Abstract
The structural and dynamical properties of the metal-free form of WT human superoxide dismutase 1 (SOD1) and its familial amyotrophic lateral sclerosis (fALS)-related mutants, T54R and I113T, were characterized both in solution, through NMR, and in the crystal, through X-ray diffraction. We found that all 3 X-ray structures show significant structural disorder in 2 loop regions that are, at variance, well defined in the fully-metalated structures. Interestingly, the apo state crystallizes only at low temperatures, whereas all 3 proteins in the metalated form crystallize at any temperature, suggesting that crystallization selects one of the most stable conformations among the manifold adopted by the apo form in solution. Indeed, NMR experiments show that the protein in solution is highly disordered, sampling a large range of conformations. The large conformational variability of the apo state allows the free reduced cysteine Cys-6 to become highly solvent accessible in solution, whereas it is essentially buried in the metalated state and the crystal structures. Such solvent accessibility, together with that of Cys-111, accounts for the tendency to oligomerization of the metal-free state. The present results suggest that the investigation of the solution state coupled with that of the crystal state can provide major insights into SOD1 pathway toward oligomerization in relation to fALS.
Keywords: amyotrophic lateral sclerosis, NMR, X-ray, mobility, H2O/D2O exchange
More than 100 different variants of human copper-zinc superoxide dismutase (Cu2Zn2 SOD) have been identified and linked to the neurodegenerative disease familial amyotrophic lateral sclerosis (fALS) by a gain-of-function mechanism (1, 2). Although the mechanism of the toxicity is unknown, aberrant SOD1 protein oligomerization has been strongly implicated in disease causation (3, 4). Several recent publications (5, 6) have presented compelling evidence that in vivo abnormal disulfide cross-linking of ALS mutant SOD1 plays a role in this oligomerization, and disulfide-linked SOD1 multimers have been detected mainly in mitochondria of neuronal tissues of SOD1-linked fALS patients and transgenic mice (7–9).
WT human SOD1 is an exceptionally stable, homodimeric 32-kDa protein, located mainly in the cytoplasm, but it is also present in the peroxisomes, the mitochondrial intermembrane space, and the nucleus of eukaryotic cells (10, 11). Each subunit of the dimer binds 1 copper and 1 zinc ion and folds as an 8-stranded Greek-key β-barrel that is stabilized by an intrasubunit disulfide bond (Cys-57, Cys-146) near the active site (12). In vivo, in the highly reducing cytoplasm environment, the existence of this intrasubunit disulfide bond points to its very low reduction potential.
In addition to the 2 cysteines involved in the formation of the intramolecular disulfide bond, 2 reduced cysteines, Cys-6 and Cys-111, are located on β-strand 1 and loop VI of WT human SOD1, respectively. Among the loops connecting the 8 β-strands, 2 have structural and functional roles. The electrostatic loop (loop VII, residues 121–144) contains charged residues that contribute to guiding the negatively-charged superoxide substrate toward the catalytic copper site. The long zinc loop (loop IV, residues 49–84) contains all of the zinc binding residues.
We have recently reported (13, 14) that oxidized WT SOD1 and several of its mutants, only when they are in the metal free form (apo), give rise, in vitro, to soluble oligomers under aerobic conditions when the proteins are kept at 37 °C and at a concentration and pH close to physiological, i.e., 100 μM and pH 7. The resulting soluble oligomers are formed by intermolecular disulfide covalent bonds, involving Cys-6 and Cys-111, and by noncovalent interactions between β-strands, forming amyloid-like structures capable of binding Thioflavin T (14). The rates of protein oligomerization are different for the various mutants, but eventually they give rise to the same type of soluble oligomeric species.
SOD1 enters as apoprotein the mitochondria, which are more oxidizing cellular compartments (15, 16). The soluble oligomeric species, formed through an oxidative process, might represent the precursor toxic species, whose existence would also suggest a common mechanism for ALS and fALS.
To investigate the mechanism for SOD1 oligomerization, the structural and dynamic features of the metal-free state of SOD1 for both WT and some ALS-related mutants were characterized both in solution, through NMR, and in the crystal, through X-ray diffraction. We found that the metal-free state is significantly disordered in the crystal for 2 pathogenic SOD1 mutants as reported for the WT protein (17, 18), at variance with that observed in the metalated state. In solution, the highly-disordered and dynamical metal-free state allows the free cysteines to become accessible for oxidation and subsequent oligomerization, at variance with what occurs in the metal-bound form.
Results and Discussion
In the present study we characterized the apo form of WT SOD1 and 2 ALS-related mutants with 2 extreme behaviors in terms of oligomerization rates: T54R oligomerizes with rates slightly slower than WT SOD1, whereas I113T has an oligomerization rate more than twice that of WT SOD1 (14).
In the crystal structures of the metal-free form of WT SOD1 and its mutants T54R and I113T, the asymmetric unit contains 2 biologically relevant dimers (A–B and C–D). Although one has a very well-defined electron density throughout the entire sequence, the other dimer has clear breaks in the electron density in the regions encompassing residues 68–78 (loop IV) and residues 125–140 (loop VII). Overall, the structures are very similar to each other. Only few minor local differences in terms of conformation, buried surface areas, and residues involved in H-bond interactions are detected (Table S1). Peculiar for apo T54R SOD1 is a mutation-related hydrogen bond, which is formed at the dimer interface, i.e., between NH2 of Arg-54 of each monomer and OD1 of Asn-19 of the other monomer in the same dimer. This interaction might lead to a partial stabilization of the dimeric state with respect to the WT protein and might correlate with the slightly slower oligomerization rates for apo T54R compared with the WT protein (14), because oligomerization presumably occurs through monomerization (19). The free Cys-6, in all 3 apo SOD1 structures, is essentially buried because of the constraints imposed by 2 H bonds with Ile-18, whereas the other free cysteine, Cys-111, has a considerably high (75%) solvent-exposed side chain (thiol group).
The 3 structures determined here clearly resemble the already available partially apo (20% zinc in 1 of the 2 dimers) WT SOD1 structure [Protein Data Bank (PDB) ID code 1HL4] (18). Similarly, the same electron density breaks were found in only 1 of the 2 dimers. On the contrary, the fully metalated (Cu2,Zn2; holo) structures of both WT SOD1 [PDB ID code 1HL5 (18)] and I113T SOD1 mutant [PDB ID code 1UXL (20)] show a well-defined electron density throughout the entire sequence for each dimer present in the asymmetric unit. The backbone rmsd between the holo and apo form of both WT and I113T SOD1 is ≈0.45 Å. The main structural differences between the holo and apo states are observed in the loops connecting the β-strands, where the electron density is broken in one of the dimers in the structures of the apo state. Similar electron density breaks were also observed for the structure of the apo state of the H46R SOD1 mutant (21), the only, up to now published structure of a completely metal-free SOD1 mutant. On the contrary, the structures of ALS-related SOD1 mutants, in the fully metalated state are very similar to each other, and particularly well ordered throughout the sequence (20–25).
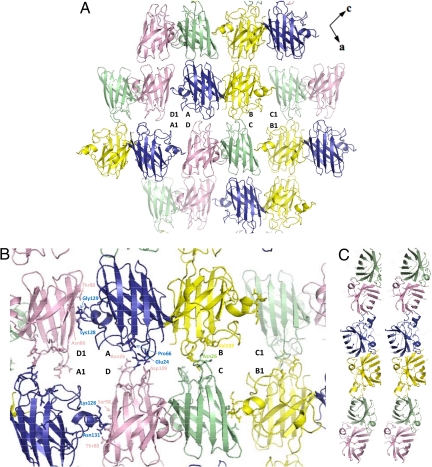
WT SOD1 and its mutants, T54R and I113T, in the apo state, form a continuous, extended arrangement of β-barrels stacked up along a direction (crystallographic b-axis) perpendicular to the dimer interface (Fig. 1A). The tetramer A+B+C+D comprising the asymmetric unit is surrounded by 4 others in the ac plane. The intermolecular contacts between the 2 dimers in the asymmetric unit are between monomers A and D and monomers B and C. The H-bonding connections are different for the 2 pairs (Fig. 1B). A H-bonding network is also present between molecules in adjacent asymmetric units, with 7 strong and 3 weak H bonds between monomers A and D1 (i.e., monomer of an adjacent asymmetric unit) and with 6 strong and 2 weak H bonds between monomers B and C1 (Fig. 1B). The view orthogonal to the crystallographic b-axis shows that the apo WT SOD1 β-barrel forms a zigzag array of filaments (Fig. 1C). This behavior is similar to that already observed for apo WT SOD1 reported by Strange et al. (18) and is common to amyloid-like fibrils (26).
Fig. 1.
Packing of molecules in apo WT SOD1. (A) The apo protein has an extended sheet of β-barrels arranged in the ac plane. Monomer A is shown in blue, monomer B is in yellow, monomer C is in light green, and monomer D is in violet. (B) Contacts between neighboring dimers: H bonds involving OD1 and ND1 of Asn-26 and the O atom of Pro-66 are symmetrically present between monomers A and D. ND1 atom of Asn-26 of monomers C forms an H bond with the O atoms of Val-103, Ile-104, and Ser-102 of monomer B. H bond is shown between OE2 atom of Glu-24 and OD2 atom of Asp-109. H-bonding network is present between molecules in adjacent asymmetric units: the O atom of Lys-128 in monomer A forms H bonds with the N and O atoms of Asn-86 of monomer D1. The O atom of Gly-129 has H bonds to the O atom of Asn-86 and to the OG1 atoms of Thr-88. The N and O atoms of Gly-130 participate in 3 H bonds: to Asn-86 O and to Ser-98 N and O atoms, respectively. Finally, the N atom of Asn-131 forms a weak H-bond to the O atom of Ser-98. (C) The view obtained after rotating A twice by 90° showing the zigzag arrangement of the constituent β-strands aligned along the long ac diagonal of the unit cell.
The comparison between the structures of WT SOD1 and ALS-related mutants was unable to shed light on the structural basis of different behaviors in oligomerization rates between WT and pathogenic mutants, although it did point at some conformational disorder as a consequence of the lack of metal ions. Therefore, a characterization of the apo state in solution of WT SOD1 and its mutants is necessary.
The 1H-15N HSQC spectra of the apo forms of both WT SOD1 and the 2 pathological mutants i.e., T54R and I113T, have reduced signal dispersion with respect to those of the metalated form (Fig. S1), indicating that some parts of the protein do not have a well-defined conformation. High protein instability and strong tendency to form high molecular weight oligomers (13) prevented us from collecting the triple resonance NMR experiments necessary to achieve a specific resonance assignment at 298 K. A protein sample analysis, aiming at finding the best compromise between protein stability toward oligomerization and line broadening effects of NMR signals, led us to acquire all of the spectra at 0.6 mM protein concentration and 288 K. In these experimental conditions, dimeric apo WT SOD1 is stable for periods long enough to collect some of the triple resonance NMR experiments necessary for sequence-specific resonance assignment even if more than one sample from the same preparation was necessary to acquire all of the experiments.
Through the NMR experiments, 68% of the backbone atoms (N, HN, and Cα) were assigned for apo WT SOD1. The unassigned peaks were all clustered in the central part of the 1H-15N HSQC spectrum, thus experiencing severe resonance overlap. They were located mainly in loops connecting the β-strands, in particular in loop IV, which contains most of the metal-binding residues. This spectral pattern was already observed in the apo state of the monomeric form of SOD1 obtained through residue mutations at the subunit–subunit interface (17). In the latter case, mutations of the 2 free cysteines (6 and 111) (as WT SOD1) prevented oligomerization, allowing to reach a much higher protein concentration that, combined with the half molecular weight, led to a more complete assignment, which was used here for comparison purposes.
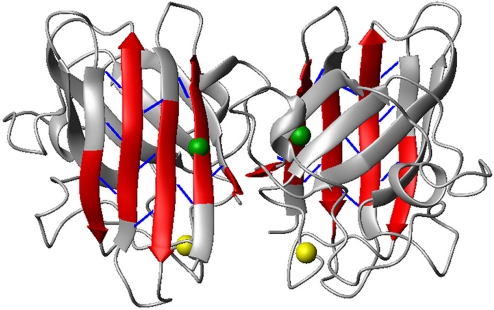
From the analysis of the assigned chemical shift resonances it appears that the secondary structural elements of the β-sheet of the SOD1 β-barrel that comprises β-strands 1, 2, 3, and 6 exist also in the metal-free state, whereas β-strands 4, 5, 7 and 8 are much shorter. NH–NH long-range NOEs are indeed present within β-strands 1, 2, 3, and 6, whereas they are mostly missing in the other β-sheet, which contains some of the metal ligand residues (Fig. 2). From the analysis of the NOESY spectrum, it also appears that most of the long-range 1H–1H NOEs, involving side-chain protons, are missing even within the secondary structural elements.
Fig. 2.
Secondary structural elements (red) based on the chemical shift index analysis for the apo WT SOD1 protein. Backbone long range NOEs (blue sticks) determined from 15N-edited NOESY spectra. The locations of the free cysteines Cys-6 and Cys-111 are represented by green and yellow spheres, respectively. The oxidation state of SOD1 cysteine residues was also investigated through 13C 1D NMR spectra (Fig. S2).
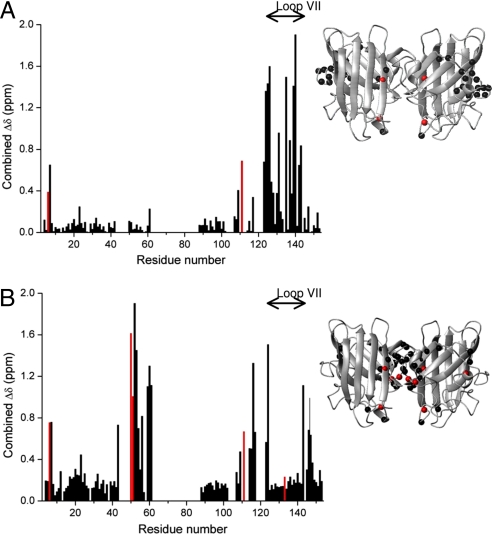
Combined chemical-shift variations of backbone amide groups between the dimeric apo SOD1 and its fully metallated form (Fig. 3A) indicate significant structural changes in loop VII, similar to that already observed for monomeric apo SOD1 (17). Furthermore, a number of NH groups in this loop show, mainly at low temperature, another set of signals with lower intensity caused by a minor conformation or a group of fast exchanging conformers in slow exchange with the rest of the conformations. Analysis of the chemical-shift variations between the monomeric and dimeric apo states of SOD1 (Fig. 3B) confirms that the absence of metal ions similarly affects the 2 forms, particularly in the electrostatic loop. The only detected differences can be ascribed to the mutation of the 2 free cysteines residues (C6A and C111S) and 3 residues (F50E, G51E, E133Q), which, by inducing protein monomerization, sizably affects the NMR signals of the residues at the dimer interface. The latter 5 mutations are present only in the “artificial” monomeric form.
Fig. 3.
Chemical shift variations comparison of monomeric and dimeric SOD1 states. (A) Combined chemical shift variations of backbone amide resonances between dimeric apo WT SOD1 and dimeric (Cu2,Zn2)-asWT SOD1. The positions where any of the structures under comparison are mutated are highlighted in red. (B) Dimeric apo WT SOD1 and monomeric apo-asWT SOD1. The positions where any of the structures under comparison are mutated are highlighted in red. The combined chemical-shift variations Δavg(HN) were calculated as [((ΔH)2 + (ΔN/5)2)/2]1/2, where ΔH and ΔN are chemical-shift differences for 1H and 15N, respectively. The locations of the largest differences (δΔ value of at least 0.3 ppm) observed are shown as black spheres on the crystal structure of the apo WT SOD1 protein (3ECU); the residues mutated in any of the structures under comparison are represented as red spheres.
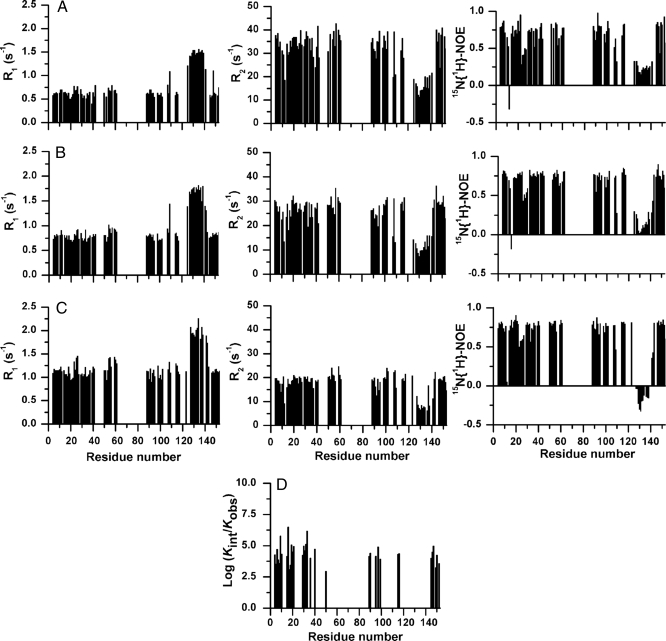
Also the dynamical properties of apo WT dimeric SOD1 are affected by the absence of the metals (Fig. 4). The 15N relaxation rates of backbone NHs, measured in the temperature range 288 to 310 K, are consistent with the protein being essentially only in the dimeric state, with its tumbling rate increasing with decreasing temperature. Residues located in the first β-sheet of the β-barrel have R1 and R2 relaxation rates and 15N{1H}–NOEs values characteristic of a structured protein (0.75 ± 0.04 s−1, 27.6 ± 0.7 s−1, and 0.76 ± 0.03, respectively at 298 K). Similar average values were found for the assigned residues of strands β7 and β8 (Fig. 4), which indeed remain significantly structured, as also confirmed by the presence of a NH–NH long-range NOE between β8 and β1 (Fig. 2).
Fig. 4.
Dynamic properties of apo WT SOD1. (A–C) Backbone 15N(H) relaxation parameters and heteronuclear 1H-15N NOEs for apo WT SOD1 at 288 K (A), 298 K (B), and 310 K (C). (D) Logarithm of protection factors for the backbone amide groups of the apo WT SOD1, log10(kint/kexp), where kint is the intrinsic exchange rate for the unprotected amide groups (34).
The relaxation rates and 15N{1H}–NOEs, which are homogeneous in the β-strand structures, are instead dramatically altered in the electrostatic loop VII. The latter has, at 298 K, lower than average R2 values, higher than average R1 values, and lower 15N{1H}–NOEs, which become even negative at 310 K, indicating that its residues experience motions faster than the overall protein tumbling, i.e., faster than a nanosecond. Internal motions in the subnanosecond time scale were also observed in loop VII of monomeric apo SOD1 (17), confirming that the absence of metal ions similarly affects the dynamical properties of both monomeric and dimeric apo forms. The overall spectral features in solution for loop VII suggest that this region, and the nonassigned residues located in the other loop regions, sample a wide range of conformations that interconvert each other very fast.
15N relaxation rates of backbone NHs were also measured for apo T54R SOD1 (Fig. S3). The results are overall very similar to the data obtained for apo WT SOD1 (Tables S2–S4), showing that, also for this mutant, the mobility of loop VII is dramatically affected by the lack of metal ions; as also confirmed by the negative values of the 15N{1H}–NOEs. The similar mobility behavior of apo WT and T54R mutant is consistent with their similar rates of oligomerization (14).
Unfortunately, the I113T mutant is too unstable, preventing the acquisition of relaxation data at temperature >288 K. At this temperature this mutant has overall the same dynamical properties as WT and T54R SOD1, being essentially structured in β-sheet 1 and β-strand 8, but showing higher backbone dynamics in loop VII where a sizable number of residues have negative 15N{1H}–NOEs values even at 288K (Fig. S4 and Table S2). At variance with WT and T54R SOD1 mutant, in I113T some residues located in other loop regions experience extensive dynamics on the subnanosecond time scale. In particular, some residues of loop V (residues 90–94), which connects β5 and β6, show a peculiar drop of the 15N{1H}–NOEs and R2 values and a corresponding increase of R1 values consistent with fast local motions (Table S2). Interestingly, β5 and β6 are located at the edge of the 2 sheets constituting the SOD1 barrel and are thought to be involved in amyloid fibril formation through intermolecular aggregation with other edge strands in other fALS-related mutants (18).
The observed local flexibility of the SOD1 apo forms with respect to the metalated ones is consistent with their solvent accessibility. Indeed, the overall number of NH protons exchanging fast with the bulk solvent, as measured from H2O/D2O exchange processes, is much higher in the apo state than in the metalated one. In the apo forms of both WT and T54R SOD1, after 20 min the samples have been dissolved in D2O, ≈60 residues were completely exchanged, whereas only ≈30 were exchanged in the metalated one. Nevertheless, hydrogen exchange protection factors of apo WT and T54R SOD1 (Fig. 4D and Fig. S3D) reveal that residues located in the secondary structure elements of the β-sheet constituted by strands β1, β2, β3, and β6 and some residues of β8 still show a significant degree of order, in agreement with the presence of a β-structure hydrogen-bond pattern. In particular, in β6 it was possible to quantify exchange rate values only for the amide protons involved in hydrogen bonds with residues located in β3, whereas the other amide protons exchanged too fast with the solvent. Overall WT and T54R SOD1 have very similar H/D exchange properties.
Particularly relevant for the oligomerization process that occurs for apo WT SOD1 at physiological conditions (13), is the solvent exposure of the free cysteines (Cys-6 and Cys-111). Although Cys-111 is highly solvent exposed in both protein forms, Cys-6 has a dramatically different solvent accessibility (Fig. S5). In the metalated form its NH is essentially buried and protected from the solvent and indeed its NH signal is still present after 5 days in D2O. On the contrary, in the apo form it almost completely disappears after only 4 h. Also, the NH signals of adjacent residues (residues 4–8, β-strand l) all disappear in 4 h, suggesting a high solvent accessibility of the region around Cys-6. The high solvent accessibility of Cys-6 and β-strand l is observed for apo T54R mutant as well, suggesting, as for apo WT SOD1, that the metalated form is rigid and solvent protected, whereas the lack of metal ions makes this region and the entire protein highly dynamic and more solvent accessible.
All of these features indicate that the apo state of SOD1 in solution is characterized by a distribution of conformations, particularly for β-strands 4 and 5 and loop VII.
Similar behavior was observed in the SOD-like protein from Bacillus subtilis (27), where its NMR properties indicate a conformational mobility for most of the protein, characterized by defined secondary-structure elements and a dynamic tertiary structure, at variance with the X-ray crystal structure of the same protein, which shows a well-ordered tertiary structure.
The overall studies here presented for the apo state of SOD1 and its mutants in solution also explain the behavior of these proteins with respect to crystallization. Crystallization trials were performed at 2 different temperatures on both the apo and metalated forms of WT SOD1 and the 2 mutants (Table S5), i.e., at 288 K, which is the temperature at which the crystals discussed above were obtained, and at 310 K, which is the temperature at which the oligomerization studies were carried out (13, 14). Crystals of the apo state of WT SOD1 and the 2 mutants can be obtained only at 288 K, whereas crystals for the metalated forms of WT SOD1 and the mutants were obtained at both temperatures. This result indicates that temperature has a major influence on the crystallization of the apo state whereas it is almost negligible on the metalated one. This overall behavior is consistent with the dynamic properties and conformational disorder of the apo state. A decrease in temperature slows down the interconversion process among the various conformations and increases the population of the most stable states, which can therefore crystallize.
The local disordered state observed for the metal-free SOD1 may appear to be in contradiction with its cooperative unfolding behavior reported by differential scanning calorimetry measurements (28). This contradiction can be, however, reconciled by the presence of a significant portion of still existing tertiary structure and extensive structural interactions at the dimer interface caused by the preserved dimeric nature.
Concluding Remarks
We have recently shown (13) that oligomerization of apo SOD1 involves oxidation of the 2 free cysteines (Cys6 and Cys111) with the formation of intersubunit disulfide bonds, thus linking a high number of protein molecules in high molecular weight species. Some have suggested that the formation of SOD1 aggregates are the consequence of both covalent disulfide cross-linking and noncovalent interactions (29), whereas others proposed that extensive disulfide cross-linking is not required for the formation of mutant SOD1 aggregates (30). Recent studies showed the importance of nonphysiological intermolecular disulfide bond between cysteines 6 and 111 in mutant SOD1 for high molecular weight aggregate formation for protein ubiquitylation and neurotoxicity, which are all dramatically reduced when these cysteines are substituted (31). In any case, there is a general agreement on the critical role played by cysteines 6 and 111, in the modulation of human SOD1 aggregation (29, 31).
We have shown that only the lack of metal ions makes SOD1 oligomerization possible (13). The reason for the dramatic different behavior of apo and metalated forms of SOD1 is now better understood. Indeed, the solvent exposure of the reduced cysteines changes dramatically from the metallated form to the apo one. Only in the latter state a free cysteine can bind another one of a different monomer to form the soluble oligomer. The crystal structures, on the contrary, are not informative on this respect as they clearly represent only one of the multiple conformations taken in solution by the protein. Consistently, the apo form of both WT and the mutants fail to crystallize at physiological temperature because of the high disorder and internal mobility.
The information obtained from the NMR spectra indicates that in solution apo WT SOD1 samples a range of conformations, which are highly disordered in some parts. Higher temperatures accelerate exchange among these conformations and could populate new ones. This behavior explains why only the disordered, locally unfolded, metal-free state has a dramatic protein flexibility that makes accessible conformations prone to oligomerize, whereas the rigid structure of the metalated protein is unable to do it.
Overall the present extensive structural and dynamical characterization of the apo state of WT SOD1 and some of its mutants showed that the lack of metal ions and the subsequent protein flexibility allows the free cysteines (in particular Cys-6) to become exposed and therefore ready to get oxidized and form the disulfide bonds that give rise to the soluble oligomers.
Materials and Methods
Protein Samples.
Apo and metallated WT, T54R and I113T SOD1 samples were expressed and purified as described in SI Text.
Crystallization, Data Collection, and Structure Solution.
Crystals of metal-free SOD1 (WT, T54R, and I113T) were obtained by using the vapor diffusion technique at 288 K from 0.1 mM protein solutions containing 0.1 M Mes (pH 6.5) or 0.1 M Hepes (pH 7.0), 20% PEG 3350. For further details on crystallization, structure solution, and refinement see SI Text and Table S6.
NMR Experiments.
NMR spectra were acquired at 288 K on an Avance 900 Bruker spectrometer equipped with a cryogenically-cooled probe. Resonance assignments of apo WT SOD1 form were performed through conventional multidimensional NMR techniques based on triple resonance experiments summarized in Table S7. Because of the instability of the apo form of WT, T54R, and I113T SOD1, >1 sample was needed to complete NMR data collection at 288 K. Samples of apo WT and T54R SOD1 were stable for ≈7 days at 288 K, 5 days at 298 K, and only 3 days at 310 K, whereas I113T SOD1 mutant was stable only for 3 days at 288 K.
The dynamic properties of the apo dimeric forms of apo WT, T54R, and I113T SOD1 were directly sampled through 15N relaxation measurements. 15N longitudinal and traverse relaxation rates and 15N{1H}–NOEs were recorded at 288, 298, and 310 K for both WT and T54R mutant at 500 MHz, using a protein concentration of ≈0.6 mM. For I113T mutant it was possible to carry out 15N relaxation measurements only at 288 K because of its very high instability. R1 and R2 relaxation rates and heteronuclear NOE values were obtained as described in Table S7.
For apo WT SOD1, the average backbone 15N longitudinal R1 and transversal R2 relaxation rates and 15N{1H}–NOEs values, calculated excluding residues 125–142, were 0.64 ± 0.04, 34.0 ± 1.6 s−1, and 0.70 ± 0.03, respectively at 288 K, 0.81 ± 0.05, 27.3 ± 1.2 s−1, and 0.72 ± 0.03 at 298 K, and 1.15 ± 0.06, 19.4 ± 0.9 s−1, and 0.75 ± 0.03 at 310 K. For apo T54R SOD1, the values were 0.66 ± 0.05 s−1, 33.0 ± 1.9 s−1, and 0.66 ± 0.05, respectively at 288 K, 0.77 ± 0.05, 28.4 ± 0.05, and 0.71 ± 0.05 at 298 K, and 1.17 ± 0.07, 20.1 ± 1.0 s−1, and 0.73 ± 0.04 at 310 K. For I113T SOD1, the values were 0.71 ± 0.05, 29.8 ± 1.5 s−1, and 0.60 ± 0.06, respectively at 288 K .
A correlation time for protein tumbling (τc) of 22.6 ± 1.9 ns at 298 K was estimated from the R2/R1 ratio excluding those residues exhibiting below-average 15N{1H}–NOEs values and those experiencing conformational processes. The result is consistent with that obtained with the HYDRONMR program for apo SOD1 being only in a dimeric state.
H2O/D2O exchange properties were analyzed on samples obtained by rapidly diluting concentrated WT and T54R SOD1 both in metallated and apo forms at pH 7.0 [which was corrected for the isotope effect as described (32)], with D2O buffer to a final D2O/H2O ratio of 0.90. H/D exchange rates were investigated through a series of 1H-15N SOFAST-HMQC experiments (33) performed from 20 min after dilution for 21 h every 9 min and then later acquired after 5 days. All experiments were carried out at 298 K. Exchange rates (kex) were determined by fitting the decay of the peaks intensities in the 1H-15N SOFAST-HMQC spectra as a function of time to a monoexponential decay (Fig. S6).
Supplementary Material
Acknowledgments.
We thank Marie-Paule Strub for providing the cysteine-auxotrophic strain BL21(DE3)cysE. This work was supported by European Commission “Understanding Protein Misfolding and Aggregation by NMR” (UPMAN) Grant LSHG-CT-2004-512052 (11/1/04-10/31/07), Ministero dell'Università e della Ricerca– Fondo per Gli Investimenti della Ricerca di Base Grant RBLA032ZM7 (12/09/05 -12/09/10), Ente Cassa di Risparmio di Firenze “Relazione varianti proteiche strutturali-malattie genetiche” and “Basi molecolari di patologie umane correlate a disfunzioni della catena respiratoria,” and Marie Curie Host Fellowship for Early Stage Research Training MEST-CT-2004-504391.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3ECU, 3ECV, and 3ECW).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809845106/DCSupplemental.
References
- 1.Valentine JS, Doucette PA, Potter SZ. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 2.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 3.Jonsson PA, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Xu G, Borchelt DR. Mapping superoxide dismutase 1 domains of non-native interaction: Roles of intra- and intermolecular disulfide bonding in aggregation. J Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng HX, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa Y, Fu R, Deng HX, Siddique T, O'Halloran TV. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc Natl Acad Sci USA. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasinelli P, et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Ohi T, Nabeshima K, Kato S, Yazawa S, Takechi S. Familial amyotrophic lateral sclerosis with His46Arg mutation in Cu/Zn superoxide dismutase presenting characteristic clinical features and Lewy body-like hyaline inclusions. J Neurol Sci. 2004;225:19–25. doi: 10.1016/j.jns.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Ferri A, et al. Familial ALS-superoxide dismutases associate with mitochondria and shift their redox potentials. Proc Natl Acad Sci USA. 2006;103:13860–13865. doi: 10.1073/pnas.0605814103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 11.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. J Biol Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 12.Bertini I, Mangani S, Viezzoli MS. In: Advanced Inorganic Chemistry. Sykes AG, editor. San Diego: Academic; 1998. pp. 127–250. [Google Scholar]
- 13.Banci L, et al. Metal-free SOD1 forms amyloid-like oligomers: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banci L, et al. SOD1 and amyotrophic lateral sclerosis: Mutations and oligomerization. Plos ONE. 2008;3:e1677. doi: 10.1371/journal.pone.0001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnesano F, et al. The unusually stable quaternary structure of human SOD1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 16.Field LS, Furukawa Y, O'Halloran TV, Culotta VC. Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J Biol Chem. 2003;278:28052–28059. doi: 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- 17.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Solution structure of Apo Cu,Zn superoxide dismutase: Role of metal ions in protein folding. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 18.Strange RW, et al. The structure of holo and metal-deficient wild-type human Cu,Zn superoxide dismutase and its relevance to familial amyothrophic lateral sclerosis. J Mol Biol. 2003;28:877–882. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 19.Rakhit R, et al. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 20.Hough MA, et al. Dimer destabilization in superoxide dismutase may result in disease-causing properties: Structures of motor neuron disease mutants. Proc Natl Acad Sci USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elam JS, et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 22.Hart PJ, et al. Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis. Protein Sci. 1998;7:545–555. doi: 10.1002/pro.5560070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardoso RM, et al. Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase. J Mol Biol. 2002;324:247–256. doi: 10.1016/s0022-2836(02)01090-2. [DOI] [PubMed] [Google Scholar]
- 24.DiDonato M, et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J Mol Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 25.Cao X, et al. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:16169–16177. doi: 10.1074/jbc.M801522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serag AA, Altenbach C, Gingery M, Hubbell WL, Yeates TO. Arrangement of subunits and ordering of β-strands in an amyloid sheet. Nat Struct Biol. 2002;9:734–739. doi: 10.1038/nsb838. [DOI] [PubMed] [Google Scholar]
- 27.Banci L, et al. A prokaryotic superoxide dismutase paralog lacking two Cu ligands: From largely unstructured in solution to ordered in the crystal. Proc Natl Acad Sci USA. 2005;102:7541–7546. doi: 10.1073/pnas.0502450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez JA, et al. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper-zinc superoxide dismutase. J Biol Chem. 2002;277:15932–15937. doi: 10.1074/jbc.M112088200. [DOI] [PubMed] [Google Scholar]
- 29.Cozzolino M, et al. Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:866–874. doi: 10.1074/jbc.M705657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karch CM, Borchelt DR. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niwa J, et al. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J Biol Chem. 2007;282:28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- 32.Glasoe PK, Long FA. Use of glass electrodes to measure acidities in deuterium oxide. J Phys Chem. 1960;64:188–190. [Google Scholar]
- 33.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Am Chem Soc. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 34.Bai YW, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins Struct Funct Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.