Here, we introduce the constant k1d−1 = W(x = 0,u = 0):
of dimension sec over cm. We also recognize that λbulk(x) = Wbulk(x,u = 0) is the distribution of relocation lengths along the DNA after a single 3-dimensional excursion.
We express our results in terms of the association rate
to the target where nbulk is the density of unbound proteins in the bulk (at nonspecific equilibrium). The nonspecific binding constant per length of DNA is Kns = neqns/nbulk. In terms of the total volume concentration of proteins ntotal = nbulk + lDNAtotalneqns, where lDNAtotal is the total length of DNA divided by the volume of the entire system, we have
The rate kon is related to the mean first arrival time for the steady-state association. Namely, konnbulk is the probability per time for protein association with the target. Thus, the (survival) probability that no protein has arrived at the target yet is Psurv(t) = exp(−konnbulkt), and the average target search time is T = 1/(konnbulk). The flux j(t) being linear in the protein concentration ntotal implies that for sufficiently low ntotal the steady state is indeed reached well before the first protein actually binds to the specific target (note that in vivo protein concentrations can be as low as nanomolar).
Results
Straight Rod Configuration.
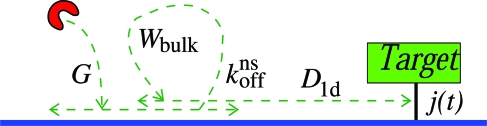
Consider first stretched DNA (see Fig. 1) corresponding to a cylinder of radius rint (the range of nonspecific interaction), with nonspecific reaction rate konns at the boundary: the flux of proteins, per length of DNA, onto the DNA is konns times the bulk concentration next to the DNA. This implies that at equilibrium of nonspecific binding, neqnskoffns = nbulkkonns such that Kns = konns/koffns. (We deviate here from the conventions of ref. 20 by only letting nbulk count “active enzymes.”)
Fig. 1.
Schematic of the search mechanisms in Eq. 2 on a piece of stretched DNA. To locate the specific binding site (target) on the DNA molecule a DNA-binding protein diffuses one-dimensionally along the DNA chain with diffusivity D1d, or it unbinds and performs a bulk excursion with diffusion constant D3d. The virgin flux G denotes proteins that have not previously bound to DNA. Note the tendency to revisit sites when immediate rebindings and thus short hops are likely.
To find the distribution Wbulk(x,t) for return to the DNA of a protein released at the DNA at x = 0 and t = 0, we solve the cylindrical diffusion equation for the probability density P(x,r,t) of the protein's position
where r measures the distance to the cylinder axis. The reactive boundary condition for the probability flux out of the cylinder boundary is
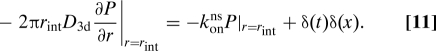 |
The initial distribution outside the cylinder is P(r,t)|t=0 = 0. From the solution of this problem we obtain Wbulk as konns P|r=rint in terms of modified Bessel functions Kν,
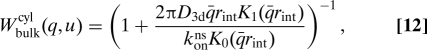 |
where we abbreviate .
With these ingredients one can evaluate numerically the search rate to reach the specific target on a straight DNA, as given by Eq. 7. However, some limits allow for an analytic approximation for the integral in Eq. 7: for konns ≪ D3d one may take 1 − λbulk(q) ≈ 1, and thus . One finds the association rate
where is the so-called sliding length of a protein during a single nonspecific binding period. Eq. 13 states that each time a protein, with rate constant konns, binds to the DNA within a distance lsl from the target it is able to “slide” onto the target. Note that in this konns ≪ D3d limit the search rate kon is independent of the DNA conformation. When konns ≪ D3d the rate of nonspecific binding will limit rebinding such that the protein will relocate by a long distance during a single 3-dimensional excursion. The part of λbulk corresponding to such long relocations does not contribute significantly to the integral in Eq. 7, which is the physical reason for the approximation leading to Eq. 13.
In the opposite limit konns ≫ D3d one may approximate 1 − λbulk(q) ≈ 2πD3d/[konns/ln(lsleff/rint)] where the length
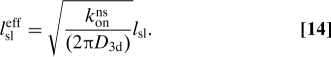 |
This produces the result
In this limit the protein can rebind quickly after unbinding. This leads to oversampling and a lowered value of kon compared to Eq. 13. lsleff can be interpreted as an effective sliding length including immediate rebindings (21). Expression 15 generalizes Smoluchowski's result Eq. 1, replacing the naked target size b by the “antenna length” entering Eq. 15. Results 7 and 12 are identical to the classical result for straight, infinitely long DNA (21). However, the approach here allows us to explicitly consider coiled DNA.
Coil with Persistence Length.
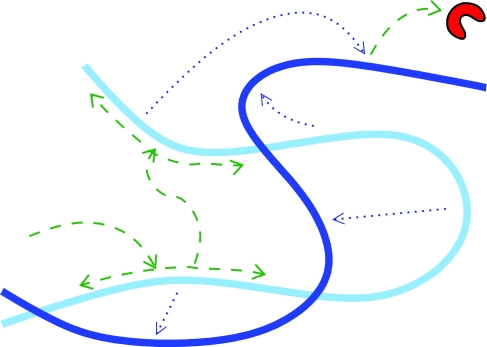
To obtain the transfer kernel Wbulk for a random coil DNA configuration (see Fig. 2) we treat the DNA as consisting locally of straight segments. We assume that the local density lDNA of DNA (length per volume) around a segment is dilute such that the typical distance between segments is much larger than rint. We then apply a superposition technique, where the probability that the protein has reacted with a segment is obtained by treating independently the interaction with each segment, and then choose among these the interaction for which binding happens first. To obtain the dynamics of the interaction of the protein with one segment of DNA we consider a large volume V around the segment of length L. Placing the protein at a uniformly random position in space relative to the DNA segment at t = 0 and denoting the probability that the protein has bound to this DNA before time t by Jsingle(t), then the probability that it has not bound to any of N pieces of DNA is (with the assumption of independent interactions) Psurvforeign(t) = [1 − Jsingle(t)]N. Taking the limit of V,N,L → ∞ for fixed lDNA = NL/V we get Psurvforeign(t) = exp[−Jcap(t)], where
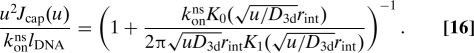 |
The bulk transfer kernel Wbulk in the coiled conformation then becomes
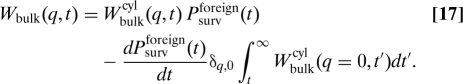 |
Here, the first term on the right-hand side represents proteins that return to the original segment without being captured by foreign segments. Those are represented by the second term, where the factor δq,0 is a result of the assumption that a capture by foreign DNA segments leads to a long relocation measured along the DNA contour.* These relocations only contribute close to q = 0 in Fourier space. Exploiting the reasoning behind the approximation that led to Eq. 13 we discard them from Wbulk when the integral in Eq. 7 is calculated. However, even numerically this integral is difficult to handle. A good approximation can be found noting that by Tauberian theorems the Laplace inversion for Jcap(u) can be divided into 3 regimes when konns ≫ D3d:
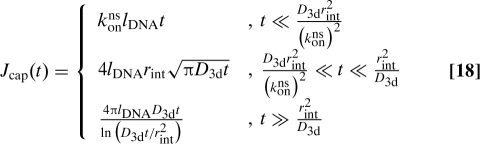 |
The first regime corresponds to reaction limited binding and is very brief for large konns. We discard this regime and ignore the slowly varying logarithm in the last regime. This allows us to approximate Psurvforeign by
Numerical analysis shows excellent agreement between Psurvforeign and Papp. The value of kcap is fixed self-consistently by the condition that the total amount of proteins returning to the original segment is the same as for Psurvforeign, i.e.,
We solved this equation numerically. Once a value for kcap is obtained the choice of Papp in Eq. 19 turns out to be very convenient. In fact, it can be written
where a = . When inserted into Eq. 17 in Laplace space we need to evaluate numerically only one integral to obtain Wbulk(q,u), namely
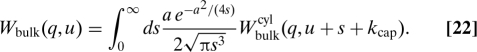 |
The term with δq,0 was already discarded here. Eq. 22 generalizes the straight rod result 12. In the limit D3d ≫ konns it is consistent with the approximation 1 − λbulk(q) ≈ 1 leading to the expected result (13) for that case.
Fig. 2.
Search rate enhancement on coiled DNA: a protein that in the case of straight DNA would perform a short hop can now, because of DNA looping, be captured by a foreign (remote) segment of DNA. This leads to a more efficient search since local oversampling is reduced by these intersegmental jumps. The DNA configuration is assumed to be dynamic such that when the protein is released from the foreign segment the latter has moved further away from the original segment. Dashed lines, motion of the protein; dotted lines, DNA configurational changes.
Discussion
A single stochastic search mechanism is usually not efficient at optimizing a search process. Thus, Brownian motion, whose fractal dimension is df = 2, in 1-dimensional and 2-dimensional geometries leads to significant oversampling in the sense that a given site is revisited many times (27, 28). Lévy motion with a jump length distribution λ(x) ≃ |x|−1−α (0 < α < 2) has fractal dimension df = α and thus a comparatively reduced degree of oversampling. However, the first arrival of Lévy motion is hampered by leapovers, i.e., frequent overshoots of the target (29). In 3-dimensional space both Brownian and Lévy motions are inefficient at locating a small target. Whereas typical chemical reactants in a liquid environment have no choice but to passively diffuse until mutual encounter (4) intermittent models combine different search mechanisms (28, 30). An important example of intermittent search models is the facilitated diffusion of transcription factors in gene regulation.
Here, we have developed a theoretical framework analyzing facilitated diffusion in the presence of intersegmental jumps whose relevance depends on the degree of DNA coiling. These results together with the experimental evidence from our single DNA experiments reported in ref. 20 demonstrate that the binding rate of DNA-binding proteins can be significantly enhanced when DNA is allowed to coil freely. This effect would be further amplified when the local density of DNA is increased beyond that of a relaxed coil, for instance, by packaging in a living cell. This is consistent with observations of increased cleavage rate of restriction enzymes in supercoiled over relaxed DNA plasmids (31).
In our derivation of the probability Psurvforeign we assumed that the density of foreign segments is uniform in space. Although this is not true for a freely fluctuating coil (the density decreases on a scale of the persistence length of the DNA) we argue that this non-uniformity is not important for evaluating the search rate: in fact, if the protein diffuses far away from the original segment it will most likely perform a long relocation measured along the DNA contour. Again it does not matter for the search rate how we bookkeep these long relocations. To estimate the density lDNA for a random relaxed coil we employ the Worm Like Chain model. A good approximation for the probability density to loop back, for a point on the chain a contour distance s away from another point on the chain, is (32)
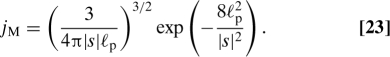 |
For a target in the middle of a chain of length L, this leads to the DNA density around the target.
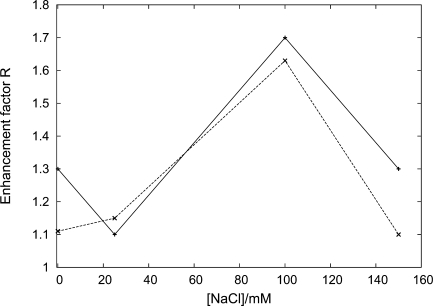
The theory presented here quantitatively describes recent experimental data from an optical tweezers setup reported in ref. 20. In that assay the search rate kon was measured at various degrees of DNA coiling for a DNA of length L = 6,538 base pairs (approximately 2.24 μ m corresponding to 42 persistence lengths). The relative increase R of kon in the maximally relaxed DNA configuration compared with kon in the stretched state is shown in Fig. 3 as a function of the NaCl concentration. R was measured to be ≈ 1.1 – 1.3 in buffers with salt concentrations of 5 mM MgCl2, and 0–25 mM for NaCl, respectively. This relative increase R compares well with the theory presented here by using estimates of lDNA based on Eq. 23 giving bp, and a value lsleff ∼ 100 bp based on a quantitative analysis (33) of experiments (34, 35), suggesting effective sliding lengths of this size. Numerical analysis of the theory presented here shows that R depends significantly only on lDNA and lsleff for realistic ranges of the remaining parameters. At physiological salt (100 mM NaCl) an increase of R ≈ 1.7 is found experimentally (20). This observation agrees well with the increased DNA density expected because of attraction between segments induced by the 5 mM divalent Mg2+ ions at this NaCl concentration, see ref. 20 for details. We are not aware of a theory to calculate lDNA under such attraction. However, the value needed to obtain R ≈ 1.7 at this salt condition appears reasonable. At even higher salt the ratio R drops again, which is consistent with a reduction in lsleff because of weaker nonspecific binding at high salt (34, 35).
Fig. 3.
Enhancement factor R of the search rate kon in the maximally relaxed DNA configuration versus the search rate kon for stretched DNA, as a function of NaCl concentration. The full line shows the values obtained from the experiment reported in ref. 20, and the dashed line represents the values from our theoretical approach. Interestingly the maximum occurs close to the physiological salt concentration of 100 mM.
We presented a model for facilitated diffusion that is useful to calculate the search rate of DNA-binding proteins for their specific binding site on a fluctuating DNA coil with variable DNA density. This model represents a convenient way to rephrase the Berg–von Hippel theory with the additional advantage that it interpolates between stretched and coiled DNA configurations. In the development of this theory a number of assumptions were made, in particular, that of sufficiently fast local relaxation of the DNA configuration. This assumption was found to be satisfied for the protein EcoRV used in ref. 20, but it will not hold universally. It would therefore be beneficial to generalize the theory to slowly fluctuating or static (quenched) DNA configurations, potentially leading to effects similar to those found in polymer models (36). Another interesting question for future work concerns the potential effect on in vivo gene regulation of the slow diffusion caused by molecular crowding (37, 38), including interesting consequences for the exchange dynamics of proteins with the DNA (39).
Acknowledgments.
We thank Tobias Ambjörnsson, Alexander Grosberg, Mehran Kardar, Leonid Mirny, and Roland Winkler for discussions. This work was partially funded by the Villum Kann Rasmussen Foundation, Deutsche Forschungsgemeinschaft, and by a Fundamenteel Onderzoek der Materie Projectruimte Grant, a Netherlands Organization for Scientific Research-Vernieuwings Impuls grant.
Footnotes
The authors declare no conflict of interest.
Cases where this assumption is justified are given by systems in which (i) the persistence length of the DNA is large such that the chemical distance along DNA loops will necessarily be larger than the effective sliding length lsleff, or (ii) the DNA attains a globular configuration, for instance, through mutual attraction bringing many chemically distant DNA segments together.
References
- 1.Alberts B, et al. Mol Biol Cell. New York: Garland Science; 2002. [Google Scholar]
- 2.Ptashne M. A Genetic Switch. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 3.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. III. Kinetic studies. J Mol Biol. 1970;53:401. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 4.von Smoluchowski M. Three presentations on diffusion, molecular movement according to Brown and coagulation of colloid particles. Physikal Zeitschr. 1916;17:557–571. [Google Scholar]
- 5.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 6.Sokolov IM, Metzler R, Pant K, Williams MC. Target search of N sliding proteins on a DNA. Biophys J. 2005;89:895–902. doi: 10.1529/biophysj.104.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys Rev Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 9.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc Natl Acad Sci USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakk A, Metzler R. In vivo non-specific binding of λ CI and Cro repressors is significant. FEBS Lett. 2004;563:66–68. doi: 10.1016/S0014-5793(04)00249-2. [DOI] [PubMed] [Google Scholar]
- 11.Bakk A, Metzler R. Nonspecific binding of the OR repressors CI and Cro of bacteriophage λ . J Theor Biol. 2004;231:525–533. doi: 10.1016/j.jtbi.2004.07.007. and references therein. [DOI] [PubMed] [Google Scholar]
- 12.Stormo GD, Fields DS. Specificity, free energy and information content in protein-DNA interactions. Trends Biochem Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 13.Bintu L, et al. Transcriptional regulation by the numbers: Models. Curr Opin Genet Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walczak AM, Onuchic JN, Wolynes PG. Absolute rate theories of epigenetic stability. Proc Natl Acad Sci USA. 2005;102:18926–18931. doi: 10.1073/pnas.0509547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blossey R, Giuraniuc CV. Mean-field versus stochastic models for transcriptional regulation. Phys Rev E. 2008;78:031909. doi: 10.1103/PhysRevE.78.031909. [DOI] [PubMed] [Google Scholar]
- 16.Kolesov G, Wunderlich Z, Laikova ON, Gelfand MS, Mirny LA. How gene order is influenced by the biophysics of transcription regulation. Proc Natl Acad Sci. 2007;104:13948–13953. doi: 10.1073/pnas.0700672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little JW, Shepley DP, Wert DW. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozbudak EM, Thattai M, Lim NH, Shraiman BI, van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 19.Bonnet I, et al. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Broek B, Lomholt MA, Kalisch SMJ, Metzler R, Wuite GJL. How DNA coiling enhances target localization by proteins. Proc Natl Acad Sci USA. 2008;105:15738–15742. doi: 10.1073/pnas.0804248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg OG, Ehrenberg M. Association kinetics with coupled 3-dimensional and one-dimensional diffusion–chain-length dependence of the association rate to specific DNA sites. Biophys Chem. 1982;15:41–51. doi: 10.1016/0301-4622(82)87015-4. [DOI] [PubMed] [Google Scholar]
- 22.Hu T, Grosberg AY, Shklovskii BI. How proteins search for their specific sites on DNA: The role of DNA conformation. Biophys J. 2006;90:2731–2744. doi: 10.1529/biophysj.105.078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppey M, Bénichou O, Voituriez R, Moreau M. Kinetics of target site localization of a protein on DNA: A stochastic approach. Biophys J. 2004;87:1640–1649. doi: 10.1529/biophysj.104.045773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomholt MA, Ambjörnsson T, Metzler R. Optimal target search on a fast-folding polymer chain with volume exchange. Phys Rev Lett. 2005;95:260703. doi: 10.1103/PhysRevLett.95.260603. [DOI] [PubMed] [Google Scholar]
- 27.Hughes BD. Random Walks and Random Environments. Vol. 1. Oxford: Oxford Univ Press; 1995. [Google Scholar]
- 28.Lomholt MA, Koren T, Metzler R, Klafter J. Lvy strategies in intermittent search processes are advantageous. Proc Natl Acad Sci USA. 2008;105:11055–11059. [Google Scholar]
- 29.Koren T, Lomholt MA, Chechkin AV, Klafter J, Metzler R. Leapover lengths and first passage time statistics for levy flights. Phys Rev Lett. 2007;99:160602. doi: 10.1103/PhysRevLett.99.160602. [DOI] [PubMed] [Google Scholar]
- 30.Bénichou O, Loverdo C, Moreau M, Voituriez R. Optimizing intermittent reaction paths. Phys Chem Chem Phys. 2008;10:7059i–7072. doi: 10.1039/b811447c. [DOI] [PubMed] [Google Scholar]
- 31.Gowers DM, Halford SE. Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling. EMBO J. 2003;22:1410–1418. doi: 10.1093/emboj/cdg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ringrose L, Chabanis S, Angrand PO, Woodroofe C, Stewart AF. Quantitative comparison of DNA looping in vitro and in vivo: Chromatin increases effective DNA flexibility at short distances. EMBO J. 1999;18:6630–6641. doi: 10.1093/emboj/18.23.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H-X. A model for the mediation of processivity of DNA-targeting proteins by nonspecific binding: Dependence on DNA length and presence of obstacles. Biophys J. 2005;88:1608–1615. doi: 10.1529/biophysj.104.052688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeltsch A, Pingoud A. Kinetic characterization of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochemistry. 1998;37:2160–2169. doi: 10.1021/bi9719206. [DOI] [PubMed] [Google Scholar]
- 35.Stanford NP, Szczelkun MD, Marko JF, Halford SE. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokolov IM, Mai J, Blumen A. Paradoxical diffusion in chemical space for nearest-neighbor walks over polymer chains. Phys Rev Lett. 1997;79:857–860. [Google Scholar]
- 37.Golding I, Cox EC. Physical nature of bacterial cytoplasm. Phys Rev Lett. 2006;96:098102. doi: 10.1103/PhysRevLett.96.098102. [DOI] [PubMed] [Google Scholar]
- 38.Dix JA, Verkman AS. Crowding effects on diffusion in solutions and cells. Annu Rev Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- 39.Lomholt MA, Zaid IM, Metzler R. Subdiffusion and weak ergodicity breaking in the presence of a reactive boundary. Phys Rev Lett. 2007;98:200603. doi: 10.1103/PhysRevLett.98.200603. [DOI] [PubMed] [Google Scholar]