Abstract
Cell surface polysaccharides have an established role as virulence factors in human bacterial pathogens. Less documented are the biosynthesis and biological functions of surface polysaccharides in beneficial bacteria. We identified a gene cluster that encodes the enzymes and regulatory and transporter proteins for the different steps in the biosynthesis of extracellular polysaccharides (EPS) of the well-documented probiotic strain Lactobacillus rhamnosus GG. Subsequent mutation of the welE gene, encoding the priming glycosyltransferase within this cluster, and comparative phenotypic analyses of wild-type versus mutant strains confirmed the specific function of this gene cluster in the biosynthesis of high-molecular-weight, galactose-rich heteropolymeric EPS molecules. The phenotypic analyses included monomer composition determination, estimation of the polymer length of the isolated EPS molecules, and single-molecule force spectroscopy of the surface polysaccharides. Further characterization of the welE mutant also showed that deprivation of these long, galactose-rich EPS molecules results in an increased adherence and biofilm formation capacity of L. rhamnosus GG, possibly because of less shielding of adhesins such as fimbria-like structures.
Bacterial surface polysaccharides are considered to be key macromolecules in determining microbe-host interactions, as they display a high degree of variety and diversity among bacterial species in terms of composition, monomer linkages, branching degree, polymer size, production level, etc. (24, 46). Since most bacteria contain more than one type of surface polysaccharides, such as lipopolysaccharides (O antigens), capsular polysaccharides (CPS), exopolysaccharides (EPS), and/or glycan chains as part of glycoproteins, the elucidation of their exact role is complex. Nevertheless, surface polysaccharides are now known to exert important functions at several stages during pathogenesis, including tissue adherence, biofilm formation, and evasion of host defenses such as phagocytosis (9, 24, 33). In addition to their role in pathogens, an important biological role for CPS and glycoproteins has also recently been shown in colonization of the gut by bacteria of the genus Bacteroides (10, 34).
Conversely, the role of surface polysaccharides in probiotic-host interactions has not yet been studied in great detail. A probiotic bacterium is defined as “a live microorganism that, when administered or ingested in adequate amounts, confers a health benefit on the host” (18). Members of the genus Lactobacillus are commonly studied for their health-promoting capacities (26, 31, 37). As polysaccharides display a high diversity among lactobacilli, they are thought to be involved in determining strain-specific properties important for probiotic action, such as adhesion, stress resistance, and interactions with specific receptors and effectors of the host defense system (13, 56). Moreover, these EPS molecules are of interest in the dairy industry for conferring textural and rheological properties to fermented products such as yogurt and soft cheese (56). Nevertheless, detailed genetic and functional studies of EPS molecules of lactobacilli are currently limited (26, 56).
Lactobacillus rhamnosus GG (ATCC 53103) is one of the probiotic strains with the largest number of proven health benefits (15). Several clinical trials have reported that L. rhamnosus GG can prevent and relieve certain types of diarrhea (22) and atopic disease (25) and reduce inflammation in some milder states of inflammatory bowel diseases (60). However, the cell surface factors or specific characteristics of L. rhamnosus GG that underlie these health benefits are largely unknown.
We recently showed by single-molecule force spectroscopy (SMFS) with specific lectin tips that the cell surface of L. rhamnosus GG wild-type cells contains two major types of cell wall-associated polysaccharides (CW-PS) (21). The longest and most abundant polysaccharides are galactose-rich and seem to correspond with the EPS molecules of L. rhamnosus GG, which were previously structurally identified by Landersjö et al. (27) using nuclear magnetic resonance spectroscopy. Additionally, shorter, yet-uncharacterized glucose-rich polysaccharides are present on the L. rhamnosus GG surface (21). In the current study, we describe the identification and annotation of the L. rhamnosus GG gene cluster that encodes the enzymes and transporter and regulatory proteins involved in the biosynthesis of long, galactose-rich EPS molecules. This was experimentally confirmed by the construction of a knockout mutant of the corresponding priming glycosyltransferase and subsequent characterization of the surface polysaccharides of wild-type and mutant strains. We also studied the specific role of these EPS molecules in adherence to mucus and gut epithelial cells and in biofilm formation by L. rhamnosus GG.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. rhamnosus GG and its derivatives were grown at 37°C in MRS (Difco) or Lactobacilli AOAC medium (Difco) in nonshaking conditions (Table 1). Escherichia coli cells, used as cloning hosts, were grown in Luria-Bertani medium with aeration at 37°C (48). When appropriate, antibiotics (Sigma-Aldrich) were used at the following concentrations: tetracycline (Tc) at 10 μg/ml, ampicillin (Ap) at 100 μg/ml, and erythromycin (Ery) at 10 μg/ml for L. rhamnosus GG and at 100 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZ ΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) supE44 λ−thi-1 girA96 relA1 | Gibco-BRL |
| LE392 | hsdR574 (rK− mK+) supE44 supF58 lacY1 [or Δ(lac1ZY)6] galK2 galT22 metB1 trpR55 λ−mcrB | ATCC 33572 |
| L. rhamnosus GG strains | ||
| Wild type | Human isolate; wild-type strain GG | ATCC 53103 |
| CMPG5351 | welE knockout mutant of strain GG; welE::tet(M) | This work |
| CMPG5354 | CMPG5351 complemented by genomic integration of pCMPG5353 | This work |
| Plasmids | ||
| pFAJ5301 | Cloning vector, pUC18 derivative, Eryr | 30 |
| pMD5057 | Plasmid from L. plantarum 5057 containing a tet(M) marker | 11 |
| pEM40 | pUC19E-derived integration vector (attB located at the 3′ end of tRNALeu locus) containing a 1.6-kb int-attP cassette of phage A2, Apr Eryr | 2 |
| pCMPG5351 | pFAJ5301 derivative used to inactivate welE gene by insertion of a tet(M) marker via double homologous recombination (for details, see text) | This work |
| pCMPG5353 | pEM40 derivative containing a fragment with the welE gene in the Ecl136II/EcoRI site (amplified with primers Pro-0722 and Pro-0723), Apr Eryr | This work |
DNA manipulations.
Routine molecular biology techniques were performed according to standard procedures (48). Restriction and modification enzymes (from New England Biolabs or Roche) were used as recommended by the manufacturer. Plasmid DNA was prepared from E. coli cells with Qiagen miniprep kits. Chromosomal DNA and plasmid DNA were isolated from L. rhamnosus GG as previously described (12).
Sequencing and annotation of the putative EPS biosynthetic cluster.
The wzb gene of L. rhamnosus GG (accession no. EF690379) was identified as previously described (32), starting from a PCR with primers based on the published wzb sequence of the closely related strain L. rhamnosus ATCC 9595 (43). This gene provided the starting point for the sequencing of the remainder of the EPS gene cluster of L. rhamnosus GG. Primers Pro-0224, Pro-0225, Pro-249, and Pro-250 (see Table S1 in the supplemental material) were designed for chromosome walking. DNA sequencing was performed using the chain termination dideoxynucleoside triphosphate method (48) (BigDye Terminator V3.1 CycleSequencing kit, ABI 3100-Avant Genetic Analyzer; Applied Biosystems). This yielded a 2.8-kb fragment including the sequences for the wzb gene and the wzr gene. This information was used to develop a digoxigenin-labeled probe with PCR primers Pro-0300 and Pro-0278 (see Table S1 in the supplemental material) based on an internal fragment of the wzr gene to screen a lambda phage genomic library of L. rhamnosus GG (λCMPG5317). This library was made with the Packagene lambda DNA packaging system (Promega) with E. coli LE392 as a host and phage EMBL3 containing inserts of ca. 15 to 20 kb of L. rhamnosus GG genomic DNA. Southern hybridization resulted in the identification of several phages containing a fragment that hybridized with the wzr probe. One phage that contained a ∼15-kb fragment upstream of wzr was selected. Phage DNA was prepared with the Qiagen lambda DNA extraction minikit and subsequently digested with SalI, BamHI, KpnI, and HindIII. The restriction fragments obtained were subcloned and sequenced. This yielded the DNA sequence of a ca. 15-kb fragment containing the genes encoding glycosyltransferases (welJIHGFE), proteins involved in dTDP-rhamnose precursor biosynthesis (rmlACB), the oligosaccharide transporter (wzx), and polymerase (wzy). To complete the EPS gene cluster, primers Pro-0708, Pro-0839, and Pro-0989 (see Table S1 in the supplemental material) were subsequently used to sequence wzd-wze with genomic DNA of L. rhamnosus GG as a template. Finally, all fragments were assembled in a ca. 20-kb contig containing the DNA sequence for the putative EPS gene cluster of L. rhamnosus GG. The Entrez nucleotide database was screened for similarities using BLASTx (1), and alignment of overlapping fragments was performed with the VectorNTI Advance 10 ContigExpress software (Informax, Oxford, United Kingdom). Annotation of the open reading frames (ORFs) was performed with the Vector NTI Advance 10 software (Informax, Oxford, United Kingdom) and the Sequin sequence submission software. Membrane-spanning regions of translated gene products were predicted using the TMpred program (23). Additionally, the Pfam database (19) was searched for conserved motifs in the translated gene products. The genes were annotated according to Péant et al. (43). These genes were named based on the bacterial polysaccharide gene nomenclature system (45).
Insertional inactivation of the welE gene (CMPG5351).
A knockout mutant of the welE gene, encoding the priming glycosyltransferase, was constructed by double homologous recombination as previously described (30). DNA fragments containing ca. 1 kb up- and downstream of welE were amplified by PCR with primers Pro-0722 and Pro-0723 (see Table S1 in the supplemental material). These fragments were cloned into the pFAJ5301 suicide vector (30). Subsequently, welE was inactivated by insertion of a tet(M) Tcr cassette from plasmid pMD5057 of Lactobacillus plantarum 5057 (11), amplified with primers Pro-221 and Pro-222 containing EcoRI sites. The tet(M) marker gene fragment was blunted and ligated into the blunted BglII site of the welE gene. The resulting suicide vector pCMPG5351 was transformed to L. rhamnosus GG wild-type cells. At 3 days after the electroporation, transformants were screened for resistance to 10 μg/ml Tc and sensitivity to 10 μg/ml Ery to select for the occurrence of a double homologous recombination event. Confirmation of DNA recombination was obtained by PCR using different primer pairs (Pro-249/Pro-0722 and Pro-0788/Pro-0789) (see Table S1 in the supplemental material). One colony showing the correct insertion pattern of the tet(M) marker and absence of the suicide vector was selected for further analyses and designated CMPG5351. The insertion of the tet(M) marker by double homologous recombination was shown to be 100% stable after 100 generations. Therefore, the mutant was grown in medium without antibiotics for all phenotypic assays, to limit possible interference of Tc with observed phenotypes.
Construction of the complemented strain (CMPG5354).
The welE knockout mutant was complemented by insertion of a wild-type copy including the genuine promoter in a single copy at another locus in the chromosome of L. rhamnosus GG. The self-integrating plasmid pEM40 (2) was used, which integrates at the tRNALeu locus in the L. rhamnosus GG genome (12). The welE gene of L. rhamnosus GG was amplified with primers Pro-0722 and Pro-0723 (see Table S1 in the supplemental material). The resulting PCR fragment was digested with the restriction enzymes Ecl136II/EcoRI and ligated in the Ecl136II/EcoRI sites of pEM40, resulting in plasmid pCMPG5353. pCMPG5353 was electroporated to the welE knockout mutant CMPG5351 for complementation, resulting in strain CMPG5354.
Isolation and characterization of CW-PS.
CW-PS were isolated and quantified as described previously (32, 51). Briefly, total CW-PS was extracted from L. rhamnosus GG cells by mild sonication followed by ethanol precipitation and dialysis against water (6,000- to 8,000-Da dialysis membrane [Spectra/Por, VWR International]). The total amount of carbohydrate was estimated by the phenol-sulfuric acid method (16). The sugar monomer composition of the isolated polysaccharides was determined according to the method of Englyst and Cummings by gas chromatography after hydrolysis and derivatization to alditol acetates (17). β-d-Allose was used as an internal standard, and calibration samples (glucose, galactose, rhamnose, and mannose) containing the expected monosaccharides were included with each set of samples. For an estimation of the molecular weights of the isolated polysaccharides, samples were analyzed by size exclusion chromatography as previously described (6). A volume of 50 μl of EPS sample (1 mg/ml) was injected, and the detection was performed with a refractive index detector. The results were compared using a dextran standard series of 8 × 104, 1.5 × 105, 2.7 × 105, 6.7 × 105, and 1.4 × 106 Da (Sigma-Aldrich). Alternatively, differences in molecular weight profiles of the isolated polysaccharides were compared on 5% polyacrylamide gels (10 by 10 cm) (Hoefer miniVE; GE Healthcare) using Tris-borate-EDTA buffer (50 mM Tris, 13 mM EDTA, 15 mM boric acid). Samples were run for 120 min at 350 V. The polysaccharides were stained with the Pro-Q Emerald 488 glycoprotein stain (Molecular Probes, Invitrogen). Gels were imaged with a Typhoon 9400 variable-mode imager (GE Healthcare) using an excitation laser at 488 nm and an emission filter at 520 nm (band-pass).
TEM and SMFS.
To observe the polysaccharides on the cell surface produced in situ, cell surface structures of L. rhamnosus GG were visualized by negative staining in transmission electron microscopy (TEM) experiments using published methods (29). Additionally, SMFS experiments were performed to detect and localize single polysaccharide molecules on live bacteria as described recently (21). Briefly, atomic force microscopy (AFM) tips functionalized with the carbohydrate-binding lectins concanavalin A (mannose and glucose specificity) and PA-I (galactose specificity) were used to scan the surfaces of wild-type L. rhamnosus GG and mutant CMPG5351 for the localization, abundance, and polymer length of mannose/glucose-rich and galactose-rich polysaccharides, respectively. Adhesion forces and rupture distances between the functionalized AFM tips and surface polysaccharides were recorded as described previously (20).
In vitro biofilm formation and adhesion assays.
In vitro biofilm formation by L. rhamnosus GG was assessed by crystal violet staining after 72 h as previously described (32). Additionally, adhesion to immobilized mucins (pig mucin type II; Sigma-Aldrich) was tested in a microtiter plate-based assay (3). Cells were labeled with N-hydroxysuccinimidobiotin (Sigma-Aldrich), followed by detection with streptavidin conjugated to alkaline phosphatase (Roche) and addition of 4-nitrophenylphosphate (Fluka) as a substrate. The percentage of adherence was calculated based on the optical density at 405 nm of the labeled adherent bacteria compared to the optical density at 405 nm of the original labeled suspension before adhesion. Adhesion was tested each time in eight replicates, and each experiment was repeated at least twice. Adhesion to bovine serum albumin (Sigma-Aldrich) was included to account for nonspecific adhesion. Finally, adhesion to Caco-2 cell line was investigated as previously described (29). The adhesion ratio (percent) was calculated by comparing the number of adherent cells to the cell number of the added original bacterial suspension (107 CFU/ml). All strains were tested in triplicate in three independent experiments.
Nucleotide sequence accession number.
The entire sequence determined in this study was submitted to the NCBI database under GenBank accession number FJ428614.
RESULTS
Identification of the EPS biosynthetic gene cluster of L. rhamnosus GG.
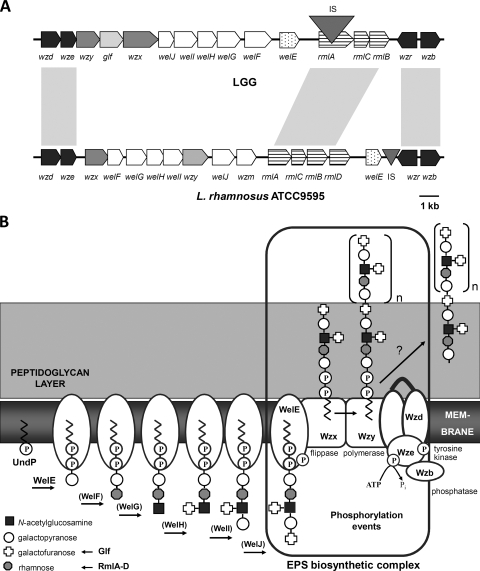
As the genome sequence of L. rhamnosus GG is not publicly available, the data for the EPS gene cluster of the phylogenetically related strain L. rhamnosus ATCC 9595 (43) were used as starting point to determine the DNA sequence of the putative L. rhamnosus GG EPS gene cluster. Following the strategy described in Materials and Methods, we obtained an L. rhamnosus GG EPS gene cluster containing 17 putative ORFs (Fig. 1A), of which 16 putatively encode proteins involved in the biosynthesis of various bacterial polysaccharides and 1 (orf1) encodes a putative transposase (Table 2). All ORFs except wzr are located in the same orientation. The EPS gene cluster of L. rhamnosus GG has a modular organization, which is typical for surface heteropolysaccharides (26). The genes putatively encoding regulatory proteins are located at both extremities of the cluster. The first two ORFs (wzd and wze) encode proteins that have predicted amino acid sequences with homology to the putative Wzd-Wze tyrosine kinase complex of L. rhamnosus ATCC 9595 (43) (Table 2). At the other end of the cluster, the wzb gene is located 145 bp downstream of wzr in reverse orientation. The Wzb protein of L. rhamnosus GG shows 99% amino acid identity with Wzb of L. rhamnosus ATCC 9595, which was recently biochemically characterized as a copper-dependent O-phosphatase (28). A regulatory role in CPS biosynthesis and polymer export has been experimentally demonstrated in streptococci for the Wzb ortholog CpsB (48% amino acid similarity) in combination with the autophosphorylating tyrosine kinase complex Wzd-Wze, designated CpsC-CpsD in streptococci (44 to 55% amino acid similarity) (4, 40-42). Wzr of L. rhamnosus GG shows homology to transcriptional regulators of the LytR-CpsA-Psr family involved in cell envelope-related functions, and it contains a short putative N-terminal cytoplasmic domain and a transmembrane domain forming a signal-anchor. Mutation of cpsA, the ortholog of wzr (encoded proteins show 61% similarity), has been shown to affect CPS production in certain streptococci (8, 40).
FIG. 1.
(A) Organization of the EPS gene cluster of L. rhamnosus GG. The organization of the EPS gene cluster of L. rhamnosus GG is compared by BLASTx analysis with that of the EPS gene cluster of the phylogenetically related strain L. rhamnosus ATCC 9595 (43). Genes encoding similar functions in EPS biosynthesis have a similar gray scaling code. Genes indicated in dark gray encode proteins putatively involved in the regulation of EPS production and polymerization. wzx and wzy (light gray) encode the putative polysaccharide transporter and polymerase, respectively. Genes indicated in white encode the putative glycosyltransferases, with wzm in L. rhamnosus ATCC 9595 encoding a putative pyruvyltransferase. Long stripes indicate genes encoding the proteins involved in the biosynthesis of the dTDP-rhamnose precursor. The lightest gray indicates the glf gene, encoding the UDP-galactopyranose mutase for the biosynthesis of the sugar nucleotide precursor for galactofuranose present in the EPS molecules of L. rhamnosus GG. The triangles indicate insertion sequence elements (IS). The insertion sequence of the L. rhamnosus GG EPS gene cluster is indicated with orf1 present on ISLrh2 in Table 2. Finally, genes sharing high nucleotide identity are linked by light gray boxes. Cutoff values for the displayed degrees of similarity are >88% identity. (B) Schematic representation of the putative steps in EPS biosynthesis by L. rhamnosus GG that are encoded within the EPS gene cluster. First, a phosphogalactosyl residue is transferred from an activated nucleotide sugar to the undecaprenyl phosphate (UndP)-lipid carrier on the cytoplasmic face of the membrane. This step is catalyzed by the membrane-associated priming glycosyltransferase WelE. Subsequently, unique glycosyltransferases WelF to -J add the remaining sugars in a sugar and glycosidic linkage-dependent manner, of which the exact order remains to be determined (indicated by brackets). The substrates for these glycosyltransferases are sugar nucleotides, which are available from existing cellular pools (e.g., UDP-galactose, UDP-N-acetylglucosamine) or synthesized by EPS-specific enzymes encoded within the EPS gene cluster (e.g., UDP-galactofuranose by the glf gene). Once an EPS subunit is completed, it needs to be translocated across the cytoplasmic membrane by a Wzx flippase, followed by linkage of the repeating units into long polysaccharides by a specific Wzy polymerase. A phosphorylation complex including a Wze autophosphorylating tyrosine kinase and a Wzb phosphotyrosine protein phosphatase is thought to be involved in the regulation of EPS biosynthesis. This figure is partly based on the model for Streptococcus pneumoniae (5), with modifications based on reference 27 and the gene cluster and phenotypic analyses presented here.
TABLE 2.
ORFs identified in the EPS gene cluster of L. rhamnosus GG
| ORF | Size (bp) | Predicted encoded function | Predicted domain(s) present in encoded ORF | Best BLASTx hit (accession no.)
|
% Amino acid identity | |
|---|---|---|---|---|---|---|
| Protein | Organism | |||||
| wzd | 920 | Auxiliary protein for polysaccharide export and chain length determination | 2 transmembrane domains | Wzd (AAW22433) | L. rhamnosus ATCC 9595 | 72 |
| wze | 756 | Autophosphorylating tyrosine-protein kinase | Walker A and B motif (nucleotide binding); C-terminal tyrosine-rich region | Wze (AAW22434) | L. rhamnosus ATCC 9595 | 88 |
| wzy | 1,221 | Repeat unit polymerase | 10 putative transmembrane helices | Wzy (CAI33441) | Streptococcus pneumoniae serotype 16a | 29 |
| glf | 1,131 | UDP-galactopyranose mutase | 1 putative transmembrane helix | Glf (YP_536435) | L. salivarius UCC118 | 73 |
| wzx | 1,437 | Repeat unit transporter | 12 putative transmembrane helices | Wzx (YP_1988153) | L. casei BL23 | 55 |
| welJ | 600 | Glycosyltransferase | No hit in Pfam database; no transmembrane helix | ZP_1274133 | L. reuteri 100-23 | 46 |
| welI | 1,137 | Glycosyltransferase | Glycosyltransferase family 1 (PF00534) | NP_784849 | L. plantarum WCFS1 | 35 |
| welH | 936 | Glycosyltransferase | Glycosyltransferase family 2 (PF00535) | NP_784850 | L. plantarum WCFS1 | 40 |
| welG | 1,017 | Glycosyltransferase | No hit in Pfam database | YP_1271955 | L. reuteri F275 | 32 |
| welF | 1,068 | Glycosyltransferase | Glycosyltransferase family 1 (PF00534) | WffU (ACA24914) | Shigella dysenteriae | 28 |
| welE | 669 | Undecaprenyl-phosphate galactose phosphotransferase; priming glycosyltransferase | Bacterial sugar transferases (PF02397 domain) | YP_1842290 | L. reuteri F275 | 61 |
| rmlAa | 795 | dTDP-glucose pyrophosphorylase | Nucleotidyl transferase | RmlA (AAW22443) | L. rhamnosus ATCC 9595 | 90 |
| orf1 | 1,494 | Putative transposase | Transposase DDE domain | YP_806463 | L. casei ATCC 334 | 62 |
| rmlC | 597 | dTDP-4-dehydro-rhamnose-3,5-epimerase | dTDP-4 dehydro-rhamnose-3,5-epimerase | RmlC (AAW22444) | L. rhamnosus ATCC 9595 | 98 |
| rmlB | 936 | dTDP-d-glucose-4,6-dehydratase | NAD-dependent epimerase/dehydratase family | RmlB (AAW22445) | L. rhamnosus ATCC 9595 | 99 |
| wzr | 891 | Transcriptional regulator of polysaccharide biosynthesis | LytR-CspA-Psr superfamily | Wzr (AAW22447) | L. rhamnosus ATCC 9595 | 97 |
| wzb | 765 | Phosphotyrosine protein phosphatase | Polymerase and histidinol phosphatase domain | Wzb (AAW22466) | L. rhamnosus RW-6541 M | 100 |
Interrupted by orf1 encoded on ISLrh2.
In addition to regulatory proteins, the EPS cluster of L. rhamnosus GG contains the genes encoding the enzymes for the actual biosynthesis of EPS. Glycosyltransferases synthesize blocks of repeating units that are linked to a lipid carrier at the inner side of the cytoplasmic membrane (Fig. 1B). Six genes (welE to -J) encoding putative glycosyltransferases are contained in the central portion of the putative EPS locus of L. rhamnosus GG. The protein encoded by the welE gene displays 61% identity with YP_001271961 from Lactobacillus reuteri F275, annotated as a galactose phosphotransferase (Table 2). The welE gene putatively encodes the priming glycosyltransferase that transfers the first sugar of each subunit of an EPS molecule of L. rhamnosus GG. welF to welJ probably encode the glycosyltransferases transferring the other sugars of the EPS subunit in an ordered and sugar- and glycosidic linkage-dependent fashion (Fig. 1B). In general, heteropolymeric polysaccharide subunits are flipped across the cytoplasmic membrane by a Wzx-type exporter and polymerized into long polysaccharides by a Wzy-type polymerase at the outer side of the cytoplasmic membrane (Fig. 1B) (13, 56, 58), which has been experimentally best studied in E. coli (57). The wzx and wzy genes of L. rhamnosus GG are located in the 5′ region of the putative EPS cluster (Fig. 1A). In contrast to the conserved Wzd/Wze and Wzb homologs, Wzx and Wzy proteins are transmembrane proteins that are specific for the associated EPS repeating unit (13, 56, 58), as reflected by their lower level of similarity among orthologs (Table 2).
The EPS cluster of L. rhamnosus GG also contains genes for the synthesis of specific nucleotide sugars that cannot be obtained from the central metabolism. The glf gene putatively encodes a UDP-galactopyranose mutase for the conversion of UDP-galactopyranose to UDP-galactofuranose (Table 2). This is in agreement with the presence of galactofuranose residues in the repeating units of the EPS molecules of L. rhamnosus GG, as previously reported (27). Genes for dTDP-rhamnose biosynthesis (rmlACB) are also located in the EPS gene cluster (Fig. 1A). However, the rmlA gene, putatively encoding the glucose-1-phosphate thymidylyl transferase, appears to be inactivated by insertion of ISLrh2. The rmlB and rmlC genes putatively encode a dTDP-glucose-4,6-dehydratase and a dTDP-4-keto-rhamnose-3,5-epimerase, respectively. No rmlD gene encoding the final-acting dTDP-rhamnose synthase was found within the L. rhamnosus GG EPS gene cluster, in contrast to the case for the previously identified EPS gene cluster of the phylogenetically related strain L. rhamnosus ATCC 9595 (43) (Fig. 1A). Nevertheless, Southern hybridization experiments suggest the presence of a complete rmlACBD operon at another locus in the L. rhamnosus GG genome distinct from the EPS gene cluster (data not shown).
Mutation of the priming glycosyltransferase reduces the total level of CW-PS.
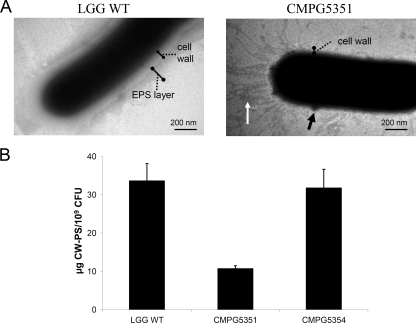
As welE putatively encodes the priming glycosyltransferase, i.e., an important control point of EPS biosynthesis, we hypothesized that deletion of this gene should result in a drastic reduction of EPS production. To test this hypothesis, a welE insertion mutant of L. rhamnosus GG, designated CMPG5351, was constructed. The growth capacity of CMPG5351 was not significantly different from that of wild-type L. rhamnosus GG. Wild-type L. rhamnosus GG and welE mutant CMPG5351 both showed a generation time of 1.6 ± 0.1 h and 1.9 ± 0.1 h in MRS and AOAC media, respectively. For subsequent analyses, cells were grown in AOAC medium, as we previously observed that this medium induces a high level of EPS production by L. rhamnosus GG (32). L. rhamnosus GG wild-type cells that are grown in AOAC medium have a typical outer layer formed by CW-PS (29). TEM analysis showed that this layer is absent in the CMPG5351 mutant, although some isolated zones with accumulation of, presumably, polysaccharides could still be detected (Fig. 2A). Additionally, fimbria-like structures appear to become exposed in welE mutant cells at the poles opposite the septum (Fig. 2A). Subsequently, total CW-PS material was extracted from L. rhamnosus GG wild-type and CMPG5351 cells as described in Materials and Methods. The quantity of CW-PS that could be extracted from welE mutant cells was ca. 3-fold lower than that from L. rhamnosus GG wild-type cells (Fig. 2B). Importantly, cis complementation of CMPG5351 with a functional welE gene containing its upstream promoter region (corresponding to strain CMPG5354) could restore CW-PS production to the wild-type level (Fig. 2B), indicating that the welE mutation is not polar and that the welE gene in the wild-type strain is transcribed from its own promoter (see also below).
FIG. 2.
Mutation of the welE gene in L. rhamnosus GG reduces the level of CW-PS. (A) TEM analysis of L. rhamnosus GG wild-type and welE mutant CMPG5351 strains grown in AOAC medium. For wild-type L. rhamnosus GG, the cell-bound EPS layer is indicated. While this layer is absent in the welE mutant CMPG5351, some patches of presumably polysaccharide accumulations could still be detected (black arrow). Additionally, fimbria-like appendages appear to become exposed in welE mutant cells (white arrow). (B) CW-PS were extracted from cell pellets from wild-type L. rhamnosus GG (WT), welE mutant CMPG5351, and the complemented strain CMPG5354. Error bars indicate standard deviations.
Mutation of the priming glycosyltransferase specifically abolishes the production of the long, galactose-rich EPS molecules.
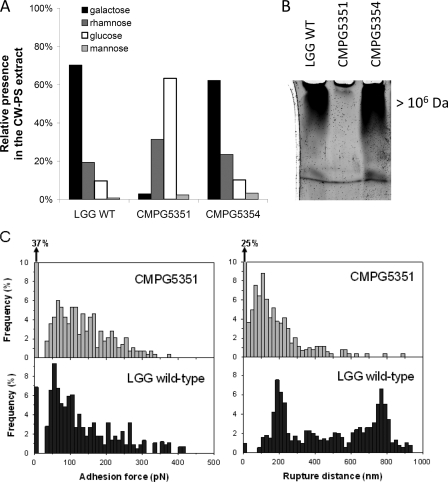
Having established that the L. rhamnosus GG welE mutant CMPG5351 contains less CW-PS than the wild-type strain, we subsequently investigated whether the welE mutation specifically affects the production of the long, galactose-rich type of L. rhamnosus GG CW-PS molecules. First, the sugar monomer composition of the CW-PS extracts was determined by gas chromatography as described in Materials and Methods. The extract of wild-type L. rhamnosus GG was found to be composed of approximately 70% galactose, 19% rhamnose, and 10% glucose (Fig. 3A). On the basis of the L. rhamnosus GG EPS structure determined by Landersjö et al. (27), this implies that the CW-PS extract of L. rhamnosus GG wild-type cells contains mainly the known galactose-rich EPS molecules. The sugar monomer composition of the CW-PS extract of the CMPG5351 mutant was drastically changed to approximately 63% glucose, 31.5% rhamnose, 3% galactose, and 2.5% mannose (Fig. 3A). This indicates that the galactose-rich CW-PS molecules are strongly reduced in the welE mutant CMPG5351, while the relative proportion of the (yet-unknown) glucose-rich CW-PS has increased. Importantly, sugar monomer analysis also showed that complementation of the welE mutation in strain CMPG5354 could fully restore synthesis of the galactose-rich type of CW-PS at the L. rhamnosus GG surface (Fig. 3A). Additionally, the size of the CW-PS molecules was estimated by gel electrophoresis of the CW-PS extracts on 5% polyacrylamide gels. These analyses revealed the absence of the largest band in the CW-PS extract of the welE mutant (Fig. 3B), corresponding to the high-molecular-mass (>1.4 × 106 Da) CW-PS molecules based on size exclusion chromatography (data not shown). Finally, SMFS for surface polysaccharides was applied on live bacteria, using functionalized AFM tips containing single lectins specific for end-standing galactose (PA-I lectin) as recently described (21). In comparison to the wild type, the welE mutant CMPG5351 showed a marked reduction in the galactose-rich CW-PS molecules (reduced adhesion frequency with the PA-1 functionalized AFM tip) and a marked reduction in polymer length (reduced elongation and rupture distance) (Fig. 3C). On the other hand, SMFS experiments with mutant CMPG5351 and concanavalin A-functionalized AFM tips showed an increase in the glucose-rich type of polysaccharides, with especially an increase in elongation and rupture length (data not shown).
FIG. 3.
Mutation of the welE gene in L. rhamnosus GG specifically attenuates the high-molecular-mass, galactose-rich EPS molecules. (A) Analysis of the sugar monomer compositions of the CW-PS extracts of wild-type L. rhamnosus GG (WT), welE mutant CMPG5351, and the complemented strain CMPG5354. The data are expressed in relative amounts, taking the total amount of detected monomeric sugars as 100%. (B) Polyacrylamide gel electrophoresis of the CW-PS extract of wild-type L. rhamnosus GG, welE mutant CMPG5351, and complemented strain CMPG5354. Equal amounts of material were loaded. The upper band corresponds to polysaccharides with a polymer size of >1.4 × 106 Da based on size exclusion chromatography (data not shown). (C) Detection of individual galactose-rich polysaccharides on wild-type L. rhamnosus GG and welE mutant CMPG5351 by SMFS with PA-1 lectin-functionalized tips. Histograms (n = 1,024) of adhesion forces (left) and rupture distances (right) are shown.
Taken together, the data on monomer composition, polymer size, and SMFS for the L. rhamnosus GG wild-type, welE mutant, and complemented welE mutant strains demonstrate that the WelE enzyme has a crucial and specific role in the biosynthesis of the high-molecular-weight, galactose-rich type of CW-PS molecules of L. rhamnosus GG, referred to as the EPS molecules of L. rhamnosus GG in this study. Moreover, abolishment of the galactose-rich EPS molecules in welE mutant CMPG5351 seems to increase the presence of the glucose-rich type of CW-PS molecules, which were described for the first time in reference 21.
Inactivation of the priming glycosyltransferase WelE results in increased biofilm formation and adhesion to mucus and epithelial cells by L. rhamnosus GG.
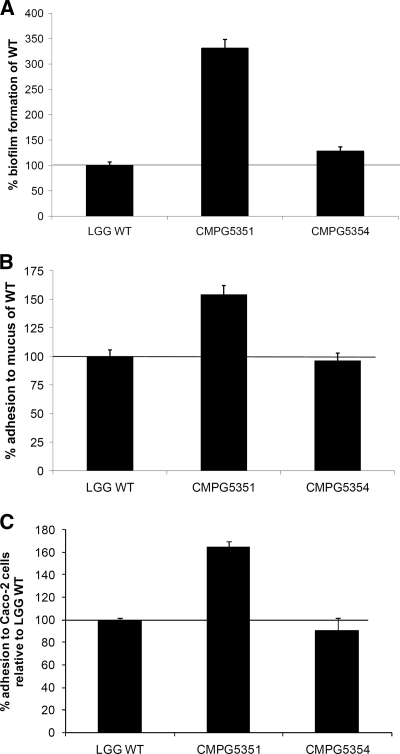
Prior experiments showed that L. rhamnosus GG has an intrinsic high biofilm formation capacity (29, 32). The welE mutant with a specific reduction in the long, galactose-rich EPS molecules allowed us to investigate the relative contribution of these EPS molecules to the biofilm-forming capacity of L. rhamnosus GG. Surprisingly, biofilm formation by the EPS mutant CMPG5351 was increased threefold compared to that by the wild type in AOAC medium (Fig. 4A). Importantly, cis complementation with a functional welE gene in CMPG5351 (strain CMPG5354), restored wild-type levels of biofilm formation (Fig. 4A). Additionally, the in vitro adherence capacity of the EPS mutant CMPG5351 was investigated with mucus as a substrate, as L. rhamnosus GG has been previously reported to display high mucus-adhering properties (52). In our assay, the welE mutant CMPG5351 grown in AOAC medium showed a ca. 1.5-fold-increased adhesion to commercially available pig gastric mucus compared to wild-type L. rhamnosus GG grown under the same conditions (Fig. 4B). Again, complementation restored the adherence capacity to wild-type levels (Fig. 4B). Finally, adhesion of wild-type L. rhamnosus GG, the welE mutant CMPG5351, and the complemented strain CMPG5354 grown in AOAC medium was studied using the human gut epithelial cell line Caco-2. A marked increase in adherence by the welE mutant could be observed, while complementation restored wild-type levels of adherence (Fig. 4C). Overall, these results indicate that the EPS-deficient mutant CMPG5351 has an increased in vitro adherence capacity to several substrates.
FIG. 4.
Biofilm formation and adhesion to mucus and Caco-2 cells by wild-type L. rhamnosus GG (WT), the EPS welE mutant CMPG5351, and complemented strain CMPG5354. (A) Biofilm formation (in AOAC medium) is expressed relative to the amount formed by wild-type L. rhamnosus GG. (B) The adhesion capacity to mucus is expressed relative to wild-type L. rhamnosus GG (set as 100%), of which ca. 22% of the added cells adhered. (C) The adhesion capacity to Caco-2 cells is expressed relative to wild-type L. rhamnosus GG, of which ca. 4% of the added cells adhered. Error bars indicate standard deviations.
DISCUSSION
In this study, we identified and annotated the EPS gene cluster of the probiotic strain L. rhamnosus GG. This cluster shows a modular organization that is characteristic for gene clusters involved in the biosynthesis of heteropolymeric EPS or CPS molecules (26, 45, 56). In comparison with the previously reported EPS gene clusters of four closely related L. rhamnosus strains (43), considerable differences can be observed, in terms of both the DNA sequences of the orthologs and the organization of the genes within the EPS gene cluster (Fig. 1A). Only the ORFs for regulatory proteins (Wzd, Wze, Wzr, and Wzb) and dTDP-rhamnose precursor biosynthesis (RmlA to -C) are highly conserved. The low level of conservation between the L. rhamnosus GG EPS gene cluster and the cluster of the other L. rhamnosus strains is in agreement with the different EPS structures of these strains: galactose-rich EPS molecules are synthesized by L. rhamnosus GG cells (27), while rhamnose-rich EPS molecules are synthesized by the L. rhamnosus ATCC 9595-related strains (54). The EPS gene cluster described in this paper seems to be unique and strain specific to L. rhamnosus GG. The mosaic organization of the EPS gene cluster of L. rhamnosus GG probably results from horizontal gene transfer and homologous recombination events, as is common for EPS/CPS gene clusters (see, e.g., reference 38). Interestingly, the rml genes within the EPS cluster of L. rhamnosus GG seem to be inactivated due to an insertion sequence element (ISLrh2 containing orf1) in rmlA, while Southern hybridization experiments suggest the presence of a complete rmlABCD operon at another locus in the L. rhamnosus GG genome.
The function of the identified EPS gene cluster in L. rhamnosus GG was confirmed by mutation of the welE gene, encoding the putative priming glycosyltransferase. Detailed phenotypic analyses of the cell wall polysaccharides (quantity, monomer composition, polymer size, and SMFS) of welE mutant CMPG5351 versus the wild type indicate a crucial and specific role for the priming galactosyltransferase WelE in the biosynthesis of the high-molecular-weight, galactose-rich EPS molecules of L. rhamnosus GG. Similarly, the priming glycosyltransferase encoded within the EPS cluster was shown to be crucial for EPS biosynthesis in the gram-positive cocci Lactococcus lactis (55) and Streptococcus thermophilus (39). However, such a key function has, to the best of our knowledge, not yet been established in gram-positive rod-shaped bacteria such as lactobacilli. Additionally, comparative phenotypic analyses of the L. rhamnosus GG wild-type and mutant CMPG5351 strains confirm the previous SMFS experiments (21) indicating that the L. rhamnosus GG surface contains, besides the long, galactose-rich type of EPS, also shorter, glucose-rich CW-PS. Of note, CW-PS quantification and monosaccharide analyses revealed that the glucose-rich CW-PS molecules become more exposed in the EPS mutant CMPG5351. As a consequence, only a ca. 3-fold decrease in the total extractable CW-PS amount in this mutant was observed, whereby the monomer composition and polymer length drastically changed. The characterization of these other CW-PS molecules and their biosynthetic genes is ongoing.
In comparison with other Lactobacillus strains, L. rhamnosus GG is known for its intrinsic high in vitro adherence capacity in several experimental setups (32, 52, 53). The construction of the L. rhamnosus GG EPS mutant CMPG5351 allowed us to study the specific contribution of the strain-specific long, galactose-rich EPS molecules to this adherence capacity. Interestingly, the EPS mutant CMPG5351 showed an increased adherence to commercially available pig mucus and Caco-2 epithelial cells. This suggests that the long, galactose-rich EPS molecules of L. rhamnosus GG are not required for its high adherence capacity. On the contrary, EPS seems to have a negative impact, possibly by shielding off adhesins. These data are in agreement with studies of streptococci showing that surface polysaccharides such as CPS can shield adhesins and reduce the adhesion capacity (35, 40, 50). Similarly, an EPS mutant of Lactobacillus johnsonii NCC533 was reported to have a slightly increased residence time in the murine gut, possibly because of enhanced exposure of adhesins (14). The characterization of the specific adhesins of L. rhamnosus GG should further substantiate this “shielding hypothesis.” Besides a putative role for the increased expression of glucose-rich polysaccharides in the adherence capacity of L. rhamnosus GG, TEM analyses suggest also an increased exposure of fimbria-like structures in the EPS mutant. Fimbriae or pili are well studied in certain bacterial species for their adhesive and biofilm-promoting properties (36), and some contain a considerable glycan chain (59). Fimbrial genes have been described for L. johnsonii NCC533 (44), but they are not common in lactobacilli for which the genome sequences have been determined to date (31). We are currently investigating further these fimbria-like structures on the L. rhamnosus GG surface.
Adhesion of probiotic strains is usually assessed in short-term assays (31). However, in most natural niches, adherent bacteria can form multicellular structures recognized as biofilm-like communities (7). Small biofilms or microcolonies have also been documented for bacteria residing in the gastrointestinal tract, and this is suggested to enhance their residence time (49). Interestingly, the EPS mutant of L. rhamnosus GG showed an increased biofilm formation capacity when grown in AOAC medium. The fact that the long, galactose-rich EPS molecules are not required for its biofilm formation is remarkable given the well-documented role for extracellular polysaccharides in biofilm matrices of various bacterial species (7). Nevertheless, these findings do not rule out that polysaccharides as such are implicated in the extracellular matrix of L. rhamnosus GG biofilms. The shorter, yet-unknown glucose-rich CW-PS molecules of L. rhamnosus GG could be important constituents of L. rhamnosus GG biofilms together with proteinaceous compounds, such as the fimbria-like structures. This would imply that the specific type of CW-PS is important for L. rhamnosus GG biofilm formation. Of interest is that a divergent role for surface polysaccharides in biofilm formation has also been shown in other bacteria, such as Pseudomonas aeruginosa (47). Future analyses of the biofilm matrix of L. rhamnosus GG are aimed to shed light on its exact composition under various conditions.
In conclusion, the current report describes the identification and annotation of the gene cluster involved in the biosynthesis of specific galactose-rich EPS molecules of the clinically well documented probiotic strain L. rhamnosus GG, as well as the functional analysis of the corresponding priming glycosyltransferase WelE. Phenotypic analysis of the welE mutant indicate that the long, galactose-rich EPS molecules of L. rhamnosus GG play an important role in determining cell surface properties such as adhesion and biofilm formation. Future studies should provide more insight into the potential role of EPS as a probiotic factor important for some of the reported health benefits of L. rhamnosus GG.
Supplementary Material
Acknowledgments
At the time of the experiments, S.L. was a research assistant of the FWO-Vlaanderen. Currently, S.L. is holding a BOF postdoctoral mandate from the K.U. Leuven. S.C.J.D.K. is a postdoctoral research fellow of the FWO-Vlaanderen. Y.D. is a senior research associate at the National Foundation for Scientific Research (FNRS). This work was partially supported by the FWO-Vlaanderen through project G.0236.07; the FNRS; the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme); and the Research Department of the Communauté Française de Belgique (Concerted Research Action).
We gratefully acknowledge David De Coster for his help with the electron microscope, Jos Desair for his help with size exclusion chromatography, Valerie Van Craeyveld for her help with the sugar monomer analysis, and Nils Verresen for his help with the Caco-2 cells. We also thank M. Alvarez, M. Danielsen, P. Augustijns, and R. Mols for kindly providing plasmids and Caco-2 cells used in this study.
Footnotes
Published ahead of print on 3 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination system of the Lactobacillus species bacteriophage A2 integrates in Gram-positive and Gram-negative bacteria. Virology 250:185-193. [DOI] [PubMed] [Google Scholar]
- 3.Beg, A. M., M. N. Jones, T. Miller-Torbert, and R. G. Holt. 2002. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem. Biophys. Res. Commun. 298:75-79. [DOI] [PubMed] [Google Scholar]
- 4.Bender, M. H., and J. Yother. 2001. CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J. Biol. Chem. 276:47966-47974. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouazzaoui, K., and G. LaPointe. 2006. Use of antisense RNA to modulate glycosyltransferase gene expression and exopolysaccharide molecular mass in Lactobacillus rhamnosus. J. Microbiol. Methods 65:216-225. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 8.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 9.Comstock, L. E., and D. L. Kasper. 2006. Bacterial glycans: key mediators of diverse host immune responses. Cell 126:847-850. [DOI] [PubMed] [Google Scholar]
- 10.Coyne, M. J., M. Chatzidaki, L. Paoletti, and L. E. Comstock. 2008. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. USA 105:13099-13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 12.De Keersmaecker, S. C. J., K. Braeken, T. L. A. Verhoeven, M. Perea Vélez, S. Lebeer, J. Vanderleyden, and P. Hols. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl. Environ. Microbiol. 72:4923-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek 76:159-184. [PubMed] [Google Scholar]
- 14.Denou, E., R. D. Pridmore, B. Berger, J. M. Panoff, F. Arigoni, and H. Brussow. 2008. Identification of genes associated with the long gut persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190:3161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doron, S., D. R. Snydman, and S. L. Gorbach. 2005. Lactobacillus GG: bacteriology and clinical applications. Gastroenterol. Clin. N. Am. 34:483-498. [DOI] [PubMed] [Google Scholar]
- 16.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 17.Englyst, H. N., and J. H. Cummings. 1984. Simplified method for the measurement of total non-starch polysaccharides by gas liquid chromatography of constituent sugars as alditol acetates. Analyst 109:937-942. [DOI] [PubMed] [Google Scholar]
- 18.FAO/WHO. 2001. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. WHO, Geneva, Switzerland.
- 19.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francius, G., D. Alsteens, V. Dupres, S. Lebeer, S. De Keersmaecker, J. Vanderleyden, H. J. Gruber, and Y. F. Dufrene. Stretching polysaccharides on live cells using single molecule force spectroscopy. Nat. Prot., in press. [DOI] [PubMed]
- 21.Francius, G., S. Lebeer, D. Alsteens, L. Wildling, H. J. Gruber, P. Hols, S. C. J. De Keersmaecker, J. Vanderleyden, and Y. F. Dufrene. 2008. Detection, localization and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano. 2:1921-1929. [DOI] [PubMed] [Google Scholar]
- 22.Guarino, A., A. Lo Vecchio, and R. B. Canani. 2009. Probiotics as prevention and treatment for diarrhea. Curr. Opin. Gastroenterol. 25:18-23. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:164. [Google Scholar]
- 24.Hooper, L. V., and J. I. Gordon. 2001. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11:1R-10R. [DOI] [PubMed] [Google Scholar]
- 25.Kalliomäki, M., S. Salminen, H. Arvilommi, P. Kero, P. Koskinen, and E. Isolauri. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076-1079. [DOI] [PubMed] [Google Scholar]
- 26.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29:393-409. [DOI] [PubMed] [Google Scholar]
- 27.Landersjö, C., Z. N. Yang, E. Huttunen, and G. Widmalm. 2002. Structural studies of the exopolysaccharide produced by Lactobacillus rhamnosus strain GG (ATCC 53103). Biomacromolecules 3:880-884. [DOI] [PubMed] [Google Scholar]
- 28.LaPointe, G., D. Atlan, and C. Gilbert. 2008. Characterization and site-directed mutagenesis of Wzb, an O-phosphatase from Lactobacillus rhamnosus. BMC Biochem. 9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebeer, S., I. J. J. Claes, T. L. A. Verhoeven, C. Shen, I. Lambrichts, J. L. Ceuppens, J. Vanderleyden, and S. C. J. De Keersmaecker. 2008. Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 74:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebeer, S., S. C. J. De Keersmaecker, T. L. A. Verhoeven, A. A. Fadda, K. Marchal, and J. Vanderleyden. 2007. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189:860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebeer, S., J. Vanderleyden, and S. De Keersmaecker. 2008. Genes and molecules of Lactobacillus supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebeer, S., T. L. A. Verhoeven, M. Perea Vélez, J. Vanderleyden, and S. C. J. De Keersmaecker. 2007. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73:6768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 34.Liu, C. H., S. M. Lee, J. M. VanLare, D. L. Kasper, and S. K. Mazmanian. 2008. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. USA 105:3951-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marco, M. L., S. Pavan, and M. Kleerebezem. 2006. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17:204-210. [DOI] [PubMed] [Google Scholar]
- 38.Mavroidi, A., D. Godoy, D. M. Aanensen, D. A. Robinson, S. K. Hollingshead, and B. G. Spratt. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J. Bacteriol. 186:8181-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minic, Z., C. Marie, C. Delorme, J. M. Faurie, G. Mercier, D. Ehrlich, and P. Renault. 2007. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J. Bacteriol. 189:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morona, J. K., D. C. Miller, R. Morona, and J. C. Paton. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189:1905-1913. [DOI] [PubMed] [Google Scholar]
- 41.Morona, J. K., R. Morona, D. C. Miller, and J. C. Paton. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J. Bacteriol. 184:577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morona, J. K., R. Morona, and J. C. Paton. 2006. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc. Natl. Acad. Sci. USA 103:8505-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Péant, B., G. LaPointe, C. Gilbert, D. Atlan, P. Ward, and D. Roy. 2005. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 151:1839-1851. [DOI] [PubMed] [Google Scholar]
- 44.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. H. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 47.Ryder, C., M. Byrd, and D. J. Wozniak. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 49.Sonnenburg, J. L., L. T. Angenent, and J. I. Gordon. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569-573. [DOI] [PubMed] [Google Scholar]
- 50.Talbot, U. M., A. W. Paton, and J. C. Paton. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect. Immun. 64:3772-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tallon, R., P. Bressollier, and M. C. Urdaci. 2003. Isolation and characterization of two exopolysaccharides produced by Lactobacillus plantarum EP56. Res. Microbiol. 154:705-712. [DOI] [PubMed] [Google Scholar]
- 52.Tuomola, E. M., A. C. Ouwehand, and S. J. Salminen. 1999. Human ileostomy glycoproteins as a model for small intestinal mucus to investigate adhesion of probiotics. Lett. Appl. Microbiol. 28:159-163. [DOI] [PubMed] [Google Scholar]
- 53.Tuomola, E. M., and S. J. Salminen. 1998. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 41:45-51. [DOI] [PubMed] [Google Scholar]
- 54.Van Calsteren, M. R., C. Pau-Roblot, A. Begin, and D. Roy. 2002. Structure determination of the exopolysaccharide produced by Lactobacillus rhamnosus strains RW-9595M and R. Biochem. J. 363:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Kranenburg, R., J. D. Marugg, I. I. vanSwam, N. J. Willem, and W. M. deVos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 56.Welman, A. D., and I. S. Maddox. 2003. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. Trends Biotechnol. 21:269-274. [DOI] [PubMed] [Google Scholar]
- 57.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39-68. [DOI] [PubMed] [Google Scholar]
- 58.Whitfield, C., and A. Paiment. 2003. Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 338:2491-2502. [DOI] [PubMed] [Google Scholar]
- 59.Wu, H., M. Zeng, and P. Fives-Taylor. 2007. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect. Immun. 75:2181-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zocco, M. A., L. Z. Dal Verme, F. Cremonini, A. C. Piscaglia, E. C. Nista, M. Candelli, M. Novi, D. Rigante, I. A. Cazzato, V. Ojetti, A. Armuzzi, G. Gasbarrini, and A. Gasbarrini. 2006. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 23:1567-1574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.