Abstract
The nucleus is the primary site of protein aggregation in many polyglutamine diseases, suggesting a central role in pathogenesis. In SBMA, the nucleus is further implicated by the critical role for disease of androgens, which promote the nuclear translocation of the mutant androgen receptor (AR). To clarify the importance of the nucleus in SBMA, we genetically manipulated the nuclear localization signal of the polyglutamine-expanded AR. Transgenic mice expressing this mutant AR displayed inefficient nuclear translocation and substantially improved motor function compared with SBMA mice. While we found that nuclear localization of polyglutamine-expanded AR is required for SBMA, we also discovered, using cell models of SBMA, that it is insufficient for both aggregation and toxicity and requires androgens for these disease features. Through our studies of cultured motor neurons, we further found that the autophagic pathway was able to degrade cytoplasmically retained expanded AR and represents an endogenous neuroprotective mechanism. Moreover, pharmacologic induction of autophagy rescued motor neurons from the toxic effects of even nuclear-residing mutant AR, suggesting a therapeutic role for autophagy in this nucleus-centric disease. Thus, our studies firmly establish that polyglutamine-expanded AR must reside within nuclei in the presence of its ligand to cause SBMA. They also highlight a mechanistic basis for the requirement for nuclear localization in SBMA neurotoxicity, namely the lack of mutant AR removal by the autophagic protein degradation pathway.
INTRODUCTION
Nuclear residing proteins are normally directed to the nucleus by a signaling sequence, a particular folding pattern and/or a post-translational modification. After they have served their function, nuclear proteins are either degraded by nuclear proteasomes or exported to the cytoplasm for degradation. A mutation within a protein, such as the expansion of a polyglutamine tract, causes it to accumulate within particular cellular compartments, as it is refractory to degradation. Nuclear accumulation of misfolded proteins is most likely due to the lack of a secondary degradation mechanism within nuclei and this accumulation of mutant protein is toxic to neurons.
Spinal and bulbar muscular atrophy (SBMA, Kennedy’s disease) is an X-linked neurodegenerative disease resulting from the expansion of a polyglutamine (polyQ)-encoding CAG tract in the 5′ end of the androgen receptor (AR) gene (1). When containing more than 40 CAG repeats, the AR causes slowly progressive proximal limb and bulbar muscle weakness, fasciculations and atrophy in men (2,3). Patients may also suffer some sensory loss (4,5) and display slight androgen insensitivity (2). While partial loss of AR function exists in SBMA, this does not represent the primary disease etiology (6,7); rather accumulation of toxic AR protein species leads to motor neuron dysfunction and death (8–10).
SBMA is one of a family of nine polyQ-expansion diseases (reviewed by 11), with a common pathological hallmark; the accumulation of misfolded and aggregated species of mutant protein in the cytoplasm or nuclei of vulnerable neurons. Although there are conflicting views in the field concerning the correlation of aggregates with disease, considerable data indicate that inclusions themselves are not toxic (12,13). Instead, species that are produced in early stages of the aggregation cascade (likely proteolyzed AR monomer and oligomer) induce toxicity. Nonetheless, the presence of inclusions in a population of neurons reveals the late stage of a pathogenic process.
The common finding of nuclear inclusions in polyQ diseases suggests a central role for the nucleus in pathogenesis. While inclusions of polyQ-expanded huntingtin are found in both the cytoplasm and nucleus, the accumulation of nuclear mutant huntingtin causes the greatest neuronal toxicity (13,14). In SCA-1 and SCA-3, inclusions of the mutant protein are found only within nuclei (15,16) and mutation of the endogenous nuclear localization signal (NLS) within each of these respective proteins, to sequester them within the cytoplasm, has proved to be neuroprotective (17,18). These findings highlight an important role for the nucleus in the toxicity induced by polyQ-expanded proteins, although the mechanistic basis for this role has remained elusive.
In SBMA, inclusions of aberrantly cleaved polyQ-expanded AR are also present primarily in nuclei (19), although neuropil accumulation of 1C2-positive material has been observed (20). In cell and rodent models of SBMA, nuclear aggregation and disease are dependent on the presence of AR ligands [testosterone or dihydrotestosterone (DHT)] (10,21–27), which drive nuclear translocation of the AR. As a type I nuclear hormone receptor transcription factor, the unliganded AR resides primarily within the cytosol, where it is associated with heat shock and accessory proteins (28,29). Upon hormone binding, the AR undergoes a conformational change that exposes its bipartite NLS, directing it to the nucleus, where it regulates transcription of its target genes.
The localization of inclusions within nuclei and the dependence of disease on androgens suggest a central role for the nucleus in SBMA. Moreover, the finding that some alternative ligands that direct AR to the nucleus also cause disease supports this idea (24,30). In a Drosophila model of SBMA, retention of a polyQ-expanded AR fragment in the cytoplasm ameliorated disease (26). However, in contrast to these results, in a cell model of SBMA, fast axonal transport was reduced by expanded AR in a hormone-independent manner (31), suggesting a cytoplasmic site of pathology, and making uncertain the role of the nucleus in disease. In this study, we sought to determine, using transgenic mouse and cell models, whether mammalian systems reveal a necessity for nuclear localization and whether nuclear localization is sufficient for disease.
Our results firmly establish that nuclear localization of polyQ-expanded AR is necessary, but not sufficient for nuclear aggregation and toxicity. Furthermore, we present evidence that the lack of access to the autophagic degradation pathway represents one explanation for the enhanced toxicity of nuclear-confined mutant AR.
RESULTS
ARdNLS112Q transgenic mice express greater levels of AR than AR112Q mice
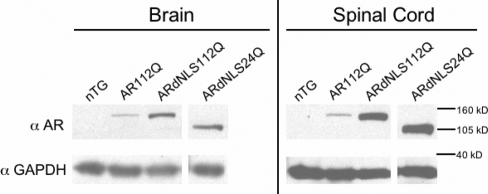
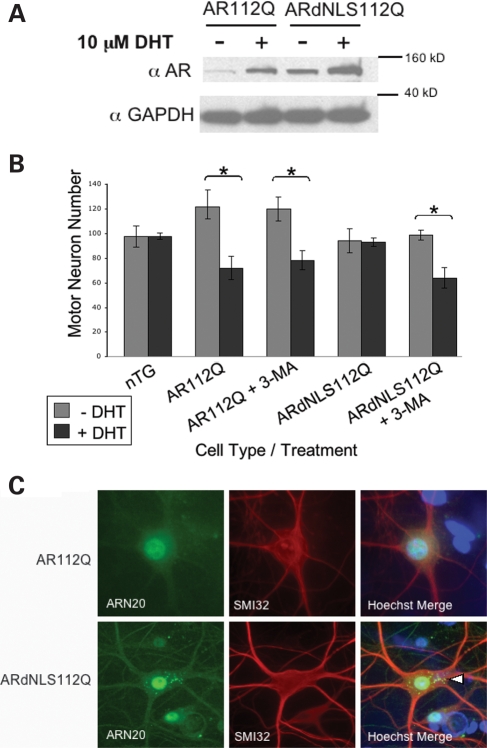
In order to understand the role of the nucleus in disease, we created transgenic mice bearing an AR with a mutation in the NLS. We previously created a transgenic mouse model of SBMA expressing full-length (human) polyQ-expanded AR (112Q) driven by the prion protein promoter (PrP) (22). The new transgenic mice (ARdNLS112Q and ARdNLS24Q) were created to express transgenic AR with a deletion of amino acids 628–640 within the bipartite NLS of the AR. A line of ARdNLS112Q was established which expresses over 2-fold more AR protein, in brain and spinal cord, than AR112Q mice (Fig. 1). In addition, ARdNLS24Q mice expressed equivalent levels of AR as ARdNLS112Q mice (Fig. 1), and both lines were used for behavioral analysis.
Figure 1.
Protein expression of ARdNLS mice. Five-week-old male mice were sacrificed and whole brain and spinal cord lysates evaluated for AR by western blot. AR protein was detected with antibody AR(N-20). GAPDH was used as a loading control. nTG, non-transgenic.
Motor deficits associated with SBMA are ameliorated in ARdNLS112Q male mice
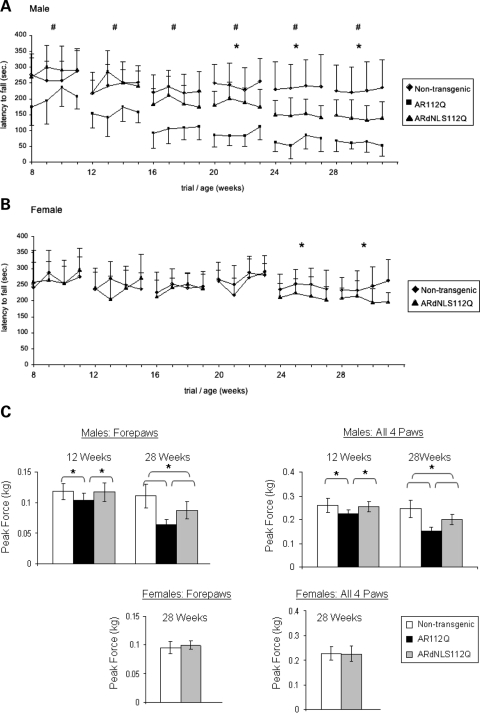
We previously determined that the rotarod assay is a sensitive measure of motor function in SBMA mice (22). Therefore, we measured latency to fall from an accelerating rotarod, as well as grip strength, every 4 weeks beginning at 8 weeks of age. This behavioral analysis of a large age-matched cohort of ARdNLS112Q transgenic males (n = 18) and AR112Q males (n = 10) revealed delayed onset and reduction of motor deficits associated with SBMA, when compared with non-transgenic littermates (n = 18). While AR112Q males showed significant and progressive deficits in maintaining themselves on an accelerating rotarod at 8, 12 and 16 weeks of age, ARdNLS112Q males performed as well as non-transgenics (Fig. 2A). At 20, 24 and 28 weeks of age ARdNLS112Q males had significantly reduced rotarod function compared with non-transgenics, but still performed substantially better than AR112Q males (Fig. 2A). Female ARdNLS112Q (n = 10) performed as well as non-transgenic littermates (n = 15) until 24 weeks of age, when they had a slight reduction in rotarod performance (Fig. 2B). ARdNLS24Q (n = 15) males did not develop any rotarod deficits up to 28 weeks of age (Supplementary Material, Fig. S1).
Figure 2.
Motor deficits associated with SBMA are ameliorated in ARdNLS112Q mice. Latency to fall from an accelerating rotarod was measured every 4 weeks from 8 to 28 weeks of age in male (A) [# = P < 0.05 between AR112Q males and both non-transgenic (nTG) and ARdNLS112 males only; * = P < 0.05 between all groups] and female (B) transgenic mice. 8 to 28 = age in weeks; Tick numbers on x-axis represent trials 1–4 for each age. Error bars represent standard deviation for each trial for each group. (C) Forepaw and all paw grip strength were measured every 4 weeks from 8 to 28 weeks of age. Error bars represent standard deviation. * = P ≤ 0.05.
Both forepaw and all paw grip strength was significantly reduced in AR112Q male mice beginning at 12 weeks of age, while both measures of grip strength of ARdNLS112Q mice were similar to non-transgenic males (Fig. 2C). At 16, 20 and 24 weeks of age (data not shown), grip strength results resembled those shown at 12 weeks of age. At 28 weeks of age, results of grip strength reflected those of rotarod analysis in that AR112Q males had significantly reduced grip strength compared with both ARdNLS112Q and non-transgenic males and ARdNLS112Q males were somewhat weaker than non-transgenic males. Female ARdNLS112Q mice showed grip strength similar to non-transgenic females up to 28 weeks of age (Fig. 2C).
AR112Q male mice fail to gain weight after 6 months of age (22). Analysis of the present cohort showed a failure of AR112Q male mice to gain weight after 28 weeks of age, while ARdNLS112Q males continued to gain weight in the same manner as non-transgenic male littermates (Supplementary Material, Fig. S2A). Female ARdNLS112Q mice had slightly greater weight gain over time compared with non-transgenic littermates (Supplementary Material, Fig. S2B). ARdNLS24Q male mice also showed no decrease in weight gain (Supplementary Material, Fig. S2C).
Additional tests of motor function, including footprint and rearing analysis, revealed similar results as rotarod and grip strength analysis (data not shown). From 8 to 16 weeks of age, ARdNLS112Q male mice demonstrated a normal gait, while AR112Q males exhibited a wider and shorter gait. From 20 to 28 weeks of age, ARdNLS112Q males had a slightly lower and wider stance compared with nTG males, but their gait was substantially better than that of AR112Q males. Analysis of vertical activity (during a 5-min period) was performed using a Versamax activity monitor (AccuScan Instruments, Columbus, OH). At ages when ARdNLS112Q males performed as well as nTG males on the rotarod, their vertical activity was also normal, while AR112Q males showed significant deficits. After 20 weeks of age, ARdNLS112Q males had decreased vertical activity compared with nTG males but increased vertical activity compared with AR112Q males. Female AR112Q mice had normal gait and vertical activity at all ages evaluated. As previously described, survival of the AR112Q mice was not substantially compromised; survival of ARdNLS112Q mice was also unchanged (data not shown).
ARdNLS112Q has delayed nuclear accumulation and forms oligomers later than AR112Q
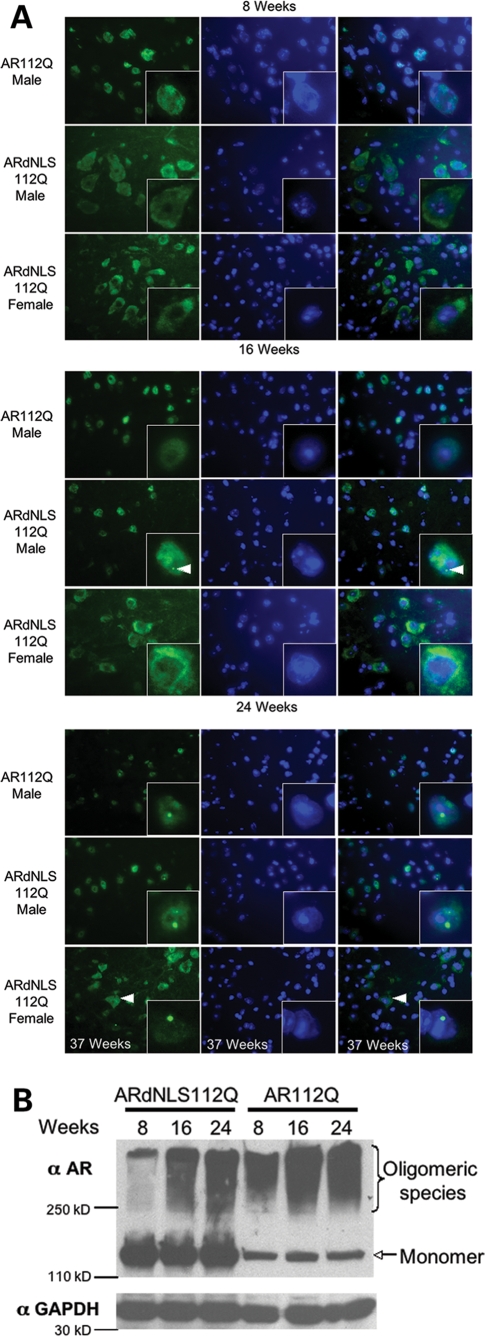
Neuropathological analysis of AR112Q, ARdNLS112Q and non-transgenic male mice was performed at 8, 16 and 24 weeks of age. In AR112Q males at 8 weeks of age, AR112Q protein was localized primarily within nuclei of spinal motor neurons (Fig. 3A and Supplementary Material, Fig. S4) and these males had significant deficits in motor function (Fig. 2A). In contrast, at this age in ARdNLS112Q mice, transgenic AR protein was observed primarily within the cytoplasmic compartment, as observed both by immuofluorescence and fractionation (Fig. 3A and Supplementary Material, Fig. S4) and males had normal motor function (Fig. 2A). In addition, western blot analysis of brain and spinal cord revealed substantially more SDS-insoluble, high-molecular weight oligomeric species of AR112Q than ARdNLS112Q at this age (Fig. 3B). At 16 weeks of age oligomeric species of AR112Q were increased (Fig. 3B), although inclusions were not detected (Fig. 3A). Sixteen week-old male ARdNLS112Q revealed accumulated mutant AR within nuclei (Fig. 3A) and a small proportion of neurons contained small punctate intranuclear inclusions (Fig. 3A). Western analysis revealed oligomeric forms of ARdNLS112Q, although these were substantially less abundant than those from AR112Q mice (Fig. 3B), despite the abundance of AR protein. By 24 weeks of age, AR112Q male mice had a considerable proportion of neurons in the anterior horn of the spinal cord with large intranuclear inclusions (Fig. 3A); as previously shown, inclusions consisted of proteolyzed AR (data not shown) (22). At this age, ARdNLS112Q males also had a significant number of neurons with large intranuclear inclusions of mutant AR (Fig. 3A); intranuclear inclusions of ARdNLS112Q were also composed of fragmented AR, as they lacked the epitopes for antibodies AR441 and ARC-19 (data not shown). Similar results were observed in cortical neurons from these animals (Supplementary Material, Fig. S3A and B). In females, ARdNLS112Q was found largely within the cytoplasm at all ages, although by 37 weeks, a small number of neurons with large nuclear inclusions were observed (Fig. 3A). In males, ARdNLS24Q had also accumulated within neuronal nuclei by 37 weeks of age, but did not form intranuclear inclusions (Supplementary Material, Fig. S5). Inclusions of ARdNLS112Q were confirmed by confocal microscopy to be contained within nuclei (data not shown). In addition, our previous studies revealed decreased immunoreactivity of unphosphorylated neurofilament heavy chain (NF-H) in soma of both spinal motor neurons and neurons of the cerebral cortex (22). This alteration was absent from neurons of ARdNLS112Q mice (data not shown).
Figure 3.
Analysis of AR aggregation in spinal cord. (A) Anterior horn from lumbar spinal cord of 8, 16 and 24 week old mice immunostained for AR (ARH280) and stained with Hoechst to reveal nuclei. Left panel, immunostaining of AR; middle panel, Hoechst staining; right panel, merged image. Arrow in image of 16 week ARdNLS112Q male indicates a small intranuclear inclusion. Arrow in image of 37 week ARdNLS112Q female indicates cytoplasmic AR. (B) Protein lysates from the same spinal cords as in (A) were prepared to evaluate oligomeric species of AR (ARH280) by western blot.
Polyglutamine-expanded ARdNLS fails to cause nuclear aggregation or toxicity in a cell model of SBMA
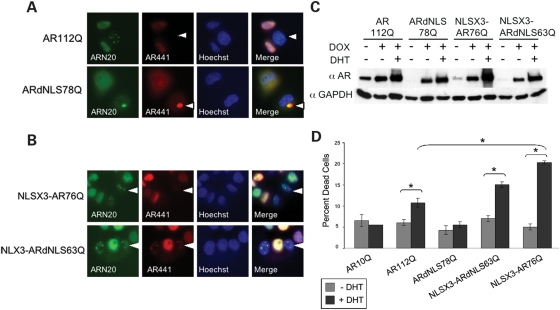
We previously created an inducible cell model of SBMA that expresses full-length human AR with 112 glutamines and forms intranuclear inclusions in response to DHT (27). Notably, as in patients’ tissue, nuclear inclusions in this model consist of proteolyzed N-terminal fragments of AR. In the present studies, we established a cell line expressing ARdNLS78Q to evaluate the metabolism of cytoplasmically retained polyQ-expanded AR in culture. In contrast to AR112Q-expressing cells, ARdNLS78Q-expressing cells showed a diffuse cytoplasmic distribution of AR in the presence of hormone (DHT), and failed to form intranuclear inclusions (Fig. 4A). Over time, these cells formed large cytoplasmic aggregates of full-length AR [detected with antibodies to the N-terminus (AR(N-20))], an internal epitope (AR441) (Fig. 4A) and the C-terminus [AR(C-19)] (data not shown). ARdNLS10Q cells also contained cytoplasmic AR in the presence of DHT and never formed nuclear or cytoplasmic aggregates (data not shown). To confirm that mutant ARdNLS78Q is capable of forming nuclear inclusions, we targeted ARdNLS to the nucleus with an exogenous NLS (NLSX3-ARdNLS63Q). This resulted in the hormone-dependent formation of nuclear inclusions of full-length AR (Fig. 4B and data not shown). In these cell lines, AR was expressed at comparable levels and was stabilized by DHT (Fig. 4C).
Figure 4.
Polyglutamine-expanded ARdNLS fails to produce intranuclear inclusions or toxicity in a cell model of SBMA. (A) Immunofluorescence of stably transfected tet-inducible PC12 cells treated with doxycycline to express either AR112Q or ARdNLS78Q and DHT for 48 h. Cells were immunostained with antibodies to the N-terminus (AR(N-20)), and an internal epitope (AR441) of the AR and stained with Hoechst to reveal nuclei. Arrow in AR112Q panel indicates intranuclear inclusions that lack the epitope for AR441. Arrow in ARdNLS78Q panel indicates cytoplasmic inclusion that contains the epitope for AR441. (B) NLSX3-AR76Q and NLSX3-ARdNLS63Q PC12 cells were treated with doxycycline to express AR in the presence of DHT and immunostained as in (A). Arrow in NLSX3-AR76Q panel indicates intranuclear inclusions that lack the epitope for AR441. Arrow in NLSX3-ARdNLS63Q panel indicates intranuclear inclusions that also contain the epitope for AR441. (C) AR112Q, ARdNLS78Q, NLSX3-AR76Q and NLS-ARdNLS63Q PC12 cells were treated with doxycycline (DOX) to induce AR in the absence or presence of DHT. Cells were harvested after 48 h and evaluated for AR protein levels via western blot analysis. (D) Analysis of PC12 cell toxicity. Cells expressing AR10Q, AR112Q and ARdNLS78Q were treated with doxycycline to express AR in the presence or absence of DHT for 12 days, and cell death determined by trypan blue uptake. Two-hundred cells were counted and the percentage of trypan blue-positive cells determined. Student’s t-tests were performed. * = P ≤ 0.05.
Hormone treatment of AR112Q-expressing cells resulted in toxicity (Fig. 4D). However, polyQ-expanded ARdNLS (ARdNLS78Q)-expressing cells (Fig. 4D) failed to die in response to hormone, indicating that nuclear localization is necessary, not only for AR aggregation, but for cell death as well. Targeting polyQ-expanded ARdNLS to the nucleus (NLSX3-ARdNLS63Q) resulted in DHT-dependent death (Fig. 4D), confirming that the deletion in the NLS does not affect the capacity of the polyQ-expanded AR to confer toxicity when localized to the nucleus. Thus, the possibility that deletion of the NLS alters AR conformation and prevents toxicity for reasons unrelated to its localization is unlikely, due to our finding that nuclear targeting confers on the mutant AR protein the capability of forming inclusions and causing toxicity.
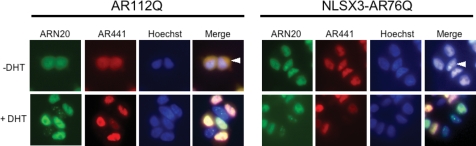
Nuclear localization of polyglutamine-expanded AR is insufficient for aggregation and toxicity
Our results indicate a requirement for nuclear localization in both the nuclear aggregation and toxicity of polyQ-expanded AR. We next sought to determine whether nuclear localization is sufficient for these events. To accomplish this, we created PC12 inducible cell lines that express an AR targeted to the nucleus in the absence of hormone (NLSX3-AR). In the absence of DHT, NLSX3-AR76Q was localized within nuclei, while AR112Q was diffusely distributed within cytoplasm and nuclei (Fig. 5). No intranuclear inclusions of NLSX3-AR76Q were observed in the absence of DHT; inclusions consisting of N-terminal AR fragments were formed exclusively in response to DHT (Fig. 5). These data indicate that nuclear localization of the polyQ-expanded AR is insufficient for nuclear aggregation. Moreover, nuclear localization is insufficient for toxicity, as NLSX3-AR76Q PC12 cells failed to die in the absence of DHT, and only did so in the presence of DHT (Fig. 4C). It was also noted that NLSX3AR, containing an even shorter polyQ-tract (76Q) than non-NLS-tagged AR (112Q), induced a greater level of toxicity, despite equivalent protein levels (Fig. 4C), consistent with a role for the nucleus in mediating toxicity. NLSX3-AR35Q cells showed no aggregation or toxicity in response to DHT treatment (data not shown).
Figure 5.
Nuclear localization of polyglutamine-expanded AR is insufficient for the formation of nuclear inclusions in a cell model of SBMA. Stably transfected tet-inducible PC12 cells were treated with doxycycline to express either AR112Q or NLSX3-AR76Q in the presence or absence of DHT for 48 h. Cells were immunostained with antibodies to the N-terminus (AR(N-20)), and an internal epitope (AR441) of AR and stained with Hoechst to reveal nuclei. White arrow in AR112Q panel indicates diffuse cytoplasmic AR in the absence of DHT. White arrow in NLSX3AR76Q panel indicates diffuse nuclear AR in the absence of DHT. Student’s t-tests were performed. * = P ≤ 0.05.
Primary motor neurons from ARdNLS112Q mice are protected from DHT-dependent toxicity by autophagy
In order to evaluate SBMA motor neuron toxicity in response to DHT, we initiated spinal cord cultures from non-transgenic, AR112Q and ARdNLS112Q transgenic mice. Monomeric levels of both AR112Q and ARdNLS112Q were increased (stabilized) in the presence of DHT; in addition, ARdNLS112Q was expressed at significantly higher levels than AR112Q (Fig. 6A). While DHT caused the loss of ∼40% of AR112Q-expressing motor neurons, it failed to cause the death of ARdNLS112Q-expressing motor neurons (Fig. 6B).
Figure 6.
Primary motor neurons from SBMA mice die in response to hormone treatment while motor neurons from ARdNLS112Q mice survive. (A) Primary motor neuron cultures were initiated from AR112Q and ARdNLS112Q transgenic mouse embryo spinal cords. Cultures were treated with or without DHT for 7 days and protein lysates evaluated by western blot for AR and GAPDH. (B) Cultures were treated as in (A) and additional ARdNLS112Q and AR112Q motor neuron cultures were treated with 3-methyladenine to inhibit autophagy. Ten random fields of immunostained (SMI32) motor neuron cultures were counted under a fluorescent Leica microscope. Counts from three separate wells for each cell line and treatment group were graphed. Student’s t-tests were performed. * =P ≤ 0.05. (C) Cultures were treated as in (A) and immunostained for AR (AR(N-20)) and neurofilament heavy chain (SMI32) to reveal motor neurons. Note the presence of cytoplasmic inclusions of ARdNLS112Q. Nuclear ARdNLS112Q protein is also observed.
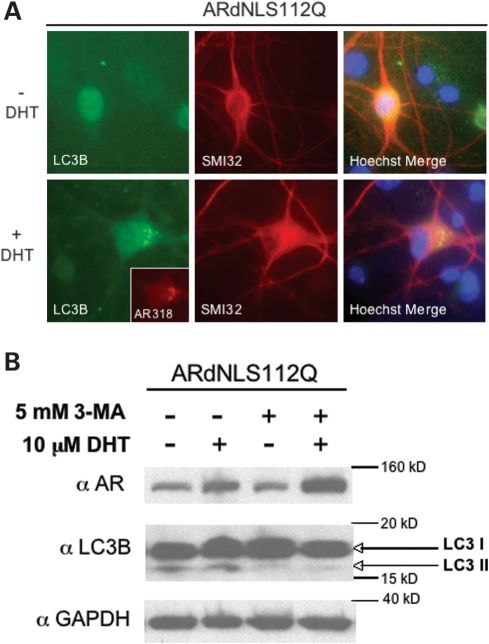
We next sought to determine the mechanism by which ARdNLS112Q motor neurons resist DHT-dependent death. Immunofluorescence staining revealed the presence of cytoplasmic puncta consisting of mutant AR in ARdNLS112Q motor neurons (Fig. 6C). With the knowledge that ARdNLS112Q enters the nucleus with reduced efficiency in the presence of hormone (Figs 3A and 4A), and that it forms cytoplasmic inclusions (Figs 3A and 6C), we considered autophagy to be a likely candidate. It is well established that activation of autophagy is neuroprotective in misfolded protein diseases (reviewed in 32). Therefore, we evaluated the essential autophagy marker LC3B (33) in primary motor neurons. Immunofluorescence analysis of LC3B in ARdNLS112Q motor neurons revealed punctate cytoplasmic staining of LC3B following treatment with DHT (Fig. 7A), indicating the activation of autophagy in these neurons. In addition, LC3B puncta were found to co-localize with AR (Fig. 7A). Punctate staining of LC3B was not detected in nTG or AR112Q motor neurons following DHT treatment (data not shown).
Figure 7.
Autophagy protects ARdNLS112Q motor neurons from DHT-dependent death. (A) Primary motor neuron cultures were initiated from ARdNLS112Q transgenic mouse embryo spinal cords. Cells were treated with or without DHT for 7 days, and immunostained for neurofilament heavy chain (SMI32) to reveal motor neurons, and LC3B (LC3B) to detect autophagosomes. ARdNLS112Q motor neuron shown was double-immunostained for AR (AR-318) and LC3B, then immunostained using SMI32. (B) Additional ARdNLS112Q motor neurons were treated with or without 3-methyladenine (3-MA), to inhibit autophagy, for the last 3 days of the 7-day treatment period with DHT. Protein lysates were analyzed by western blot with antibodies to AR (AR(N-20)), LC3B and GAPDH.
Given the suggestion that autophagy was activated in ARdNLS112Q motor neurons, we determined the role of autophagy in the resistance of ARdNLS112Q motor neurons to DHT-dependent death. To this end, we treated spinal cord cultures with 3-methyladenine (3-MA), a well-known inhibitor of autophagy (34). 3-MA failed to cause toxicity of non-transgenic motor neurons (data not shown); moreover, it did not enhance DHT-dependent toxicity of AR112Q-expressing motor neurons (Fig. 6B). In contrast, 3-MA induced DHT-dependent death of ARdNLS112Q motor neurons (Fig. 6B). Biochemical analysis of protein extracts from these cultures showed a large increase in the monomeric form of ARdNLS112Q following treatment with DHT and 3-MA (Fig. 7B), well above the stabilization of AR seen with DHT alone. In addition, the active form of LC3B (LC3B II) was decreased in the presence of 3-MA (Fig. 7B), validating the inhibitory effects of 3-MA on autophagy.
DHT-dependent death of motor neurons from AR112Q mice is prevented by activation of autophagy
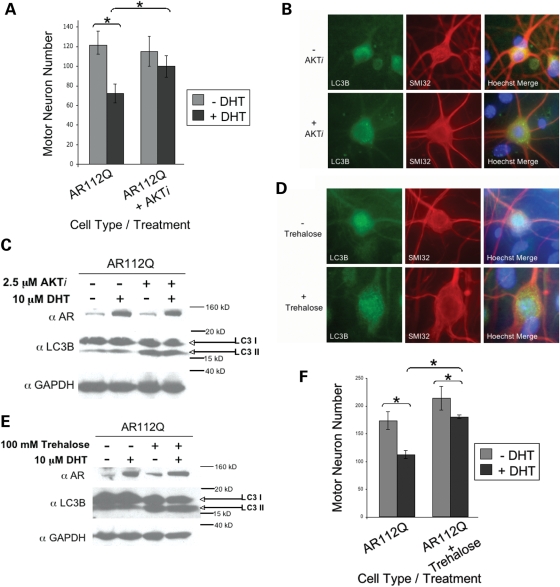
The observation that endogenous autophagy can protect motor neurons from DHT-dependent death when polyQ-expanded AR is retained within the cytoplasm (ARdNLS112Q) confirms the importance of this degradation pathway in clearing misfolded cytoplasmic proteins. We next sought to determine whether pharmacologic activation of autophagy could rescue nuclear polyQ-expanded AR (AR112Q)-expressing motor neurons from DHT-dependent death. We used an AKT inhibitor (AKTi) to activate autophagy in spinal cord cultures from our SBMA (AR112Q) mice. Previous studies demonstrated the ability of the AKT inhibitor, phenoxazine, to activate autophagy in primary neurons expressing mutant huntingtin (Tsvetkov and Finkbeiner, unpublished results). Treatment of AR112Q motor neurons with AKTi for the last 3 days of a 7-day DHT treatment resulted in a substantial rescue from DHT-dependent death (Fig. 8A). As expected, AKTi-treated motor neurons contained cytoplasmic puncta of LC3B (Fig. 8B). Moreover, western analysis of AKTi-treated cultures revealed a significant increase in the active form of LC3B (LC3B II) (Fig. 8C). Non-transgenic motor neuron cultures also showed an increase in LC3B II following treatment with AKTi (data not shown). To confirm these findings, we evaluated another activator of autophagy, trehalose, which was previously shown to activate mTOR-independent autophagy (35) and relieve the neurotoxicity of polyQ-expanded huntingtin (36,37). Treatment of AR112Q spinal cord cultures with trehalose resulted in the formation of LC3B-positive cytoplasmic puncta (Fig. 8D), an increase in LC3B II (Fig. 8E) and rescue from DHT-dependent death (Fig. 8F). Non-transgenic cultures also showed increased LC3B II levels following trehalose treatment (data not shown). No effect on monomeric levels of AR112Q by either autophagy-inducing regimen was observed (Fig. 8C and E).
Figure 8.
DHT-dependent death of motor neurons from SBMA mice is prevented by activation of autophagy. (A) Primary motor neuron cultures were initiated from AR112Q transgenic mouse embryo spinal cords. Cells were treated with or without DHT for 7 days, in the presence or absence of an AKT inhibitor (AKTi) for the last 3 days. Counts from three separate wells for each cell line and treatment group were graphed. AKTi treatment rescued AR112Q motor neurons from DHT-dependent death. (B) Cultures were immunostained for neurofilament-heavy chain (SMI32) to reveal motor neurons and immunostained for LC3B to reveal autophagosomes. (C) Cells treated in parallel to those described in (A) were harvested and protein lysates analyzed by western blot with antibodies against AR (AR(N-20)), LC3B and GAPDH. (D) AR112Q motor neurons were treated with trehalose for the last 3 days of a 7-day treatment period with or without DHT. Motor neurons were immunostained for neurofilament-heavy chain (SMI32) and LC3B. (E) Cells treated as in (D) were harvested and protein lysates analyzed by western blot for AR (AR(N-20)), LC3B and GAPDH. (F) Motor neurons were counted as in (A) following trehalose treatment, and trehalose was found to protect AR112Q motor neurons from DHT-dependent death. Student’s t-tests were performed. * = P ≤ 0.05.
DISCUSSION
A critical role for the nucleus in polyglutamine disease has emerged in recent years. In SBMA, this is evidenced by the presence of inclusions of polyQ-expanded AR within nuclei and the dependence of disease upon androgens, which enable the AR to translocate into the nucleus following binding. Previous studies of Drosophila and mammalian cell culture models to delineate the role of nuclear versus cytoplasmic AR in SBMA have raised questions due to conflicting results (26,31). To clarify the importance of the nucleus in SBMA using mammalian systems, we created transgenic mouse and cell models that express polyQ-expanded AR with a deletion in a portion of its bipartite NLS (amino acids Δ628–640; ARdNLS112Q), to reduce its androgen-dependent nuclear transit. We hypothesized that nuclear localization of the mutant AR is essential for disease and that cytoplasmic retention of mutant AR would be neuroprotective in these models.
We observed that DHT-dependent polyQ-induced toxicity was ameliorated in three mammalian models of SBMA. First, even temporary retention of polyQ-expanded AR within the cytoplasm ameliorated motor deficits in male transgenic mice. At 8 weeks of age, when ARdNLS112Q was localized within the cytoplasm, male mice were completely normal, while AR112Q male mice, with exclusively nuclear AR, displayed substantial motor deficits. With age, older male mice accumulated nuclear ARdNLS112Q, despite mutation of the NLS. This nuclear localization was also observed in male ARdNLS24Q mice, but not in female transgenic mice, demonstrating that ARdNLS is capable of hormone-dependent nuclear translocation, albeit with substantially reduced efficiency. Only when ARdNLS112Q had accumulated within nuclei and formed both oligomeric and aggregated species did male mice begin to display signs of motor deficits, consistent with the previous demonstration that oligomeric AR species precede disease symptoms (38). However, despite the eventual nuclear localization and aggregation of mutant ARdNLS112Q protein, male ARdNLS112Q mice exhibited substantially improved motor function. These results indicate that (i) retention of a significant portion of polyQ-expanded AR within the cytoplasm is sufficient to both delay and ameliorate disease and (ii) nuclear localization enhances the formation of oligomeric AR species that precede motor deficits. In addition to the amelioration of motor deficits in mice by cytoplasmic AR retention, motor neurons from ARdNLS112Q mice were resistant to DHT-dependent death. Finally, our studies in PC12 cells indicate that the mutant AR must enter the nucleus both for nuclear aggregation and toxicity. Therefore, nuclear localization is essential for polyQ-expanded AR to elicit its primary toxic effects.
Complete and efficient nuclear localization of polyQ-expanded AR (AR112Q) caused early, severe and progressive motor deficits in male mice; these deficits were significantly worse than those eventually observed in older ARdNLS112Q mice, which exhibited aggregated nuclear AR. Nuclear accumulation ARdNLS112Q in male mice was somewhat surprising, based on our data in PC12 cells, but it was also not completely unexpected. A similar, but more substantial, deletion of the AR NLS (Δ628–657) allowed partial nuclear entry upon androgen binding (39). These results suggest that an alternative hormone-dependent signal may be utilized in the absence of a functional bipartite NLS. It is also important to note that the ARdNLS112Q likely translocated to the nucleus as full-length monomer rather than as a proteolyzed fragment. In support of this, we observed substantial levels of full-length ARdNLS112Q by western analysis at ages when this protein was visualized within nuclei by immunofluorescence. In addition, we observed the localization of normal ARdNLS24Q within nuclei of male mice in the absence of pathologic inclusions, confirming that full-length ARdNLS is capable of eventual nuclear translocation. It is curious that, despite higher levels of ARdNLS112Q protein and its eventual accumulation within nuclei, ARdNLS112Q mice developed only modest motor impairments with age. Data from PC12 cells expressing expanded ARdNLS demonstrate that this mutant AR is benign when retained within the cytoplasm, but causes substantial toxicity when directed to the nucleus with an exogenous NLS. Thus, in the case of the mice, it may be that simply delaying onset of disease by reducing nuclear transit of mutant AR minimizes its overall impact on neuronal function.
Our finding that ARdNLS112Q formed inclusions earlier than AR112Q was somewhat surprising. The formation of ARdNLS112Q inclusions may be due to the higher levels of the protein, once it has accumulated within nuclei, compared with AR112Q. We also observed that, although ARdNLS112Q formed inclusions earlier than AR112Q (at 16 weeks, with oligomers also present at this time), AR112Q formed oligomers much earlier (8 weeks of age) than ARdNLS112Q. The efficient nuclear localization of AR112Q likely resulted in the earlier formation and sustained presence of oligomers and thus earlier and more substantive disease.
The requirement in SBMA for nuclear mutant AR localization defined by our transgenic mouse and cell culture studies led us to evaluate if nuclear localization is sufficient for disease. Our cell studies revealed that nuclear localization alone is not sufficient for disease, and that androgen binding by the AR is essential for its aberrant metabolism and ability to induce toxicity. Targeting of a polyQ-expanded AR with a shorter polyglutamine tract (76Q) to the nucleus led to toxicity in a hormone-dependent manner. Moreover, we observed enhanced toxicity of this protein over normally trafficked AR112Q, despite the shorter polyglutamine length, confirming the importance of nuclear localization in toxicity.
In SBMA, nuclear inclusions consist of an N-terminal fragment(s) of AR (19,22,27). Fragmented polyQ-expanded proteins have been documented by numerous groups, and may be a result of normal or aberrant protease cleavage, or inefficient processing by the proteasome. These fragments have been shown to be refractory to degradation (40) and are more toxic than intact, full-length, polyglutamine-expanded proteins (22,41–45). In our present studies, the cytoplasmic retention of polyQ-expanded AR led to the formation of large cytoplasmic inclusions that contained full-length AR, unlike the nuclear inclusions of patients’ tissue, which contain only N-terminal AR species (19). When the mutant expanded ARdNLS was directed to the nucleus with an exogenous NLS, intranuclear inclusions were detected that contained the epitope for antibody AR441. It is unclear whether there is any fragmented AR within these aggregates or whether complete loss of the AR441 epitope would occur with more time. Our aggregation studies were carried out after 2 days of hormone treatment, while toxicity was evaluated after 12 days of hormone treatment. In mice, nuclear accumulated ARdNLS112Q was found to form intranuclear inclusions of fragmented AR, and thus we hypothesize that fragmented AR represents the most toxic species. In all, these data suggest that nuclear localization of polyQ-expanded AR is a prerequisite for its proteolysis and nuclear accumulation. This feature places the nucleus at a central point of pathology, the aberrant cleavage of mutant AR to a form that is both toxic and aggregation-prone.
While our studies place the location of mutant AR toxicity in the nucleus, the mechanism by which the polyglutamine-expanded AR confers toxicity within the nucleus is unclear. While AR transcriptional activity is not required for toxicity (24), transcriptional dysregulation occurs in the presence of the mutant AR (46,47). In addition, proteasome function is impaired in mutant AR-expressing cells (our unpublished results) and flies (48). Mitochondrial dysfunction has also been described in the face of nuclear mutant AR (49), concomitant with the altered transcription of genes involved in mitochondrial function. In addition to representing a major site for the toxic cellular sequelae of expanded-polyglutamine AR, the nucleus also represents a major site of altered metabolism of the mutant AR. One of the distinctive features of nuclear mutant AR when compared with cytoplasmic AR is the host of AR post-translational modifications, protein–protein interactions and structural AR changes that occur in response to hormone binding (50,51). Our current and future studies will address alterations in these pathways and their role in nuclear polyQ-expanded AR toxicity.
Upon determining that a critical role for the nucleus exists in SBMA pathogenesis, we investigated the mechanistic basis for the neuroprotective role of cytoplasmic retention of polyQ-expanded AR. Previous studies have revealed failure of the proteasome to efficiently and appropriately degrade misfolded proteins, specifically those containing polyQ expansions (40), unless chaperone-mediated therapies are initiated (52–59). In addition, it has become increasingly clear that the ubiquitin proteasome system is reduced in neuronal nuclei compared with the cytoplasmic compartment (60,61), suggesting one explanation for the differential toxicities conferred by misfolded proteins in these two locations. However, another important and emerging feature of cytoplasmic localization of misfolded proteins is their availability to activate a second method of degradation, the autophagic/lysosomal pathway, which has been shown to degrade polyQ-expanded proteins (62). When pharmacologically activated, autophagy can effectively degrade misfolded proteins and is neuroprotective (reviewed by 32,35).
Our studies of cultured, transgenic motor neurons revealed that ARdNLS112Q motor neurons failed to die in response to DHT (Fig. 6). The observation of LC3 puncta indicates that autophagy was activated in these motor neurons. Furthermore, the inhibition of autophagy led to DHT-dependent toxicity, indicating that the cytoplasmic mutant AR is capable of causing toxicity when autophagy is inhibited. Finally, the increase in mutant AR protein upon autophagy inhibition supports our conclusion that the mutant AR is, at least in part, degraded by this pathway. While inhibition of autophagy caused a substantial increase in DHT-dependent toxicity in ARdNLS112Q motor neurons, it had no effect on the toxicity of AR112Q motor neurons (Fig. 7B) or of non-transgenic motor neurons (data not shown), indicating that endogenous autophagy does not substantially modulate toxicity in AR112Q motor neurons, in which AR112Q is confined to the nucleus. Thus, the results of our studies shown here establish that the differential toxicity of nuclear versus cytoplasmic mutant AR can be explained, in part, by the differential activation of, and AR degradation by, autophagy.
The data presented here reveal that cytoplasmically retained polyQ-expanded AR (ARdNLS112Q) can be degraded by autophagy, protecting motor neurons from DHT-dependent death. The high levels of ARdNLS112Q protein, even in the face of robust and efficient autophagic degradation, are consistent with the increased transgene copy number in ARdNLS112Q mice. Despite this increased protein, however, ARdNLS112Q mice showed reduced motor symptoms. Thus, the increased ARdNLS112Q protein in the cytoplasm represents a form that is less toxic than nuclear-confined AR. Whether this form is non-toxic due to its lack of amino-terminal fragment-producing proteolysis or to other aspects of AR metabolism that occur within the nucleus is an active area of investigation. In all, our observations indicate one mechanism by which cytoplasmic retention of polyQ-expanded AR is neuroprotective; the mutant protein is available to be degraded by autophagy. In accordance, nuclear localization of polyQ-expanded AR likely limits its access to the autophagic pathway and thus is one mechanism by which this localization contributes to its toxic effects within motor neurons.
The potent neuroprotective effects of autophagy in ARdNLS112Q motor neurons led us to evaluate whether enhanced activation of the autophagic pathway would protect neurons from a nuclear localized polyQ-expanded protein, AR112Q. Pharmacologic induction of both mTOR-dependent and -independent pathways of autophagy rescued AR112Q motor neurons from DHT-dependent death. This intervention, however, had no effect on monomeric levels of AR112Q. This lack of an effect on AR112Q levels is similar to findings of a previous study in which autophagy was ineffective at eliminating nuclear inclusions of mutant protein (63), but is contrary to results in a fly model of SBMA, in which HDAC6 over-expression (which enhances autophagy) led to lower steady-state levels of monomeric and aggregated polyQ-expanded AR (48). It may be that, while monomeric AR was unchanged in our study, oligomeric and nuclear aggregated forms of AR112Q were altered; these species were not evaluated in our spinal cord culture model due to difficulties with their detection. This would be in keeping with earlier studies showing that nuclear aggregates may be dynamic in nature (64–66). Alternatively, the effects of autophagy on motor neuron viability may be independent of direct effects on mutant AR. It may be that activation of autophagy alleviates proteasomal inhibition induced by mutant AR, in turn enhancing cell viability, as described by Pandey et al. (48). It may also be that autophagy plays a more general role, relieving proteotoxic stress induced by polyQ-expanded nuclear AR, perhaps by promoting the autophagic degradation of misfolded metastable proteins (67).
In all, these findings indicate that hormone binding and nuclear localization are essential for the polyQ-expanded AR to aggregate and induce toxicity within motor neurons. Therefore, nuclear hormone-dependent AR events will be key in understanding the specific modifications, interactions and metabolic products responsible for causing disease. Although hormone withdrawal has proved neuroprotective in mouse models of SBMA, its effects in SBMA patients have yet to be firmly established. Moreover, it is expected that therapies directed at the specific events that lead to the formation of a toxic AR species within motor neurons will prove to be more beneficial to patients and cause less side effects, as they will allow for normal AR function that is otherwise interrupted by hormone withdrawal. The studies herein highlight a need to focus on the nucleus in SBMA, as well as on the autophagic pathway when developing these therapies.
MATERIALS AND METHODS
ARdNLS and NLSX3-AR inducible PC12 cell lines
Site-directed mutagenesis (Quick Change II XL, Stratagene) was performed on a pTRE plasmid (Clontech, Mountain View, CA), previously engineered to contain full-length human AR cDNA, bearing 10 or 112 CAGs, to delete the nucleic acids encoding amino acids 628–640 of the AR (within the NLS) (ARdNLS). Mutation and CAG repeat length were confirmed by sequence analysis.
NLSX3-AR was created as follows: The SV40 NLS in triplicate (NLSX3) was PCR-amplified from pShooter™ pEF/myc/nuc (Invitrogen, Carlsbad, CA) vector, and an EcoRI restriction digest site engineered on both the 5′ and 3′ ends. An NheI restriction was also engineered just upstream of the EcoRI site at the 5′ end. The PCR product was cloned into plasmid pCMVAR (16-CAG)ΔHA (9) (EcoRI site is just 5′ of the CTG start of the AR cDNA). The pCMV-NLSX3-AR (16-CAG)ΔHA was then digested with NheI and NarI, and pTRE-AR (112-CAG) was linearized with NheI and partially digested with NarI. The NLSX3-AR fragment from pCMVAR(16-CAG)ΔHA was ligated to pTRE-AR(112-CAG) (containing full length AR), resulting in pTRE-NLSX3-AR(112-CAG). ARdNLS was then cloned into this construct using NruI and BstBI. All constructs were sequenced to verify mutation and CAG length.
Stable transfections of Tet-On PC12 cells (Clontech) were performed using LipofectAMINE Plus (Invitrogen) with a plasmid conferring hygromycin resistance (pTK-hygromycin). Stable transformants were selected with 200 µg/ml hygromycin. Single colonies were isolated and expanded and screened for doxycycline-inducible AR protein expression by slot blot and western blot analysis using AR(N-20) antibody (Santa Cruz, Santa Cruz, CA). AR expression levels were adjusted with various doxycycline concentrations to achieve protein levels equivalent to AR112Q PC12 cells. Genomic DNA was extracted from each clone to verify mutation and CAG length via sequence analysis. Cells were maintained in normal growth media [Dulbecco’s modified Eagle’s medium with 10% heat-inactivated horse serum, 5% fetal bovine serum, 2 mm L-glutamine, 100 units/ml penicillin/streptomycin, 200 µg/ml hygromycin (Invitrogen) and 100 µg/ml G418 (Mediatech, Manassas, VA)] at 37°C, 10% CO2.
Treatment of inducible PC12 cell lines
Stable Tet-On PC12 cell lines were treated with doxycycline to express AR for various times and with various concentrations of DHT in charcoal-stripped serum-containing cell-culture media.
PC12 cell toxicity assay
Stable Tet-On PC12 cell lines (AR10Q, AR112Q, ARdNLS78Q, NLSX3-AR76Q and NLSX3-ARdNLS63Q) were treated with doxycycline to express equivalent levels of AR, in the absence and presence of 10 nM DHT for 12 days. At the end of treatment, cells were harvested and stained with trypan blue. Two hundred cells were counted and the percentage of trypan blue-positive cells determined. Significance was determined with Student’s t-test.
ARdNLS PC12 cell and transgenic mouse constructs
The human AR gene, bearing either 24 CAGs (normal) or 112 CAGs (expanded), was previously cloned into the prion protein promoter (PrP) construct deleted of coding sequences (22). The deleted portion of the nuclear localization sequence of the AR (deleted of nucleic acids encoding amino acids 628–640) was cloned into the NruI and BstBI sites of both the 24 CAG and 112 CAG containing PrP-AR constructs. DNAs were linearized and the plasmid backbone (pBS) removed by digestion with NotI, gel-purified and injected into fertilized oocytes (C57Bl/6), by the Kimmel Cancer Center Transgenic Facility at Thomas Jefferson University. Both ARdNLS and AR112Q mice were maintained on a C57Bl/6 background (Charles River, Wilmington, MA). Founders were screened by genotyping tail clips and brain and spinal cord lysates from 5-week-old male mice were analyzed for ARdNLS protein expression compared with those from age-matched AR112Q SBMA male mice. Additionally, CAG repeat length was determined by sequence analysis of PCR products (Laragene, Inc., Los Angeles, CA).
Genotyping mice
DNA from mice was prepared from tail or ear biopsies using Red Extract-N-Amp Kit (Sigma). Transgenic animals were identified by PCR of the human AR: forward primer from the PrP promoter region (5′-ACTGAACCATTTCAACCGAGC-3′) coupled with a reverse primer from the AR sequence 5′ to the CAG repeat (5′AGGTGCTGCGCTCGCGGCCTCT-3′).
Western blot analysis
Freshly dissected tissue was flash-frozen in liquid nitrogen. Frozen sections were pulverized in a mortar and pestle on dry ice and homogenized in either 10 volumes of Triton-DOC buffer (1% sodium deoxycholate and 0.5% Triton X-100 in PBS with protease inhibitors) or RIPA buffer (50 mM Tris–HCl, pH 8.0, 0.15 M NaCl, 0.1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitors). PC12 cells and cells from primary spinal cord cultures were lysed with Triton-DOC buffer. All lysates were sonicated three times for 10 s using a Branson cup sonifier. A portion of tissue lysates in RIPA was centrifuged at 15 000g for 5 min at 4°C for detection of oligomeric species of AR (38). A DC protein assay (BioRad, Hercules, CA) was performed to determine protein concentration and lysates were electrophoresed by SDS–PAGE and transferred to 0.45 µm PVDF (Immobilon-P). Western hybridization was performed using the following antibodies: AR(N-20), GAPDH (1:1000) (Santa Cruz Biotechnology) and LC3B (1:500) (NB600-1384) (Novus Biologicals, Littleton, CO). Detection was performed with ECL (Amersham Biosciences, Arlington Heights, IL).
Behavioral analysis
Every 4 weeks, beginning at 8 weeks of age, an age-matched cohort of ARdNLS24Q males, AR112Q males, ARdNLS112Q males, ARdNLS112Q females, non-transgenic males and non-transgenic females was subject to various measures of motor function. Mice were tested during the light phase of a 12 h light/dark cycle for their latency to fall off a steadily accelerating rotarod (4–40 rpm over 10 min) (Ugo Basile, Comerio, VA-Italy). During the first week of testing, mice were tested four times per day for 3 consecutive days. The first 2 days constituted a learning period and data collected on the third day were used for analysis. For subsequent testing sessions, mice were only subjected to rotarod for 1 day, as statistical analysis revealed no loss of statistical power under this regimen (unpublished data). Mice were allowed a rest period of at least 15 min between testing sessions. Scores were analyzed by two-way repeated measures ANOVA with a Tukey post hoc analysis using SigmaStat 3.0 software (SPSS Inc., Chicago, IL). A grip strength meter (Columbus Instruments, Columbus, OH) was used to measure the force exerted by a mouse as it was pulled across a grid by its tail. Grip strength was measured for forepaws only or hindpaws and forepaws together. Six measures were taken for both measures of grip strength and the lowest and highest scores for each animal dropped. An average for each animal was used for statistical analysis. Significance was determined with two-way repeated measures ANOVA and a Tukey post hoc analysis (SigmaStat).
Primary motor neuron cultures
Dissociated spinal cord cultures were established according to Roy et al. (68). In brief, spinal cords were dissected on ice from 13.5-day-old embryos in dissection media (0.1% dextrose, 2% sucrose, 1.4 mm NaCl, 5.4 mm KCl, 0.17 mm Na2HPO4, 22 µM KH2PO4, 9.9 mm HEPES). Genotyping was performed from a tail biopsy. Transgenic AR112Q, ARdNLS112Q or non-transgenic spinal cords were pooled separately, plated for culture and incubated for 3 weeks in media conditioned by glial culture from 13.5-day-old non-transgenic brain [MEM, 35 mm NaHCO3, 0.5% dextrose, 1% N3, 10 nM 2.5 S NGF (added after conditioning)]. During the development of this culture system, motor neurons were identified using antibodies to choline acetyltransferase, neuron-specific enolase and neurofilament heavy chain (SMI32). Motor neurons were identified to have much larger cell bodies relative to other spinal neurons and large, tapering, highly branched dendrites with a fibrillar appearance. In our experiments presented here, SMI32 immunoreactivity and morphology were used to identify motor neurons. Three weeks after initiation, cultures were treated with or without 10 µM DHT for 7 days. Additional reagents/drugs were administered for the last 3 days of the 7 day treatment period [5 mm 3-Methyladenine (3-MA), 100 mm Trehalose (Sigma), 2.5 µM AKT inhibitor X [10-(4′-N-diethylamino)butyl)-2-chlorophenoxazine] (AKTi) (Calbiochem, San Diego, CA)]. Three wells of each motor neuron culture line and treatment group were immunostained as described in what follows. Motor neurons were determined by SMI32 stain and morphology and counted. Significance was determined by a Student’s t-test.
Immunofluorescence
Dissected fresh whole brain and spinal cord were frozen in OCT, then sectioned with a cryostat (7 µm). Tissue sections, motor neuron cultures and PC12 cells were fixed with 4% paraformaldehyde for 10 min, washed in PBS, permeabilized with 0.3% Triton X-100 for 15 min (cells only), blocked in 1.5% goat serum (Jackson ImmunoResearch, West Grove, PA) for 20 min and incubated for 60 min in primary antibody diluted in 1.5% goat serum. Tissue sections or cells were washed in PBS and incubated for 30 min with secondary antibodies (FITC- or Texas Red-conjugated) (Jackson ImmunoResearch, West Grove, PA), washed in PBS, incubated for 10 min with Hoechst (2 µg/ml), washed in PBS and mounted in Vectashield (Vector Laboratories, Burlingame, CA). Fluorescence was visualized with a Leica (Leica Microsystems GmbH, Wetzlar, Germany) microscope; images were captured with a Leica camera and compiled with IP Lab software (BD Biosciences, Rockville, MD). Antibodies used include AR(N-20), ARH280, AR441 (1:100) (Santa Cruz), AR-318 (Vector Laboratories Burlingame, CA), SMI32 (1:1,000) (Sternberger Monoclonal, Baltimore, MD) and LC3B (NB600-1384) (1:200) (Novus Biologicals).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (NS047381 and NS32214 to D.E.M.); (2NS045191 and 2P01AG022074 to S.F.); (DK07705 supporting H.L.M.); The Taube-Koret Center for Huntington's Disease Research (S.F.); and a Milton Wexler Award and Fellowship from the Hereditary Disease Foundation (A.T.).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Carlisle Landel, Ph.D., Director, Transgenic and Gene Targeting Facility at Thomas Jefferson University for creation of transgenic mice and for thoughtful discussions. We also thank Heather Durham, Ph.D., Montreal Neurological Institute, McGill University, Montreal, for helpful advice on the initiation of spinal cord cultures.
Conflict of Interest statement. None declared.
REFERENCES
- 1.La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;353:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy W.R., Alter M., Sung J.H. Progressive proximal spinal and bulbar muscular atrophy of late onset: A sex-linked recessive trait. Neurology. 1968;18:671–680. doi: 10.1212/wnl.18.7.671. [DOI] [PubMed] [Google Scholar]
- 3.Sobue G., Hashizume Y., Mukai E., Hirayama M., Mitsuma T., Takahashi A. X-linked recessive bulbospinal neuronopathy: a clinicopathological study. Brain. 1989;112:209–232. doi: 10.1093/brain/112.1.209. [DOI] [PubMed] [Google Scholar]
- 4.Antonini G., Gragnani F., Romaniello A., Pennisi e.M., Morino S., Ceschin V., Santoro L., Cruccu G. Sensory involvement in spinal-bulbar muscular atrophy (Kennedy’s disease) Muscle Nerve. 2000;2:252–258. doi: 10.1002/(sici)1097-4598(200002)23:2<252::aid-mus17>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Li M., Sobue G., Doyu M., Mukai E., Hashizume Y., Mitsuma T. Primary sensory neurons in X-linked recessive bulbospinal neuropathy: histopathology and androgen receptor gene expression. Muscle Nerve. 1995;3:301–308. doi: 10.1002/mus.880180306. [DOI] [PubMed] [Google Scholar]
- 6.Quigley C.A., Friedman K.J., Johnson A., Lafreniere R.G., Silverman L.M., Lubahn D.B., Brown T.R., Wilson E.M., Willard H.F., French F.S. Complete deletion of the androgen receptor gene: definition of the null phenotype of the androgen insensitivity syndrome and determination of carrier status. J. Clin. Endo. Metab. 1992;74:927–933. doi: 10.1210/jcem.74.4.1347772. [DOI] [PubMed] [Google Scholar]
- 7.Thomas P.S., Jr, Fraley G.S., Damian V., Woodke L.B., Zapata F., Sopher B.L., Plymate S.R., La Spada A.R. Loss of endogenous androgen receptor protein accelerates motor neuron degeneration and accentuates androgen insensitivity in a mouse model of X-linked spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2006;14:2225–2238. doi: 10.1093/hmg/ddl148. [DOI] [PubMed] [Google Scholar]
- 8.Diamond M.I., Robinson M.R., Yamamoto K.R. Regulation of expanded polyglutamine protein aggregation and nuclear localization by the glucocorticoid receptor. Proc. Natl Acad. Sci. USA. 2000;97:657–661. doi: 10.1073/pnas.97.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merry D.E., Kobayashi Y., Bailey C.K., Taye A.A., Fischbeck K.H. Cleavage, aggregation, and toxicity of the expanded androgen receptor in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 1998;7:693–701. doi: 10.1093/hmg/7.4.693. [DOI] [PubMed] [Google Scholar]
- 10.Stenoien D.L., Cummings C.J., Adams H.P., Mancini M.G., Patel K., DeMartino G.N., Marcelli M., Weigel N.L., Mancini M.A. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum. Mol. Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 11.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 12.Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 13.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 14.Benn C.L., Landles C., Li H., Strand A.D., Woodman B., Sathasivam K., Li S.H., Ghazi-Noori S., Hockly E., Faruque S.M., et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum. Mol. Genet. 2005;20:3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt T., Landwehrmeyer G.B., Schmitt I., Trottier Y., Auburger G., Laccone F., Klockgether T., Volper M., Epplen J.T., Schols L., et al. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 1998;8:669–679. doi: 10.1111/j.1750-3639.1998.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skinner P.J., Koshy B.T., Cummings C.J., Klement I.A., Helin K., Servadio A., Zoghbi H.Y., Orr H.T. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 17.Bichelmeier U., Schmidt T., Hubener J., Boy J., Ruttiger L., Habig K., Poths S., Bonin M., Knipper M., Schmidt W.J., et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J. Neurosci. 2007;28:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 19.Li M., Miwa S., Kobayashi Y., Merry D.E., Yamamoto M., Tanaka F., Doyu M., Hashizume Y., Fischbeck K.H., Sobue G. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann. Neurol. 1998;44:249–254. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 20.Adachi H., Katsuno M., Minamiyama M., Waza M., Sang C., Nakagomi Y., Kobayashi Y., Tanaka F., Doyu M., Inukai A., et al. Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain. 2005;57:236–251. doi: 10.1093/brain/awh381. [DOI] [PubMed] [Google Scholar]
- 21.Becker M., Elke M., Schneikert J., Krug H.F., Cato A.C.B. Cytoplasmic localization and the choice of ligand determine aggregate formation by androgen receptor with amplified polyglutamine stretch. J. Cell Biol. 2000;149:255–262. doi: 10.1083/jcb.149.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chevalier-Larsen E.S., O’Brien C.J., Wang H., Jenkins S.C., Holder L., Lieberman A.P., Merry D.E. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci. 2004;24:4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darrington R.S., Butler R., Leigh P.N., McPhaul M.J., Gallo J.M. Ligand-dependent aggregation of polyglutamine-expanded androgen receptor in neuronal cells. Neuroreport. 2002;13:2117–2120. doi: 10.1097/00001756-200211150-00025. [DOI] [PubMed] [Google Scholar]
- 24.Katsuno M., Adachi H., Doyu M., Minamiyama M., Sang C., Kobayashi Y., Inukai A., Sobue G. Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy. Nat. Med. 2003;9:768–773. doi: 10.1038/nm878. [DOI] [PubMed] [Google Scholar]
- 25.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 26.Takeyama K., Ito S., Yamamoto A., Tanimoto H., Furutani T., Kanuka H., Miura M., Tabata T., Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 27.Walcott J.L., Merry D.E. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J. Biol. Chem. 2002;277:50855–50859. doi: 10.1074/jbc.M209466200. [DOI] [PubMed] [Google Scholar]
- 28.Bohen S.P., Kralli A., Yamamoto K.R. Hold ‘em and fold ‘em: chaperones and signal transduction. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. [DOI] [PubMed] [Google Scholar]
- 29.Caplan A.J., Langley E., Wilson E.M., Vidal J. Hormone-dependent transactivation by the human androgen receptor is regulated by a dnaJ protein. J. Biol. Chem. 1995;270:5251–5257. doi: 10.1074/jbc.270.10.5251. [DOI] [PubMed] [Google Scholar]
- 30.Furutani T., Takeyama K., Tanabe M., Koutoku H., Ito S., Taniguchi N., Suzuki E., Kudoh M., Shibasaki M., Shikama H., et al. Human expanded polyglutamine androgeon receptor mutants in neurodegeneration as a novel ligand target. JPET. 2005;315:545–552. doi: 10.1124/jpet.105.087643. [DOI] [PubMed] [Google Scholar]
- 31.Morfini G., Pigino G., Szebenyi G., You Y., Pollema S., Brady S.T. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat. Neurosci. 2006;7:907–916. doi: 10.1038/nn1717. [DOI] [PubMed] [Google Scholar]
- 32.Ravikumar B., Sarkar S., Rubinsztein D.C. Clearance of mutant aggregate-prone proteins by autophagy. Methods Mol. Biol. 2008;445:195–211. doi: 10.1007/978-1-59745-157-4_13. [DOI] [PubMed] [Google Scholar]
- 33.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;2:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seglen P.O., Gordon P.B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA. 1982;6:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkar S., Rubinsztein D.C. Huntington’s disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;17:4263–4270. doi: 10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 36.Sarkar S., Davies J.E., Huang Z.Q., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;8:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka M., Machida Y., Niu S., Ikeda T., Jana N.R., Doi H., Kurosawa M., Nekooki M., Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 2004;2:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 38.Li M., Chevalier-Larsen E.S., Merry D.E., Diamond M.I. Soluble androgen receptor oligomers underlie pathology in a mouse model of SBMA. J. Biol. Chem. 2007;5:3157–3164. doi: 10.1074/jbc.M609972200. [DOI] [PubMed] [Google Scholar]
- 39.Simental J.A., Sar M., Lane M.V., French F.S., Wilson E.M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 40.Holmberg C.I., Staniszewski K.E., Mensah K.N., Matouschek A., Morimoto R.I. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abel A., Walcott J., Woods J., Duda J., Merry D.E. Expression of expanded repeat androgen receptor produces neurologic disease in transgenic mice. Hum. Mol. Genet. 2001;10:107–116. doi: 10.1093/hmg/10.2.107. [DOI] [PubMed] [Google Scholar]
- 42.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 43.Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A., et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 44.Hodgson J.G., Agopyan N., Gutekunst C.A., Leavitt B.R., LePiane F., Singaraja R., Smith D.J., Bissada N., McCutcheon K., Nasir J., et al. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 45.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;6:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 46.Lieberman A.P., Harmison G., Strand A.D., Olson J.M., Fischbeck K.H. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 2002;11:1967–1976. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 47.Luthi-Carter R., Strand A.D., Hanson S.A., Kooperberg C., Schilling G., La Spada A.R., Merry D.E., Young A.B., Ross C.A., Borchelt D.R., et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington’s disease mouse models reveal context-independent effects. Hum. Mol. Genet. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- 48.Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 49.Ranganathan S., Harmison G.G., Meyertholen K., Pennuto M., Burnett B.G., Fischbeck K.H. Mitochondrial abnormalities in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2008;18:27–42. doi: 10.1093/hmg/ddn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Black B.E., Vitto M.J., Gioeli D., Spencer A., Afshar N., Conaway M.R., Weber M.J., Paschal B.M. Transient, ligand-dependent arrest of the androgen receptor in subnuclear foci alters phosphorylation and coactivator interactions. Mol. Endocrinol. 2004;18:834–850. doi: 10.1210/me.2003-0145. [DOI] [PubMed] [Google Scholar]
- 51.Faus H., Haendler B. Post-translational modifications of steroid receptors. Biomed. Pharmacother. 2006;9:520–528. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- 52.Adachi H., Katsuno M., Minamiyama M., Sang C., Pagoulatos G., Angelidis C., Kusakabe M., Yoshiki A., Kobayashi Y., Doyu M., et al. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J. Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adachi H., Waza M., Tokui K., Katsuno M., Minamiyama M., Tanaka F., Doyu M., Sobue G. CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular transgenic mouse model. J. Neurosci. 2007;19:5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey C.K., Andriola I.F., Kampinga H.H., Merry D.E. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 55.Chai Y., Koppenhafer S.L., Bonini N.M., Paulson H.L. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J. Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cummings C.J., Mancini M.A., Antalffy B., DeFranco D.B., Orr H.T., Zoghbi H.Y. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nature Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 57.Cummings C.J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H.T., Dillmann W.H., Zoghbi H.Y. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 58.Ishihara K., Yamagishi N., Saito Y., Adachi H., Kobayashi Y., Sobue G., Ohtsuka K., Hatayama T. Hsp105alpha suppresses the aggregation of truncated androgen receptor with expanded CAG repeats and cell toxicity. J. Biol. Chem. 2003;278:25143–25150. doi: 10.1074/jbc.M302975200. [DOI] [PubMed] [Google Scholar]
- 59.Katsuno M., Sang C., Adachi H., Minamiyama M., Waza M., Tanaka F., Doyu M., Sobue G. Pharmacological induction of heat-shock proteins alleviates polyglutamine-mediated motor neuron disease. Proc. Natl Acad. Sci. USA. 2005;102:16801–16806. doi: 10.1073/pnas.0506249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tydlacka S., Wang C.E., Wang X., Li S., Li X.J. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J. Neurosci. 2008;28:13285–13295. doi: 10.1523/JNEUROSCI.4393-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou H., Cao F., Wang Z., Yu Z.X., Nguyen H.P., Evans J., Li S.H., Li X.J. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J. Cell Biol. 2003;163:109–118. doi: 10.1083/jcb.200306038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 63.Iwata A., Christianson J.C., Bucci M., Ellerby L.M., Nukina N., Forno L.S., Kopito R.R. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chai Y., Shao J., Miller V.M., Williams A., Paulson H.L. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl Acad. Sci. USA. 2002;14:9310–9315. doi: 10.1073/pnas.152101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S., Nollen E.A., Kitagawa K., Bindokas V.P., Morimoto R.I. Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 66.Stenoien D.L., Mielke M., Mancini M.A. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 2002;4:806–810. doi: 10.1038/ncb859. [DOI] [PubMed] [Google Scholar]
- 67.Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;5766:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 68.Roy J., Minotti S., Dong L., Figlewicz D.A., Durham H.D. Glutamate potentiates the toxicity of mutant Cu/Zn-superoxide dismutase in motor neurons by postsynaptic calcium-dependent mechanisms. J. Neurosci. 1998;18:9673–9684. doi: 10.1523/JNEUROSCI.18-23-09673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.