Preface
To ensure its faithful duplication, chromosomal DNA must be precisely duplicated in each cell cycle, with no sections left unreplicated and no sections replicated more than once. Eukaryotic cells achieve this by dividing replication into two non-overlapping phases. During late mitosis and G1, replication origins are ‘licensed’ for replication by loading the Mcm2-7 proteins to form a pre-replicative complex (pre-RC). Mcm2-7 proteins are then essential for initiating and elongating replication forks during S phase. Recent data have provided biochemical and structural insight into the process of replication licensing, and the mechanisms that regulate it during the cell cycle.
Introduction
In order for the cell division cycle to produce two daughter cells that inherit a perfect copy of the genetic material originally present in the mother cell, it must accomplish two difficult tasks: the chromosomal DNA must first be precisely duplicated, with no errors, deletions or duplications, and then the two copies must be precisely segregated to the two daughter cells. The accuracy of these events is particularly crucial to multicellular organisms, where any changes to the genome could potentially give rise to cancers which threaten the life of the entire organism. This review will concentrate on the first of these linked problems: how eukaryotic cells ensure that their chromosomal DNA is precisely duplicated during S phase of the cell cycle.
A large number of replication origins (typically from ~103 – 105) (see BOX 1) are used by eukaryotes to ensure that the entire genome is fully replicated. But these origins must be regulated very strictly. How does the cell know whether or not it has already replicated a section of DNA in S phase? Eukaryotes have solved this problem by providing a marker to distinguish replicated from unreplicated DNA. The fact that replicated DNA differs from unreplicated DNA was first suggested by the classic cell fusion experiments of Rao and Johnson1. In hybrids of G1 and G2 cells, the unreplicated G1 nucleus passes directly into S-phase, whilst the DNA of the G2 nucleus (which has already been replicated) does not replicate again until after the hybrid cell has passed through mitosis. The G2 nucleus is therefore refractory to undergoing further replication. Subsequent work using cell-free extracts of Xenopus eggs refined this idea and suggested a model whereby replication origins were “licensed” for replication during late mitosis and G1, but the licence was removed as the DNA was replicated2. Dividing the process of DNA replication into two non-overlapping phases (one phase permissive for the licensing of DNA replication and a second phase that is permissive for the initiation of replication but not for licensing) can potentially explain how cells ensure the precise duplication of chromosomal DNA in a single cell cycle.
Detailed experimental support for the licensing model has now been obtained, which shows that it comprises four essential features3,4. First, replication origins are licensed by stably binding complexes of the mini-chromosome-maintenance 2-7 proteins (Mcm2, 3, 4, 5, 6 and 7). Mcm2-7 proteins form an essential component of the “pre-replicative complex” (pre-RC) of proteins found at replication origins during G1 phase. Second, the binding of Mcm2–7 proteins to origin DNA is essential for the origin to be able to initiate a pair of replication forks. Third, the licensing of origins and the loading of Mcm2-7 onto DNA is restricted to late mitosis and G1 of the cell cycle. Fourth, Mcm2-7 proteins are displaced from origins as DNA replication is initiated, probably by travelling ahead of the replication fork. The licence is therefore never associated with replicated DNA.
These features can explain why DNA replicates only once in each cell cycle (Fig 1). Recent results also indicate that the presence or absence of licensed origins has even deeper implications for cell biology, as it might play a role in determining the proliferative capacity of cells3.
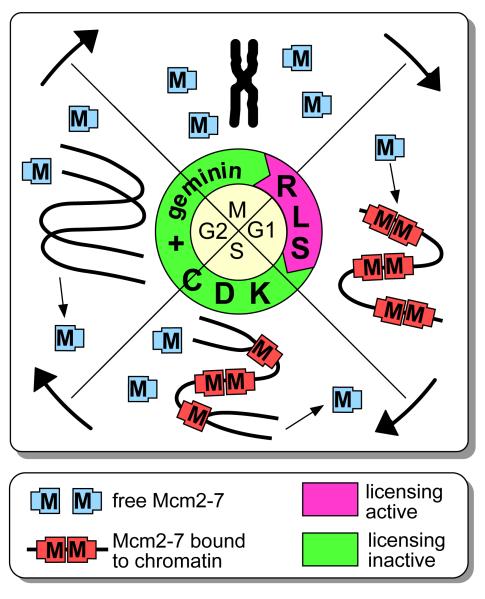
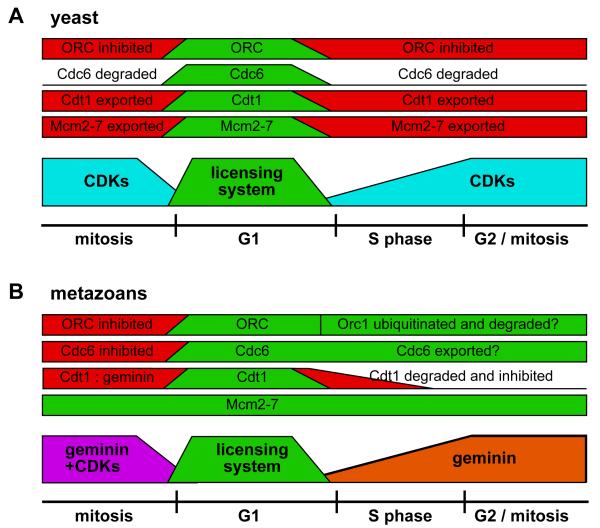
Figure 1.
Regulated loading and unloading of Mcm2-7 during the cell cycle.
A small segment of chromosomal DNA that encompasses 3 replication origins is shown. At the end of mitosis (M), the replication licensing system (RLS) is activated, which causes Mcm2-7 (the asymmetric rectangles marked ‘M’) to be loaded onto potential replication origins (origin licensing). The licensing system is turned off at the end of G1 by inhibition by CDKs and/or geminin. During S phase, the Mcm2-7 complexes are displaced from replicated DNA by moving ahead of the replication fork, and are removed from DNA at fork termination. In this way, replicated DNA cannot undergo further initiation events until passage through mitosis.
Understanding the function of Mcm2-7
The Mcm2–7 complex consists of 6 closely related proteins that are highly conserved throughout the eukaryotic kingdom (forming the MCM/P1 family). Each protein has a relatively highly conserved C-terminus which shows sequence homology to hexameric DNA helicases and contains an ATPase motif. A range of complexes have been described containing different combinations of the Mcm2-7 proteins. A purified hexamer containing Mcm4, 6 and 7 has been shown to have limited 3′ to 5′ DNA helicase activity in vitro5. However, only the heterohexamer that contains all 6 Mcm2-7 proteins can support licensing in Xenopus egg extract6. In vitro, all 6 proteins are required for maximum ATPase activity, and the ATPase motifs of all 6 proteins are required for viability in S. cerevisiae7. Chromatin immunoprecipitation has revealed that Mcm proteins are associated with replication forks as they elongate along chromosomal DNA8, and inhibition of Mcm2-7 function during S phase causes a rapid cessation of DNA synthesis9-11, indicating that Mcm2-7 function is required for fork progression as well as initiation. These observations all suggest that Mcm2-7 functions as a helicase that unwinds DNA ahead of the replication fork. The Mcm2-7 helicase is therefore likely to be displaced from replicated DNA by virtue of its moving ahead of the replication fork. The displacement of the replication licence from replicated DNA is one of the major features (point 4 above) of the licensing factor model.
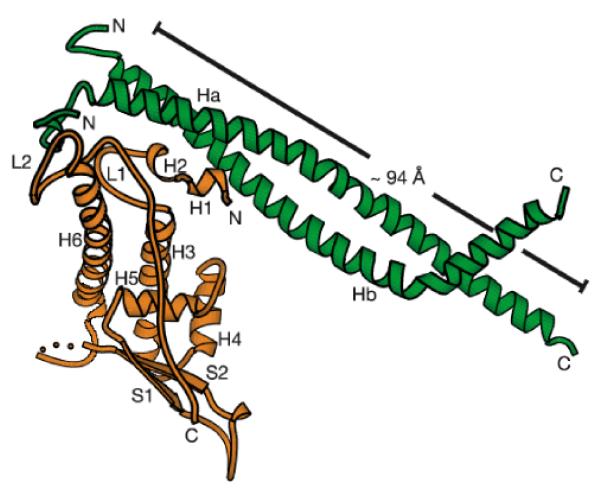
The archaea possess proteins with high sequence homology to eukaryotic MCM/P1 proteins. The single MCM protein from Methanobacterium thermoautotrophicum (MtMCM) forms dodecamers that display DNA helicase activity in vitro12-14. A crystal structure of the N-terminus of the MtMCM has recently been reported15. This reveals a double hexameric structure, with a large positively charged channel running through the centre that is wide enough to surround double-stranded DNA (FIG. 2a-c). The C-terminal helicase domain that is missing from the crystal structure could potentially form an extra ring facing away from the dimer interface. This conclusion in supported by electron microscopy of the full-length MtMCM which shows a bilobed hexameric structure with a large central channel16 Modelling indicated that the N-terminal domain fits well into the central part of the structure, whereas the helicase-containing C-terminus could form the extra lobe (Fig 2d).
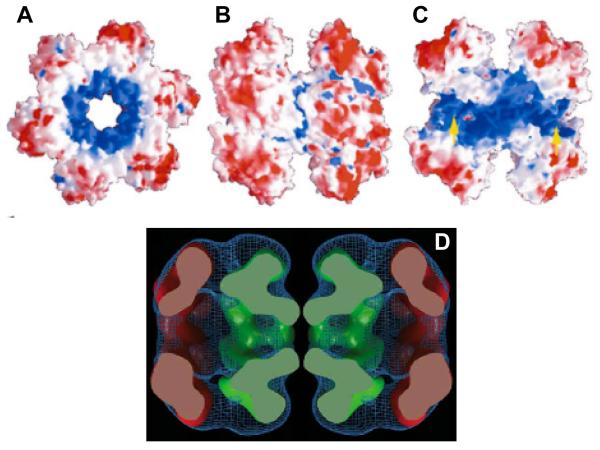
Figure 2.
Crystal structure of the N terminus of the MtMCM.
A - C. Different views of the N terminus of the MtMCM dodecamer, highlighting the central channel that runs through it. Positive charges are shaded in blue, negative charges in red. A, End view. B, Side view. C, Same view as B, but with the two front-most monomers removed to reveal the central channel. Yellow arrows show the side channels passing between exterior and interior. Reproduced from reference15. D A side view of the electron density of full-length MtMCM, aligned to approximately correspond to the size view of the crystal structure shown in B. In green, crystal structure of the N terminus has been fitted into the electron density. In red is fitted a hexameric model of the core AAA+ domain of RuvB, which has significant homology to the C-terminus of the Mcm2-7 proteins. Reproduced from reference16.
This structure has interesting implications for the function of the eukaryotic Mcm2-7 proteins and their role in replication licensing. As can be seen from FIG. 1, it is crucial that the binding of Mcm2-7 to DNA is both stable and regulated. Mcm2-7 complexes that are loaded onto DNA in late mitosis must remain associated through G1 and into S phase. It is also crucial that once the licensing system is turned off in late G1, there is no illegitimate association of Mcm2-7 with DNA. These problems can be overcome if the loading of Mcm2-7 onto DNA represents the Mcm2-7 hexamer encircling the DNA. This would lead to a stable association between Mcm2–7 and the DNA, which could be tightly controlled by regulating the activity of the loader which would be required to open up the hexamer and clamp it around DNA. The MtMCM structure therefore provides an explanation for two of the fundamental features (points 1 and 3 above) of the licensing model — the stability and temporal control of Mcm2–7 protein binding.
The MtMCM structure is highly congruent with the helicase domain of the SV40 large T antigen, the crystal structure of which has recently been determined17. T antigen is the replicative helicase for SV40 DNA. Like MtMCM, T antigen forms a bilobed double hexamer with a large central channel. Interestingly, T antigen contains “exit channels” through which single-stranded DNA might be extruded as the helicase progresses, and features consistent with this can be seen in MtMCM16. Li et al propose a mechanism by which T antigen unwinds DNA as it is replicated (FIG. 3). T antigen encircles both strands of template DNA prior to initiation, which for Mcm2-7 would correspond to the licensed but uninitiated state. On initiation, one of the single-strands is extruded through the side channels, whilst the other single strand exits the helicase at the hexamer-hexamer interface. This model is consistent with previous models based on the extruded ‘rabbit ears’ of single-stranded DNA that had been observed when T antigen interacts with origin DNA in vitro18.
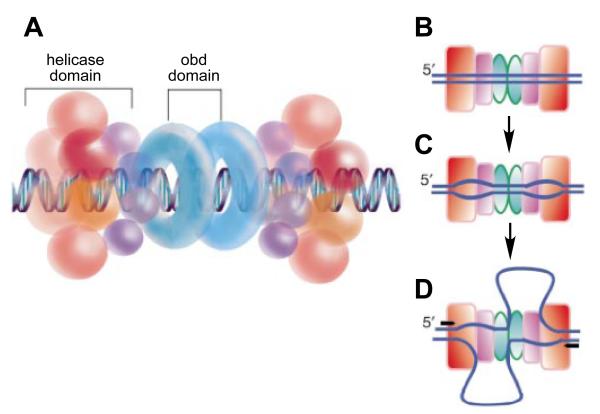
Figure 3.
Model for SV40 T antigen function at replication initiation.
Model for the function of the SV40 T antigen based on the crystal structure of the helicase domain. A, A double hexamer of the SV40 T antigen, comprising a bilobed helicase domain and an origin binding domain (obd), is shown encircling origin DNA. B, C, D, Model for the possible extrusion of unwound DNA through a side channel of the helicase domain to unwind the two forks bidirectionally. A, T antigen binds and encircles origin DNA. B, Origin DNA is unwound, possibly by the two T antigen hexamers rotating relative to one another. C, A conformational change to form the fully initiated complex may involve extrusion of one of the single strands through an exit channels in the helicase domain, with the other single strand being extruded at the hexamer-hexamer interface. Reproduced from reference17.
The ‘Mcm Paradox’
There are some observations, however, that do not fit neatly into a model of Mcm2-7 functioning as a simple replication fork helicase. First, whereas one might have thought that only one double hexamer needs to be loaded onto each replication origin, in fact the number in eukaryotes seems much higher, with 10 – 40 Mcm2-7 hexamers present at each origin19-22. The loading of Mcm2-7 onto chromatin depends on the Origin Recognition Complex (ORC; see below), which is thought to bind specifically to replication origins. ORC recruits two further proteins – Cdc6 and Cdt1 – which together facilitate the loading of Mcm2-7. However, studies in HeLa cells and Xenopus extracts indicate that Mcm2-7 is also loaded onto sites on the chromosome that are distant from ORC22-26. ATP hydrolysis by S. cerevisiae ORC is specifically required for loading the subsequent Mcm2-7 complexes after the first ones have been loaded onto DNA27, which provides further evidence that multiple copies of Mcm2-7 are loaded onto each origin. The distribution of Mcm2-7 on chromatin therefore does not match the expected distribution of a simple replicative helicase, which should be loaded at replication origins and at only two copies per origin. A second discrepancy is that although some Mcm2-7 colocalizes with replication forks8, the bulk of the chromatin-bound protein is not associated with sites of DNA synthesis28-30.
There are a number of possible explanations for this paradoxical distribution of Mcm2–7 on chromosomes. One possibility is that Mcm2-7 does not act as a typical DNA helicase, but instead acts as a fixed pump that forces DNA into immobilized replication forks where DNA synthesis occurs31. Because the DNA will be twisted as it is pumped, this will provide the energy to unwind the DNA ahead of the DNA polymerases, obviating the need for a helicase at the fork. However, for this to happen, the DNA between the Mcm2-7 pump and the DNA polymerases must be somehow protected from topoisomerase-mediated relaxation, which would dissipate the stored-up energy and allow the DNA to rewind.
An alternative possibility is that although a small fraction of Mcm2–7 is required to licence and initiate the replication of DNA, the bulk of the Mcm2-7 is not involved in DNA synthesis, either having some completely different function32,33 or only being required for some contingency. Consistent with the idea that most of the Mcm2–7 is not normally involved in DNA synthesis is the observation that near-maximal replication rates can be achieved in Xenopus with only 1-2 heterohexamers of Mcm2-7 per replication origin21,22,34. Although Xenopus Cdc6 is required for the loading of Mcm2-7 onto DNA, its affinity for replication origins decreases once the first Mcm2-7 hexamers have been loaded onto each origin34. This plausibly ensures that under conditions where the extent of Mcm2-7 loading is limited, each origin gets the minimum allocation for it to support initiation. But what is the function of the excess Mcm2-7 hexamers? One proposal is that the excess Mcm2-7 might activate intra-S phase checkpoints. Human Mcm7 interacts with ATRIP, a protein that binds and activates the ATR kinase which is involved in checkpoint signalling when DNA replication is disturbed35. When Mcm7 levels were slightly reduced so that replication rates were not significantly reduced, checkpoint signalling pathways were also disrupted35.
The presence of excess Mcm2-7 at replication origins might explain a puzzling feature of eukaryotic replication origins, which in many circumstances form diffuse initiation sites that are not precisely specified by DNA sequence36. If the Mcm2-7 helicase has a similar mode of action to SV40 T antigen, then the site of assembly of a back-to-back Mcm2-7 dimer on DNA could potentially specify a replication origin. It is possible that the 10–40 Mcm2-7 hexamers loaded onto each origin could form a number of back-to-back dimers, each of which could provide a different site at which initiation could occur. This might provide redundant origins for use if various components of the replication machinery were to fail.
The mechanism of licensing and Mcm2-7 protein loading
The loading of Mcm2-7 onto DNA that occurs as an origin is licensed is likely to involve the opening of the Mcm2-7 ring to allow it to encircle the DNA. Other protein components of the pre-RC are required to load Mcm2–7 onto chromatin. One of the first events is the binding of ORC to DNA (FIG. 4). ORC in turn recruits two further proteins – Cdc6 and Cdt1 – which are all required for the loading of Mcm2-7 and the functional licensing of the origin3,4,37. This process has been reconstituted on Xenopus sperm chromatin using purified ORC, Cdc6, Cdt1 and Mcm2-7, in addition to a chromatin remodelling protein, nucleoplasmin, which is required for the initial binding of ORC to DNA38.
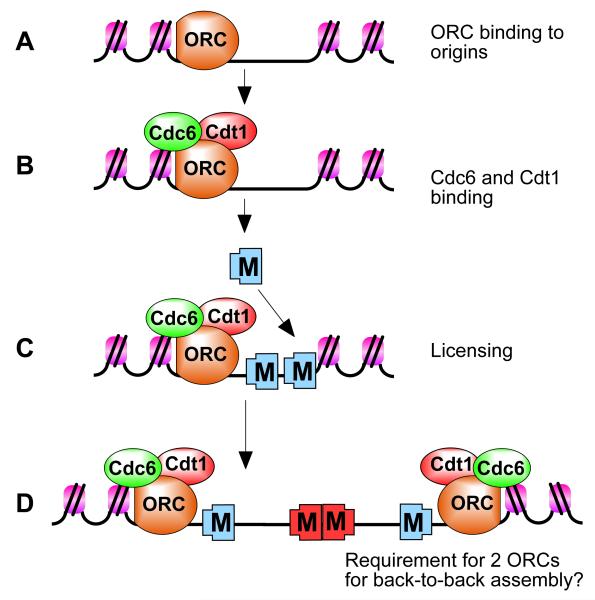
Figure 4.
Stepwise assembly of pre-replicative complex (pre-RC) proteins during origin licensing.
A, ORC is first recruited to the replication origin. B, ORC recruits Cdc6 and Cdt1. C, ORC, Cdc6 and Cdt1 act together to load multiple Mcm2-7 hexamers onto the origin, which licenses the DNA for replication. D, Initiation-competent complexes are probably formed by the back-to-back assembly of two Mcm2-7 complexes. Since ORC is asymmetrical, this might require a second ORC molecule in the opposite orientation to load Mcm2-7 in the opposite orientation.
Little is known about this reaction, but the available evidence indicates that ORC, Cdc6 and Cdt1 act together as a clamp loader to open up the Mcm2-7 ring and load it around DNA37. Alternatively, the ORC, Cdc6 and Cdt1 molecules might facilitate the assembly of the Mcm2-7 hexamer from the many subcomplexes of Mcm proteins that have been described. Consistent with these models, ORC, Cdc6 and Cdt1 are only required for the loading of Mcm2-7 onto DNA (licensing), but are not required for the continued association of Mcm2-7 on DNA once this has occurred39-42. Both ORC and Cdc6 are ATPases, and the licensing reaction is dependent on hydrolysable ATP38. ATP hydrolysis could plausibly provide the energy to break open the hexameric ring of Mcm2-7 to allow it to reform around DNA. Cdt1 can directly bind components of the Mcm2-7 hexamer43-45, and so the role of Cdt1 might be to recruit Mcm2-7 to the DNA. Cdc6 must already be bound to chromatin before Cdt1 can join the complex in an active form, consistent with Cdt1 having a role in recruiting Mcm2-7 to a clamp-loader that comprises ORC and Cdc646. Geminin, a major inhibitor of the licensing system in metazoans (see below) binds tightly to Cdt147,48, potentially blocking its ability to bind Mcm2-749. As both ORC and the Mcm2-7 hexamer are asymmetric complexes, it is possible that the orientation of ORC on DNA determines the orientation that Mcm2-7 is loaded onto DNA. If, as discussed above, initiation requires two back-to-back Mcm2-7 complexes, it might then be necessary to have two ORC-Cdc6-Cdt1 complexes in different orientations on the DNA to achieve this (FIG. 4d). Alternatively, Mcm2-7 might be loaded as preformed double-hexamers.
The components of the pre-RC may also have functions other than supporting origin licensing. Several lines of evidence indicate that ORC has an important role in generating transcriptionally silent chromatin in a process that is independent of its role in DNA replication50. Several reports also suggest that Cdc6 is involved in the activation of checkpoint pathways within S phase or in the progression from G2 into mitosis34,51,52.
Regulation of the Licensing System
To prevent the possibility of replicated origins becoming re-licensed during S phase, it is important that the ability to license new replication origins is down-regulated before entry into S phase. Because ORC, Cdc6 and Cdt1 are required for the loading of Mcm2-7 onto DNA, but are not required for the continued association of Mcm2-7 on DNA, down-regulation of their activity at the end of G1 is an effective way of preventing the re-licensing of replicated DNA.
Regulation of licensing in yeast
Early work in fission yeast (Schizosaccharomyces pombe) showed that continued activity of cyclin-dependent kinases (CDKs; see BOX 2) in S phase and G2 is necessary to prevent re-replication of DNA. Down-regulation of either Cdk153 or the mitotic cyclin Cdc1354 in G2 permitted extensive re-replication of DNA in a single cell cycle. This implied that re-loading of Mcm2-7 occurs in G2 cells if CDK activity is ablated. Similar experiments in budding yeast (Saccharomyces cerevisiae) showed that pre-RC assembly (origin licensing) is inhibited by Cdk1 activity55. This suggested an intellectually satisfying model where low CDK level at the beginning of the cell cycle (late mitosis and early G1) permit origin licensing whilst rising CDK levels at the end of G1 both prevent further licensing and at the same time promote the initiation of replication56.
In yeasts, CDKs appear to have multiple redundant effects on preventing re-licensing of replicated DNA, targeting all of the components of the licensing system3,4. One of the major substrates of CDK regulation in S. cerevisiae is Cdc6 (or the S. pombe homologue Cdc18), which is targeted for degradation following phosphorylation by CDKs at the G1-S phase transition57-59. Cdc6/Cdc18 levels are also regulated at the transcriptional level to give maximum expression in late mitosis and G13,4. In addition, the ability of ORC to support re-replication is directly inhibited by CDK phosphorylation60,61. In both fission and budding yeast, CDKs are directly recruited to ORC bound to origin DNA, and this helps maintain ORC in an inactive state during S phase and G262,63. CDKs can also bind directly to Cdc6, which may also contribute to the prevention of re-replication64.
In S. pombe, Cdt1 levels peak in late mitosis and early G1 and may be controlled by CDK-dependent transcription and proteolysis in a manner similar to Cdc6/Cdc18. This is likely to be important in preventing re-replication of DNA, since overexpression of Cdt1 enhances re-replication induced by overexpression of SpCdc18/Cdc665. In S. cerevisiae, CDKs promote the nuclear export of Cdt1 and Mcm2-7 during S phase, G2 and early mitosis, thus preventing them from gaining access to chromosomal DNA66,67. Experiments in S. cerevisiae showed that in order for significant re-replication to occur, all these different CDK-dependent controls must be inactivated. Partial over-replication was only seen when unphosphorylatable mutants of orc2 and orc6 were combined with non-degradable Cdc6 and constitutively nuclear Mcm2-7 and Cdt160. These experiments suggest that in yeasts, direct inhibition of components of the licensing system by CDKs prevents the relicensing of replicated DNA.
Regulation of licensing in metazoans
There is evidence that CDK levels are also important for regulating the licensing system in metazoans4. For example, fluctuations of cyclin E levels are required to drive endoreduplication cycles in Drosophila68,69. However, the role of CDKs in inhibiting the licensing system in metazoans is less clear-cut than it is in yeasts (Fig 5B), and it seem likely that CDK levels only indirectly affect the activity of the licensing system. ORC from mammalian somatic cells is a labile complex. The Orc1 subunit is loosely associated with the Orc2, 3, 4, 5 subcomplex and is targeted for degradation in S phase by an SCF dependent polyubiquitination reaction that may depend on CDK activity70-72. Alternatively, in some cell lines, Orc1 appears to be stable in S phase but is mono-ubiquitinated and/or phosphorylated as S phase progresses73,74. It has been suggested that this Orc1 cycle contributes to the regulation of the licensing system, but experimental evidence for this is still lacking. Unlike the situation in yeast, metazoan Cdc6 appears to be bound to chromatin throughout much of the cell cycle, including G234,75,76. Excess Cdc6 is exported out of the nucleus during S phase as a consequence of CDK activity, but there is no evidence that this plays a part in preventing re-replication of DNA77-80. Only in mitosis is there clear evidence in metazoans of a direct inhibitory effect of high CDK levels on the licensing system47,81.
Figure 5.
Cell cycle regulation of the licensing system in yeasts and metazoans.
The activity of components of the licensing system during the cell cycle of yeasts (A) and metazoans (B) is shown. In the lower part of each figure, the licensing system is shown active (green) only in G1. In yeasts, licensing is inhibited at other times by CDKs (blue), whilst in metazoans, licensing is inhibited in S phase and G2 by geminin (orange) and in mitosis by a combination of geminin and CDKs (purple). Above this, the activity of different pre-RC components (ORC, Cdc6, Cdt1 and Mcm2-7) is shown: green for active, red for inhibited.
Instead, the major route by which metazoans prevent licensing during S phase and G2 is by downregulation of Cdt1 activity. This is brought about both by degradation of the Cdt1 protein and by activation of a Cdt1 inhibitory protein called geminin. Cdt1 is degraded at the end of G1 and early S phase (Fig. 5b) in a process that depends on the activity of SCF-class ubiquitin ligases48,82-86. In human cells, a Skp2-containing SCF interacts with Cdt1, dependent on prior phosphorylation of Cdt1 by Cdk2 or Cdk485,87-89. CDK-dependent degradation of Cdt1 has also been observed in Xenopus90. There are, however, Skp2-independent pathways that can still degrade Cdt1, because mutants of Cdt1 that are not phosphorylated by CDK and do not interact with Skp2 are still degraded normally at the onset of S phase (ref91 and D. Takeda and A. Dutta, unpublished results). In Caenorhabditis elegans, a related CUL-4 ubiquitin ligase is required to downregulate Cdt1 levels at the end of G186. Loss of CUL-4 leads to Cdt1-dependent re-licensing and replication86. Similarly, overexpression of Cdt1 in human, Drosophila or Xenopus led to significant re-replication, an effect that was also enhanced by co-expression of Cdc679,90-94. In Xenopus, simultaneous loss of geminin and stabilisation of Cdt1 is sufficient to cause extensive re-licensing and re-replication92.
In addition to being degraded during S phase, Cdt1 is stabilised during G2 and M by binding to geminin, which appears to protect Cdt1 from ubiquitin-mediated degradation81. This stabilisation by geminin allows Cdt1 levels to build up in an inactive form, ready for its function in late mitosis. If only geminin-bound Cdt1 is protected from degradation, this ensures that the quantity of Cdt1 does not exceed the amount that can be inhibited by geminin.
Geminin inhibits Cdt1 by binding to it and provides the major inhibitor of licensing during S phase and G247,48,84,95. Geminin activity is down-regulated in late mitosis, either through proteolysis95 or by posttranslational inactivation such as CDK-dependent ubiquitination84,96, and this allows activation of the licensing system as cells exit from metaphase. At the end of G1, geminin is then stabilised and/or re-activated, thus preventing any further licensing. The importance of geminin in preventing re-replication is demonstrated by the observation that the loss of geminin in Drosophila or human tissue culture cells leads to extensive re-replication of DNA83,97-99. Geminin has been reported to be a regulator of transcription factors, and so misregulation of transcription could contribute to the re-replication100,101. However, overexpression of Cdt1 can by itself stimulate re-replication, suggesting that Cdt1 is the relevant target of geminin for the prevention of re-replication79,90-94. Taken together, these results suggest that in most metazoan cells the major feature preventing the re-licensing of replicated DNA is the absence of Cdt1 activity, which can occur despite any potential CDK-dependent inhibitory phosphorylation of ORC, Cdc6 and Mcm2-7 proteins.
Several structural studies have recently illuminated how geminin might inhibit Cdt1 function. Geminin forms an elongated dimer through the interaction of its coiled-coil domains49,102-104. The crystal structure of the central region of Cdt1 complexed with a fragment of geminin has been solved49 (Fig 6). One molecule of Cdt1 interacts asymmetrically with the geminin dimer forming contacts between highly conserved amino acids in helix H1 and surface residues on the coiled-coil domain of both subunits of geminin. A second unstructured loop immediately N-terminal to the coiled-coil domain of geminin makes a second contact with Cdt1 that is essential for the inhibition of DNA replication. A structure-function study carried out with full length Cdt1 indicated that the N-terminal 160 residues of Cdt1 that were not present in the co-crystal also interact with the N-terminal unstructured loop of geminin103. In addition, an electron microscopic study indicated that geminin dimers may associate with each other to form tetramers105. These studies indicate that the geminin-Cdt1 interaction might interfere with Cdt1-Mcm2-7 interaction by steric hindrance and thus interfere with the loading of Mcm2-7.
Figure 6.
Structure of a geminin : Cdt1 complex.
Structure of a complex between the central regions of geminin (tGeminin, in green) and Cdt1 (tCdt1, in orange). The N-terminal ten residues are structured in the tGeminin monomer that binds to tCdt1 but are disordered in the other tGeminin monomer, indicating that this region might undergo induced folding after binding to tCdt1. Comparison with the structure of geminin on its own103 suggests that the kink at the C terminal end of geminin may be a distortion due to a packing interaction. Reproduced from reference49.
Checkpoint pathways and re-replication
When the licensing system is artificially misregulated to allow cells to inappropriately re-relicense their DNA, re-replication of DNA is observed60,79,92,97-99. This indicates that replication licensing is the major system preventing re-replication of DNA in a single cell cycle. However, when re-replication occurs, various checkpoint pathways are activated. Checkpoint kinases – comprising the upstream ATM and ATR kinases and the Chk1 and Chk2 downstream kinases – are typically activated when DNA is damaged or if problems are detected during progression through the cell cycle. Although the precise mechanism is not understood, it seems likely that checkpoint activation in response to misregulation of the licensing system is a consequence of some structural problem occurring during re-replication (such as a fork running into the back of another fork), rather than re-licensing itself106.
Overexpression of Cdt1 or depletion of geminin (which induces re-replication) activates ATM/ATR-Chk1/Chk2 dependent checkpoint pathways that lead to an arrest of the cell-cycle and/or apoptosis79,92,97-99. Cells overexpressing Cdt1 and Cdc6 utilize p53 as the major effector downstream from the ATM/ATR checkpoint kinases, while cells depleted of geminin utilize the checkpoint kinase Chk1 as the major effector. The reason for this differential use of effectors is unclear but could suggest a role of either Cdc6 or of geminin in determining how the checkpoint kinases are directed to their substrates34. Consistent with this idea, Cdc6 has been reported to be involved in checkpoint activation independent of it its role in licensing34,51,52,99. The activation of p53, of course, leads to apoptosis in addition to the arrest of the cell-cycle. The cells also undergo apoptosis (but in a p53-independent reaction) if the Chk1-dependent G2 arrest seen following geminin depletion is over-ridden. Thus, metazoans are spared the ill consequences of re-replication by at least two back-up pathways: checkpoint induced cell-cycle inhibition and/or apoptosis.
In addition to being degraded during S phase, Cdt1 is also degraded in response to DNA damaging agents. Treatment of human or Drosophila tissue culture cells with ionising radiation led to an almost complete loss of Cdt1 protein107. This Cdt1 degradation occurred in cells during G1 and depended on the CUL4 ubiquitin ligase, but not on ATR or ATM-dependent checkpoint pathways. Human tissue culture cells treated with UV also undergo Cdt1 degradation108. In contrast to the findings with ionising radiation, the sensitivity of UV-induced Cdt1 degradation to various checkpoint kinase inhibitors suggested that the ATR kinase activity is involved. These results suggest that downregulation of the licensing system is an additional route by which cells can prevent DNA replication in the face of DNA damage.
Co-ordination of Licensing and Proliferative Potential
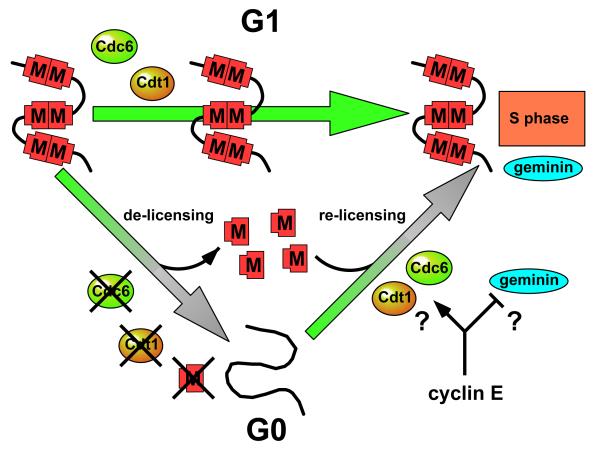
When cells withdraw from the cell cycle, either temporarily into G0 (quiescence), or permanently as a consequence of terminal differentiation or senescence, Cdc6, Cdt1 and Mcm2-7 are down-regulated and degraded, leaving only ORC at the origins (FIG. 7; reviewed in Ref. 3). At first sight it seems surprising that unreplicated DNA becomes unlicensed on exit from the cell cycle. One possibility is that this provides a barrier to prevent non-proliferating cells from re-entering the cell division cycle. Indeed, an early change that is seen in pre-malignant lesions is the presence of Mcm2-7 in cells which do not normally express them109. As the loss of the licensed state represents a profound change to the replicative potential of the cell as it exits from G1 into G0, it provides an attractive functional definition of exit from the proliferative state3,110.
Figure 7.
The possible role of cyclin E in promoting re-licensing of DNA on exit from G0. Mcm2-7 (red shapes ‘M’) are shown on a small segment of chromosomal DNA in a cell either passing directly through G1 (top arrow) or passing into G0 and then into S phase (lower arrows). During passage through G1, Mcm2-7 loaded onto DNA in late mitosis remains stably bound to DNA. Geminin is activated in late G1, just prior to entry into S phase. From G1, cells also have the option of entering quiescence (the G0 state). On entry into G0 from G1, the DNA is de-licensed and Mcm2-7, Cdc6 and Cdt1 are degraded. This might provide a barrier that prevents the inappropriate proliferation of these cells. On exit from G0 into S phase, quiescent cells must relicense their DNA, and cyclin E is required for this re-licensing. Cyclin E might be required to drive the re-synthesis or activation of Mcm2-7, Cdc6 or Cdt1, or cyclin E might be required to temporarily suppress geminin activity, which is also resynthesized prior to entry into S phase.
The transcription of CDC6, CDT1 and MCM2-7 all appear to be responsive to the E2F family of transcription factors which are activated on exit from G0111-113. E2F-driven transcription is normally repressed by members of the Rb family, which is in turn repressed by CDK-dependent phosphorylation (primarily by cyclin E-Cdk2 and cyclin D-Cdk4/6). Cyclin E knockout embryos (cyclin E1-/- cyclin E2-/-) show virtually normal proliferation and developmental capacity. However, they fail to make trophoblast giant cells and megakaryocytes, both cell types that need to endoreduplicate to attain a >2N DNA content114,115. Mouse embryo fibroblasts (MEFs) derived from the cyclin E knockout embryos were unable to re-enter S phase after being driven into G0 by serum starvation or contact inhibition. In particular, these quiescent cyclin E knockout cells failed to load Mcm2 onto chromatin despite the normal loading of ORC and Cdc6114,115. These studies indicate that whereas cyclin E is dispensable for licensing to occur after exit from normal mitosis, it is essential for licensing at other times – either on exit from G0 or during endoreduplication cycles. Consistent with this idea, work in Drosophila has also shown a requirement for cyclin E in Mcm2-7 loading during endoreduplication cycles68,69,116.
The picture is further complicated by the observation that Cdk2 knockout mice – which lack the only known kinase partner of cyclin E – are viable, and produce MEFs that can exit normally from serum starvation117,118. Although the precise role of CDKs in activating the licensing system is unclear, it is possible that during normal cell cycle progress, a range of CDKs (including cyclin B-Cdk196) play redundant roles in activating the licensing system. On exit from G0 fewer CDKs may be available – whilst cyclin A-Cdk1 may be able to replace cyclin E-Cdk2 in Cdk2 knockout cells, cyclin A-Cdk2 may not be able to do so in cyclin E knockout cells115.
What is the essential role of cyclin E in licensing during exit from G0? One possibility is that cyclin E is required for the synthesis of Cdt1 (ORC and Cdc6 levels were normal in the cyclin E-/- MEFs). Another possibility is that cyclin E is directly or indirectly required for the activation of one of the pre-RC components when cells exit from G0. Consistent with this suggestion, an in vitro system that supports the replication of nuclei isolated from cells exiting from G0 shows a requirement for cyclin E in loading Mcm2-7 proteins onto chromatin119. One step that might require CDK activity is the inactivation of geminin. On exit from metaphase in Xenopus egg extracts, geminin is inactivated as a consequence of CDK-dependent ubiquitination96. An analogous reaction requiring cyclin E for the inactivation of geminin could be postulated on exit from G0, However, geminin expression is also low in quiescent cells120,121, and this explanation would require geminin levels to rise prior to the completion of licensing when cells re-enter the cell cycle (FIG. 7), a point that has so far not been studied.
As licensing must be complete before cells enter S phase, it is plausible that cells possess a ‘licensing checkpoint’ that blocks entry into S phase until licensing is complete. In support of this idea, primary cells that over-expressed constitutively active geminin arrested in a G1-like state with hypophosphorylated Rb and showing no evidence of attempts to initiate DNA replication122,123. By contrast, a variety of cancer-derived cell lines progressed into an unsuccessful S phase and ultimately underwent apoptosis. The nature of the ‘licensing checkpoint’ seen in primary cells is currently unclear, but may provide a useful route by which to induce cancer-specific cell-killing.
Conclusions and Perspectives
Over the past few years, the basic mechanisms that ensure once-per-cycle replication of chromosomal DNA have been elucidated. Although the regulation seems to differ between yeasts and metazoans, in both groups it centres upon restricting the rebinding of Mcm2-7 to replicated DNA, thus conforming to the original licensing factor hypothesis. In yeasts, the re-licensing of replicated DNA is prevented by high CDK activity which represses different pre-RC components late in the cell cycle. In contrast, metazoans prevent the re-licensing of replicated DNA largely by inhibiting Cdt1 activity, which in turn may largely be due to the presence of active geminin. With the understanding of these basic pathways in place, the time is now ripe to address a number of further questions. It will be of great interest to understand the biochemical events occurring when Mcm2-7 are loaded onto DNA. Are Mcm2-7 loaded as hexamers or as double hexamers? Are there any features in the DNA sequence or chromatin that direct ORC to specific sites on the chromosomes? What is the relationship between the sites where ORC is bound, the sites where Mcm2-7 is bound, and the sites where replication forks actually initiate? The answers to these questions may also shed light on the ‘Mcm paradox’: why there is such an excess of Mcm2-7 over replication origins. It will also be of great interest to understand how the licensing system is regulated when cells enter and exit quiescence. What roles do CDKs play in reactivating the licensing system on exit from quiescence? Are there checkpoint or feedback systems monitoring the success of this process? Is the unlicensed state of quiescent cells important for preventing illegitimate re-entry into the cell cycle? Do cancer cells acquire errors in these checkpoint and feedback pathways, and do such errors lead to genomic instability? Answering these and other questions will help to integrate our knowledge of cell cycle control with the biophysical events occurring during the process of DNA replication and with the genomic instability that is the hallmark of many cancers.
Supplementary Material
Acknowledgements
Thanks to Margret Michalski-Blow for assembling the Glossary. This work was supported by Cancer Research UK grant C303/A3135 and NIH grants CA60499 and CA89406.
References
- 1.Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 3.Blow JJ, Hodgson B. Replication licensing - defining the proliferative state? Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishitani H, Lygerou Z. DNA replication licensing. Frontiers in Bioscience. 2004;9:2115–2132. doi: 10.2741/1315. [DOI] [PubMed] [Google Scholar]
- 5.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 6.Prokhorova TA, Blow JJ. Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem. 2000;275:2491–2498. doi: 10.1074/jbc.275.4.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 8.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S- cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 9.Labib K, Tercero JA, Diffley JFX. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 10.Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–76. doi: 10.1038/sj.emboj.7600369. Epub 2004 Aug 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shechter D, Ying CY, Gautier J. DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J Biol Chem. 2004;279:45586–93. doi: 10.1074/jbc.M407772200. Epub 2004 Aug 23. [DOI] [PubMed] [Google Scholar]
- 12.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl. Acad. Sci. USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shechter DF, Ying CY, Gautier J. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J. Biol. Chem. 2000;275:15049–15059. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher RJ, et al. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nature Structural Biology. 2003;10:160–167. doi: 10.1038/nsb893. The crystal structure of an archaeal MCM reveals a dodecameric complex providing a positively-charged central channel capable of accommodating double-stranded DNA.
- 16.Pape T, et al. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. A 3-dimensional reconstruction of an archaeal MCM shows a structure with a central channel, consistent both with the MCM crystal structure of Fletcher et al and the structure of SV40 T antigen (Li et al).
- 17.Li D, et al. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. The crystal structure of SV40 T antigen shows hexamers organised into two tiers that enclose a positively-charged channel capable of accommodating double-stranded DNA.
- 18.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkhart R, et al. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur. J. Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- 20.Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahbubani HM, Chong JP, Chevalier S, Thömmes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards MC, et al. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 23.Ritzi M, et al. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 1998;273:24543–24549. doi: 10.1074/jbc.273.38.24543. [DOI] [PubMed] [Google Scholar]
- 24.Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- 25.Harvey KJ, Newport J. CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol. Cell. Biol. 2003;23:6769–6779. doi: 10.1128/MCB.23.19.6769-6779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danis E, et al. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. Shows that initiation sites in Xenopus egg extracts can be induced by creating a transcription domain, possibly by inducing histone acetylation at that site.
- 27.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 28.Madine MA, Khoo CY, Mills AD, Musahl C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- 29.Krude T, Musahl C, Laskey RA, Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J. Cell Sci. 1996;109:309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- 30.Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskey RA, Madine MA. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4:26–30. doi: 10.1038/sj.embor.embor706. Proposes a solution to the ‘MCM paradox’ whereby Mcm2-7 act at a distance from the replication fork unwinding DNA by a rotary pump mechanism.
- 32.Yankulov K, et al. MCM proteins are associated with RNA polymerase II holoenzyme. Mol. Cell. Biol. 1999;19:6154–6163. doi: 10.1128/mcb.19.9.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dziak R, Leishman D, Radovic M, Tye BK, Yankulov K. Evidence for a role of MCM (mini-chromosome maintenance)5 in transcriptional repression of sub-telomeric and Ty-proximal genes in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:27372–27381. doi: 10.1074/jbc.M301110200. [DOI] [PubMed] [Google Scholar]
- 34.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. Shows that the affinity of Xenopus Cdc6 for chromatin drops once the origin has loaded the first Mcm2-7 hexamers, potentially providing a mechanism to ensure that all origins are licensed.
- 35.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–55. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- 38.Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- 44.Yanagi K, Mizuno T, You Z, Hanaoka F. Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J. Biol. Chem. 2002;277:40871–40880. doi: 10.1074/jbc.M206202200. [DOI] [PubMed] [Google Scholar]
- 45.Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J. Biol. Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- 46.Tsuyama T, Tada S, Watanabe S, Seki M, Enomoto T. Licensing for DNA replication requires a strict sequential assembly of Cdc6 and Cdt1 onto chromatin in Xenopus egg extracts. Nucl. Acids Res. 2005;33:765–775. doi: 10.1093/nar/gki226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wohlschlegel JA, et al. Inhibition of eukaryotic replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 49.Lee C, et al. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature. 2004;430:913–917. doi: 10.1038/nature02813. Shows the crystal structure of a complex between portions of Cdt1 and geminin, and suggests how the binding of geminin hinders the association of Mcm2-7 with the C-terminus of Cdt1.
- 50.Leatherwood J, Vas A. Connecting ORC and heterochromatin: why? Cell Cycle. 2003;2:573–575. [PubMed] [Google Scholar]
- 51.Clay-Farrace L, Pelizon C, Santamaria D, Pines J, Laskey RA. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J. 2003;22:704–712. doi: 10.1093/emboj/cdg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murakami H, Yanow SK, Griffiths D, Nakanishi M, Nurse P. Maintenance of replication forks and the S-phase checkpoint by Cdc18p and Orp1p. Nat. Cell Biol. 2002;4:384–388. doi: 10.1038/ncb789. [DOI] [PubMed] [Google Scholar]
- 53.Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature. 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- 54.Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 55.Dahmann C, Diffley J, Nasmyth K. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 56.Diffley JF. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes and Development. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 57.Jallepalli PV, Brown GW, MuziFalconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65(cdc18) by CDK phosphorylation. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol. Biol. Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drury LS, Perkins G, Diffley JFX. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- 60.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 61.Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wuarin J, Buck V, Nurse P, Millar JB. Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell. 2002;111:419–431. doi: 10.1016/s0092-8674(02)01042-5. [DOI] [PubMed] [Google Scholar]
- 63.Wilmes GM, et al. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. Shows that in S. cerevisiae, Orc6 can bind the Clb5 cyclin, and that blocking the interaction permits re-replication of DNA.
- 64.Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–23. doi: 10.1038/nature03024. Epub 2004 Oct 20. [DOI] [PubMed] [Google Scholar]
- 65.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 66.Labib K, Diffley JFX, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- 68.Follette PJ, Duronio RJ, O’Farrell PH. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol. 1998;8:235–238. doi: 10.1016/s0960-9822(98)70089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su TT, O’Farrell PH. Chromosome association of minichromosome maintenance proteins in Drosophila endoreplication cycles. J. Cell Biol. 1998;140:451–460. doi: 10.1083/jcb.140.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J. Biol. Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 71.Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J. Biol. Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 72.Mendez J, et al. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- 73.Li CJ, DePamphilis ML. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 2002;22:105–116. doi: 10.1128/MCB.22.1.105-116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Molecular and Cellular Biology. 2004;24:5875–5886. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coverley D, Pelizon C, Trewick S, Laskey RA. Chromatin-bound Cdc6 persists in S and G(2) phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 2000;113:1929–1938. doi: 10.1242/jcs.113.11.1929. [DOI] [PubMed] [Google Scholar]
- 76.Mendez J, Stillman B. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saha P, et al. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by Cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaziri C, et al. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. Shows that overexpression of Cdt1 and Cdc6 can cause extensive re-replication in human cells, and that as a consequence, a ATM/ATR and p53-dependent checkpoint pathways are activated.
- 80.Alexandrow MG, Hamlin JL. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol. 2004;24:1614–1627. doi: 10.1128/MCB.24.4.1614-1627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ballabeni A, et al. Human Geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. Describes the effect of geminin in protecting Cdt1 from proteasome-mediated degradation on exit from mitosis, and shows that CDK inhibition during mitosis is sufficient to promote premature licensing.
- 82.Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- 83.Quinn LM, Herr A, McGarry TJ, Richardson H. The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 2001;15:2741–2754. doi: 10.1101/gad.916201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hodgson B, Li A, Tada S, Blow JJ. Geminin becomes activated as an inhibitor of Cdt1/RLF-B following nuclear import. Curr. Biol. 2002;12:678–683. doi: 10.1016/s0960-9822(02)00778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. Provides evidence that down-regulation of Cdt1 late in the cell cycle involves SCF-dependent ubiquitination of phosphorylated Cdt1.
- 86.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. Shows that the C. elegans CUL-4 ubiquitin ligase is responsible for down-regulating Cdt1 late in the cell cycle, and that failure of this process can cause re-replication of DNA.
- 87.Liu E, Li X, Yan F, Zhao Q, Wu X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 2004;279:17283–17288. doi: 10.1074/jbc.C300549200. [DOI] [PubMed] [Google Scholar]
- 88.Nishitani H, Lygerou Z, Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 2004;279:30807–30816. doi: 10.1074/jbc.M312644200. [DOI] [PubMed] [Google Scholar]
- 89.Sugimoto N, et al. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 2004;279:19691–19697. doi: 10.1074/jbc.M313175200. [DOI] [PubMed] [Google Scholar]
- 90.Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005;19:114–26. doi: 10.1101/gad.1255805. Epub 2004 Dec 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomer M, May NR, Aggarwal BD, Kwok G, Calvi BR. Drosophila double-parked is sufficient to induce re-replication during development and is regulated by cyclin E/CDK2. Development. 2004;131:4807–18. doi: 10.1242/dev.01348. Epub 2004 Sep 01. [DOI] [PubMed] [Google Scholar]
- 92.Li A, Blow JJ. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 2005;24:395–404. doi: 10.1038/sj.emboj.7600520. Shows that in Xenopus, activation of the licensing system on exit from metaphase depends on the ubiquitination of geminin which renders it incapable of inhibiting Cdt1.
- 93.Maiorano D, Krasinska L, Lutzmann M, Mechali M. Recombinant cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr Biol. 2005;15:146–53. doi: 10.1016/j.cub.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 94.Yoshida K, Takisawa H, Kubota Y. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells. 2005;10:63–73. doi: 10.1111/j.1365-2443.2005.00815.x. [DOI] [PubMed] [Google Scholar]
- 95.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 96.Li A, Blow JJ. Non-proteolytic inactivation of geminin requires CDK-dependent ubiquitination. Nature Cell Biology. 2004;6:260–267. doi: 10.1038/ncb1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mihaylov IS, et al. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 2002;22:1868–1880. doi: 10.1128/MCB.22.6.1868-1880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Melixetian M, et al. Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 2004;165:473–482. doi: 10.1083/jcb.200403106. This paper, along with that of Zhu et al, shows that in human cells, ablation of geminin is sufficient to induce re-replication of DNA, and that as a consequence p53-independent checkpoint pathways are activated that block subsequent entry into mitosis.
- 99.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. See Melixetian et al.
- 100.Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 101.Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 102.Benjamin JM, Torke SJ, Demeler B, McGarry TJ. Geminin has dimerization, Cdt1-binding, and destruction domains that are required for biological activity. J. Biol. Chem. 2004;279:45957–45968. doi: 10.1074/jbc.M407726200. [DOI] [PubMed] [Google Scholar]
- 103.Saxena S, et al. A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol. Cell. 2004;15:245–258. doi: 10.1016/j.molcel.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 104.Thepaut M, et al. Crystal structure of the coiled-coil dimerization motif of geminin: structural and functional insights on DNA replication regulation. J. Mol. Biol. 2004;342:275–287. doi: 10.1016/j.jmb.2004.06.065. [DOI] [PubMed] [Google Scholar]
- 105.Okorokov AL, et al. Molecular structure of human geminin. Nature Structural and Molecular Biology. 2004;11:1021–1022. doi: 10.1038/nsmb835. [DOI] [PubMed] [Google Scholar]
- 106.Green BM, Li JJ. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol Biol Cell. 2005;16:421–32. doi: 10.1091/mbc.E04-09-0833. Epub 2004 Nov 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. Shows that ionising radiation induces the degradation of Cdt1 in a process depending on the CUL-4-ubiquitin ligase and the COP9-signalosome.
- 108.Kondo T, et al. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J. Biol. Chem. 2004;279:27315–27319. doi: 10.1074/jbc.M314023200. [DOI] [PubMed] [Google Scholar]
- 109.Williams GH, et al. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc. Natl. Acad. Sci. USA. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stoeber K, et al. DNA replication licensing and human cell proliferation. J. Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- 111.Leone G, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ohtani K, Tsujimoto A, Ikeda M, Nakamura M. Regulation of cell growth-dependent expression of mammalian CDC6 gene by the cell cycle transcription factor E2F. Oncogene. 1998;17:1777–1785. doi: 10.1038/sj.onc.1202105. [DOI] [PubMed] [Google Scholar]
- 113.Ohtani K, et al. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene. 1999;18:2299–2309. doi: 10.1038/sj.onc.1202544. [DOI] [PubMed] [Google Scholar]
- 114.Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. Shows two specific defects in mice lacking both cyclins E1 and E2: a failure to form various endoreduplicated cell types and an inability of cells to relicense DNA during progression from G0 into S phase.
- 115.Parisi T, et al. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Su TT, O’Farrell PH. Chromosome association of minichromosome maintenance proteins in Drosophila mitotic cycles. J. Cell Biol. 1997;139:13–21. doi: 10.1083/jcb.139.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ortega S, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 118.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr. Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 119.Coverley D, Laman H, Laskey RA. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 2002;4:523–528. doi: 10.1038/ncb813. [DOI] [PubMed] [Google Scholar]
- 120.Wohlschlegel JA, Kutok JL, Weng AP, Dutta A. Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am. J. Pathol. 2002;161:267–273. doi: 10.1016/S0002-9440(10)64178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xouri G, et al. Cdt1 and geminin are down-regulated upon cell cycle exit and are over-expressed in cancer-derived cell lines. Eur. J. Biochem. 2004;271:3368–3378. doi: 10.1111/j.1432-1033.2004.04271.x. [DOI] [PubMed] [Google Scholar]
- 122.Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Feng D, Tu Z, Wu W, Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Research. 2003;63:7356–7364. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.