Abstract
When chromosomes are aligned and bioriented at metaphase, the elastic stretch of centromeric chromatin opposes pulling forces exerted on sister kinetochores by the mitotic spindle. Here we show that condensin ATPase activity is an important regulator of centromere stiffness and function. Condensin depletion decreases the stiffness of centromeric chromatin by 50% when pulling forces are applied to kinetochores. However, condensin is dispensable for the normal level of compaction (rest length) of centromeres, which probably depends on other factors that control higher-order chromatin folding. Kinetochores also do not require condensin for their structure or motility. Loss of stiffness caused by condensin-depletion produces abnormal uncoordinated sister kinetochore movements, leads to an increase in Mad2(+) kinetochores near the metaphase plate and delays anaphase onset.
INTRODUCTION
Centromeric chromatin is a special region of chromosomes that has important mechanical and signaling functions in mitosis (Pidoux and Allshire, 2005; Ekwall, 2007; Cheeseman and Desai, 2008; Vagnarelli et al., 2008). In metaphase, pulling forces generated by interactions between spindle microtubules (MTs) and kinetochores are opposed by tension produced by centromeric chromatin stretch. Centromere and kinetochore tension and stretch are important for maintaining chromosome alignment (McIntosh et al., 2002), stabilizing kinetochore microtubule (kMT) attachments (Nicklas and Koch, 1969), spindle checkpoint signaling (Musacchio and Salmon, 2007; McEwen and Dong, 2009), and also for the back-to-back orientation of sister kinetochores (Loncarek et al., 2007). At least three independent factors have roles in the establishment of centromeric tension in metaphase: sister chromatid cohesion (Yeh et al., 2008), the elastic properties of chromatin (Houchmandzadeh et al., 1997; Almagro et al., 2004; Marko, 2008), and the higher order structure of the centromeric chromatin.
Condensin is important for the architecture of mitotic chromosome arms (Coelho et al., 2003; Hudson et al., 2003; Hirota et al., 2004; Hirano, 2006), but it also localizes to centromeres (Saitoh et al., 1994; Gerlich et al., 2006), where condensin I, but not condensin II was reported to have a role in stabilizing the structure (Gerlich et al., 2006). It has recently been suggested that condensin could have a role in regulating the elastic behavior of centromeric chromatin. One study found that condensin I–depleted Drosophila chromosomes were unable to align at a metaphase plate, had distorted kinetochore structures, and lost elasticity of their centromeric chromatin (Oliveira et al., 2005). However a similar study in human cells reported that although loss of condensin I caused kinetochores to undergo abnormal movements, these movements were bidirectional (e.g., reversible; Gerlich et al., 2006).
Even after the publication of those results, the regulation and functional significance of centromere stretch remained unknown. An elegant study in budding yeast went on to find that chromatin structure sets the rest length of the centromere, but does not regulate its stretch (Bouck and Bloom, 2007). Here, we analyzed the movements of kinetochores, pericentromeres, and distal chromosome arms during metaphase in DT40 cells bearing a conditional knockout of SMC2, an essential subunit of condensin I and II. Our results reveal that condensin ATPase activity is required to regulate centromere stretch and that loss of condensin results in a mitotic delay accompanied by an increased number of Mad2-positive kinetochores on chromosomes aligned at the metaphase plate.
MATERIALS AND METHODS
Cell Culture, Transfections, and Drug Treatments
The SMC2 conditional knockout cell line and SMC2:CENP-H:GFP_H2B:mRFP were cultured as previously described (Hudson et al., 2003; Vagnarelli et al., 2006). The SMC2 conditional knockout cell line containing a human X-derived minichromosome was described previously (Vagnarelli et al., 2006).
The constructs GgMad2-green fluorescent protein (GFP) and GFP-hMCAK were obtained from T. Fukagawa (National Institute of Genetics, Tokyo) and from J. Swedlow (University of Dundee, United Kingdom), respectively. Stable transfectants were selected in 0.5 μg/ml puromycin or 25 μg/ml blasticidin for 10 d.
The construct GFP-GgCENPA was obtained by cloning GgCENPA into pEGFPC1 with a 17-amino acid linker.
The cell lines lacO:lacI-GFP in SMC2 conditional knockout background were described elsewhere (Vagnarelli et al., 2006). The lacO:lacI-GFP CEN integration was used to prepare stable cell lines expressing the rescue construct SMC2-SBP wild-type or the mutant form of SMC2-SBP S1086R.
For Hec1 RNA interference (RNAi) a 21-mer oligonucleotide (ccagacugaggaagaaauudtdt) covering bases 1404-1424 downstream of the start codon of Gallus gallus Hec1 cDNA was used. A 21-mer oligonucleotide (cguacgcggaauacuucgadtdt) with no significant homology to any known chicken mRNA in the databases was used as control. The lacO:lacI-GFP cell line was grown in the presence of doxycycline for 8 h and then transfected with 10 μM of each small interfering RNA using the Nucleofector system (Amaxa, Cologne, Germany) and plated in complete medium plus doxycycline. The experiments were analyzed between 26 and 30 h after repression.
To calculate the interkinetochore distances in the absence of MTs, cells were treated for 2 h with colcemid at 0.5 μg/ml. The analysis of the lacO CEN movements in the presence of different drugs was conducted as follows. Cells were incubated for 2 h with 20 μM MG132 and then prepared for live cell imaging. For the nocodazole experiments cells were treated with 0.5 μg/ml nocodazole while imaging. ICRF-159 treatments were carried out for 3 h with 10 μg/ml the drug.
For MT drug experiments, cells were treated with 0.5 μg/ml nocodazole (Sigma, St. Louis, MO) or 40 nM paclitaxel (Taxol; Sigma) for the period of time indicated in the text.
Live Cell Imaging
Digital images were collected with a cooled CCD camera (Orca ER; Hamamatsu, Bridgewater, NJ) coupled to a Yokogawa spinning disk confocal unit (CSU10; Perkin Elmer, Norwalk, CT), which was attached to an inverted microscope (TE300; Nikon, Melville, NY) with a 100× 1.4 NA plan-Apochromatic differential interference contrast objective. Image acquisition and microscope shutter were controlled by Metamorph software (Universal Imaging, West Chester, PA) on a PC computer. Stage temperature was maintained at ∼37°C using an air curtain incubator (ASI 400; Nevtek, Burnsville, VA). Fluorescence images were acquired at 488 nm at a single focal plane with an exposure time of 1500 ms every 4 s.
Image Analysis and Kymographs
The movement of paired lacO:lacI-GFP integrations was followed using the Track Objects tool in Metamorph after image calibration; whenever any of the spots was out of focus, the time point was removed from the raw movie. After exporting the coordinates (x,y) of the LacI spots to Excel (Microsoft, Redmond, WA), the angle between a line drawn through both spots, and the horizontal axis was calculated for each image in the stack. A custom-written program in MatLab (MathWorks, Natick, MA; created by J. Gatlin) was used to align and rotate the raw image stacks based on the calculated angle and the position of one of the two LacI spots. The aligned stack was then exported back to Metamorph where the Kymograph tool was used to build kymographs.
The distance of paired lacO:lacI-GFP integrations in live cell imaging movies was determined using the Measure Distance tool in Metamorph, and the values were exported to Excel. The distance values and the time were plotted in a line graph in Excel.
The measurements of the lacO integration in fixed samples were carried out on a microscope (Model IX-70; Olympus) controlled by DeltaVision Softworks (Delta Data Systems, Cornwells Heights, PA) using the SoftWorks tool “Measure Distance.” The cells were selected according to the following criteria: 1) perfectly aligned metaphase plates and 2) both lacO integrations visible in the same focal plane. Fluorescence images were acquired at 488 nm on a single focal plane, and the measurements were determined.
The distance between the centroids of Hec1 and CENP-A was obtained by image calibration and line profile analyses (Metamorph) on projected three-dimensional (3D) stacks. After obtaining the graph, the distance between the highest peaks for Hec1 and GFP-CENP-A was calculated.
The spindle pole separation measurements were carried out on cells immunostained for γ-tubulin with DNA visualized using DAPI. Measurements were only considered in cells where the metaphase plate was clear and both γ-tubulin stained poles were in the same focal plane. These selected cells were imaged in a single focal plane at 568 nm, and the distance was measured.
Quantification of the CENP-H:GFP Molecules
The quantification was determined by quantitative fluorescence of CENP-H:GFP (in cells where GFP was knocked into the endogenous single copy CENP-H gene) relative to the copy number of 8 Ndc80 molecules per budding yeast kinetochore according to the method previously described (Joglekar et al., 2006).
Indirect Immunofluorescence and Microscopy
Immunostaining in DT40 cells was conducted as previously described (Vagnarelli et al., 2006). Antibody incubation was done in 1% BSA-phosphate-buffered saline (PBS) for 1 h at 37°C. The following antibodies were used: mouse anti-α-tubulin at 1:1000 (Sigma), mouse anti-γ-tubulin at 1:1000 (Sigma), anti-phospho H3T3 (Abcam, Cambridge, MA) at 1:200, anti-Hec1 (T. Fukagawa, National Institute of Genetics, Tokyo) at 1:200, anti-BubR1 at 1:500, anti-CENP-H at 1:200 (T. Fukagawa), anti-CENP-A at 1:2000 (T. Fukagawa), fluorescence-labeled secondary antibodies 1:200 (Jackson ImmunoResearch Laboratories, West Grove, PA).
TEEN experiments were performed as previously described (Hudson et al., 2003 and see text). 3D data sets were collected with a DeltaVision system (Applied Precision, Issaquah, WA) and deconvolved using the standard algorithms in SoftWorX software. Images were either used as single planes or as quick projections.
Electron Microscopy
DT40 cells were attached to concanavalin A–coated grided coverslips. The cells were fixed with 2.5% electron microscopy (EM) grade glutaraldehyde in phosphate-buffered saline (PBS) and in some cases 1% Triton was added to the primary fixative to aid in visualizing kinetochore MTs. Metaphase cells were identified by fluorescence light microscopy and located on the finder grid. Cells were postfixed with osmium tetroxide, tannic acid, and uranyl acetate, dehydrated in a grade ethanol series and propylene oxide, and flat-embedded in Epon. Same-cell correlative light microscopy (LM)/EM methods were used to find metaphase cells after embedment. Serial sections 100 and 150 nm thick were cut, stained, and imaged on a Zeiss 910 transmission electron microscope (Thornwood, NY).
RT-PCR and Immunoblotting Analysis
To screen for SMC2ON/OFF lacO cell lines expressing the rescue construct SBP-smc2WT or the mutant form SBP-smc2S1086R cells were grown in absence (SMC2ON) or the presence (SMC2OFF) of doxycycline and harvested for RNA extraction. Total RNAs were reverse-transcribed using oligo-dT primers and Superscript reverse transcriptase (Invitrogen-BRL, Carlsbad, CA) under conditions recommended by the manufacturer. Amplification was carried out with the Taq DNA Polymerase (Roche, Indianapolis, IN) using either a specific forward primer located in the 5′UTR of the Gg SMC2 sequence (5′-ttcactgagggctcccttcg-3′) or a specific primer for the SMC2-tetO transgene (5′-aatggcattgaataacgg-3′). The reverse primer for both was specific for the Gg SMC2 sequence (5′-aatggcattgaataacgg-3′). RT-PCR products were then resolved in a 0.8% agarose gel, and expression of the transgene was detected by the appearance of fragments of 190 base pairs, whereas the SMC2-tetO driven cDNA generated a 120-base pairs fragment.
Immunoblotting was conducted as previously described (Ruchaud et al., 2002) using anti-SMC2 (1:500), anti-SBP (1:300), or anti-β tubulin (1:10,000), followed by horseradish peroxidase–conjugated secondary antibodies (1:10,000, Amersham Pharmacia Biotech, Piscataway, NY), and results were visualized using the enhanced chemiluminescence protocol (Amersham).
Chromosome Mis-segregation Analysis
An SMC2ON/OFF cell line containing a 2.7-Mb X-derived minichromosome was analyzed at 30 h after the addition (or not) of doxycycline. Cytochalasin D (Sigma) at 6 μg/ml was added 5 h before fixation in order to maintain the products of each segregation event within a common cytoplasm. Fixed cells were subjected to FISH with an X-specific α-satellite probe as previously described (Vagnarelli et al., 2006).
RESULTS
We previously described a chicken DT40 B lymphocyte cell line in which the single copy SMC2 gene is disrupted, and cells are kept alive by an SMC2 cDNA expressed under control of a tetracycline-repressible promoter (Hudson et al., 2003). In cells grown with doxycycline for 30 h (SMC2OFF), no SMC2 protein or mRNA was detected by immunoblotting or RT-PCR, respectively (Hudson et al., 2003; Vagnarelli et al., 2006; see Supplementary Figure S1, a and b).
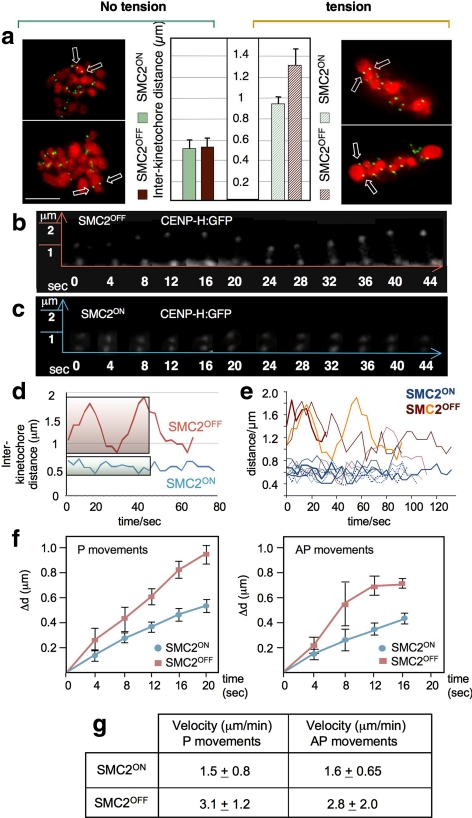
In SMC2OFF cells, mitotic chromosome condensation still occurs; however, the chromosome arms only reach 60% the normal level of compaction (Vagnarelli et al., 2006). In those cells, the average distance between sister kinetochores at metaphase in fixed preparations was 1.4 times greater than that in SMC2ON cells (Figure 1a, right). However, after MT de-polymerization with colcemid, interkinetochore distances were indistinguishable between SMC2ON and SMC2OFF cells (Figure 1a, left). Thus, condensin is not required for compaction of the heterochromatin between sister kinetochores, i.e., to set the “rest length” of the centromeric chromatin. However, those normally compacted centromeres lacking condensin respond abnormally to MT pulling forces. These findings confirm and extend a previous study in HeLa cells in which a MT-dependent abnormal mobility of kinetochores in metaphase was observed after condensin I RNAi (Gerlich et al., 2006).
Figure 1.
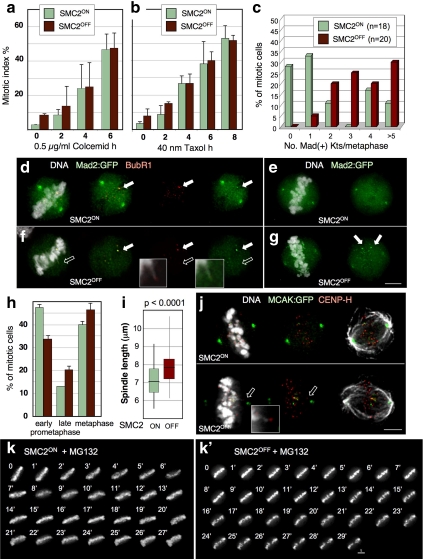
Condensin affects centromere stiffness but not rest length. (a) Interkinetochore distance of SMC2ON and SMC2OFF metaphase cells in the absence (+ colcemid) and presence of MTs. Arrows indicate sister kinetochore pairs. For the graph, n = 100. Green, CENP-H:GFP; red, H2BmRFP. Scale bar, 5 μm. (b and c) Stills from movies of a CENP-H:GFP cell line showing kinetochore movements (time lapse, 4 s): (b) SMC2OFF; (c) SMC2ON. (d) Quantification of kinetochore movements in SMC2ON (blue) and SMC2OFF (red) cells. The shaded areas show the regions of the curves represented in b and c. (e) Examples of typical kinetochore movements in SMC2ON (blue lines) and SMC2OFF (red lines) during metaphase. Time lapse, 4 s. (f) Velocities of poleward (P) and away from the pole (AP) kinetochore movements in SMC2ON (blue lines) and SMC2OFF(red lines) cells during metaphase. (g) Average velocities of poleward (P) and away from the pole (AP) kinetochore movements in SMC2ON and SMC2OFFcells during metaphase. For SMC2ON kinetochores, 30 P and 32 AP movements were analyzed; for SMC2OFF kinetochores, 12 P and 10 AP movements were analyzed.
Condensin Depletion Affects the Dynamic Behavior But Not the Structure of Kinetochores in Mitosis
The core kinetochore protein CENP-H:GFP expressed from its own endogenous promoter was used to follow kinetochore movements in living cells. CENP-H is encoded by a single copy essential gene present on the Z sex chromosome. In vertebrate mitotic cells, the oscillations of sister kinetochores of bioriented chromosomes—poleward (P movements) and away from the pole (AP movements)—are coordinated (Skibbens et al., 1993). This coordination maintains elevated tension between sister kinetochores as they oscillate back and forth across the spindle equator (Waters et al., 1996). In SMC2OFF cells, the movements were uncoordinated, and single kinetochores underwent extended P “excursions,” moving out from the bulk of the chromosome by up to ∼2 μm, trailing a thin chromatin thread (Figure 1, b–e; Supplementary Movies 1 and 2). This extensive centromere stretching was reversible. Thus, as in HeLa cells (Gerlich et al., 2006), condensin is not required for the elasticity of DT40 centromeric chromatin. When nocodazole was added, kinetochore excursions ceased, and the interkinetochore distance of bioriented chromosomes in SMC2OFF cells decreased to the control rest length of SMC2ON cells observed in fixed preparations (Supplementary Movie 3).
Kinetochore velocities measured during P excursions were 2.1 times faster in SMC2OFF cells than in wild type (3.1 ± 1.2 vs. 1.5 ± 0.8 μm/min). The compensatory antipoleward (AP) returns were also 1.8 times faster (2.8 ± 2.0 vs. 1.6 ± 0.65 μm/min; Figure 1, f and g). These somewhat higher velocities probably reflect a decreased resistance (increased compliance) of the centromeric chromatin to spindle forces. However, a lack of pauses or less frequent switching between directions of motion relative to the rate of image capture when filming in the absence of condensin could also contribute to the apparent increased velocity.
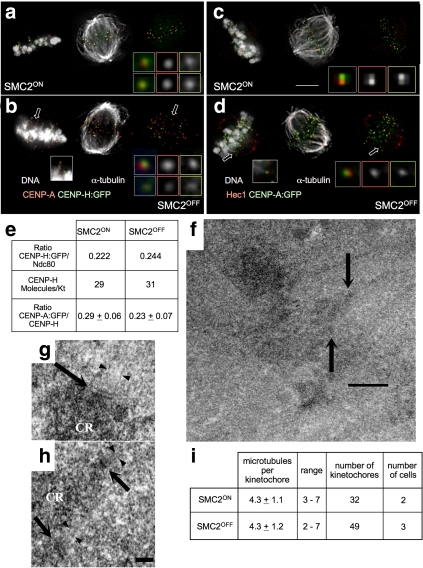
Several lines of evidence indicate that condensin depletion selectively effects the inner centromeric chromatin but not the specialized structures or chromatin of the DT40 kinetochore. 1) CENP-H and CENP-A (markers for the inner kinetochore) localize as discrete spots that are not distorted in kinetochores undergoing excursions (Figure 2, a and b). CENP-A is a modified histone H3 specific for inner kinetochore chromatin (Warburton et al., 1997; Marshall et al., 2008), and CENP-H purifies with CENP-A containing mono-nucleosomes in vitro (Foltz et al., 2006). 2) Although we cannot exclude that levels of some kinetochore proteins may differ in condensin-depleted kinetochores, loss of condensin had no detectable effect on the absolute number of CENP-H molecules per kinetochore (29 in SMC2ON; 31 in SMC2OFF) measured by quantitative fluorescence (Figure 2e) relative to the amount of Ndc80 at budding yeast kinetochores (Joglekar et al., 2006). 3) The ratio of CENP-A to CENP-H was unaltered in the presence and absence of condensin (Figure 2e). 4) Localization of Hec1 in the outer kinetochore plate (DeLuca et al., 2005) relative to CENP-A was unchanged by condensin depletion. The average distance between centroids of CENP-A-GFP and Hec1 staining was 60 nm for SMC2ON kinetochores and 66 nm for kinetochores undergoing P excursions in SMC2OFF cells (Figure 2, c and d). 5) EM revealed normal plates, even in kinetochores undergoing P excursions (Figure 2, f–h). 6) DT40 kinetochore plates had an unexpectedly small number of associated kMTs (∼4), but this was the same in the presence and absence of condensin (Figure 2i). Indeed, with a number of MTs per kinetochore similar to that in S. pombe (Ding et al., 1993), DT40 cells may provide a system that is naturally more sensitive to factors that influence kMT interactions than other vertebrate kinetochores, which have roughly fivefold more MTs per kMT fiber. In summary, neither the inner nor the outer kinetochore showed detectable structural alterations in DT40 cells depleted of condensin.
Figure 2.
Kinetochore structure is preserved in the absence of condensin. (a and b) Relative localization of CENP-A and CENP-H:GFP in SMC2ON (a) and SMC2OFF (b) metaphase cells. Inset in b: kinetochore excursion (arrow) stained for CENP-A (red) and CENP-H:GFP (green). (c and d) Colocalization of Hec1 and CENP-A:GFP in SMC2ON (c) and SMC2OFF cells (d). Inset in d, kinetochore excursion (arrow) stained for Hec1 (red) and CENP-A:GFP (green). Scale bar, 5 μm. (e) Absolute numbers of CENP-H and relative numbers of CENP-A are conserved in SMC2ON and SMC2OFF kinetochores.(f) Low-magnification EM image of an SMC2OFF metaphase cell. Upper arrow, kinetochore excursion; lower arrow, kinetochore at the metaphase plate. (g) Kinetochore from a SMC2ON cell: inner and outer plates (arrow) and associated MTs (arrowheads). (h) Higher magnification of the two SMC2OFF kinetochores indicated in panel f. The outer plate (arrow) and attached MTs (arrowheads) of both kinetochores are indicated. Scale bars, (f) 1 μm; (h) 200 nm. (i) The number of MTs per kinetochore is the same in SMC2ON and SMC2OFF cells. This number was determined from chromosomes aligned at metaphase in serial EM sections of SMC2ON and SMC2OFF cells.
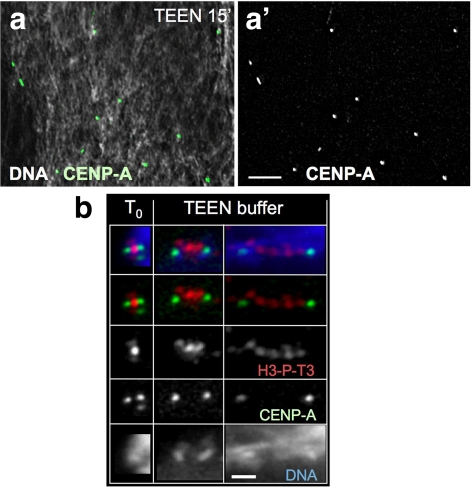
These results suggested that vertebrate centromeres have a robust kinetochore platform consisting of a CENP-A chromatin core beneath that is elastic inner centromeric chromatin that links sister kinetochores together. Such a two-domain model was further supported by in vitro experiments in which wild-type chromosomes were placed in a buffer (10 mM triethanolamine:HCl, pH 8.5, 10 mM NaCl, and 1 mM EDTA) designed to be inefficient at neutralizing the excess negative charges on the DNA that remain in chromatin. In this buffer, chromatin higher-order structure unfolds to beads-on-a-string nucleosomes (Earnshaw and Laemmli, 1983), and mitotic chromosome structure was completely disrupted (Figure 3a; Earnshaw and Laemmli, 1983; Hudson et al., 2003). Under these conditions, CENP-A–containing chromatin retained its condensed morphology, whereas the inner centromeric heterochromatin progressively unfolded (Figure 3b). With longer incubations, the CENP-A–containing chromatin also unraveled into strings of dots, consistent with it being a specialized chromatin domain.
Figure 3.
CENP-A:GFP containing chromatin is more resistant to unfolding than centromeric heterochromatin. (a–a′) After 15 min in TEEN buffer the majority of CENP-A signals are still compact (dot-like). Scale bar, 5 μm. (b) Kinetochore domains of mitotic chromosomes containing CENP-A:GFP (green) are more resistant to unfolding in low ionic strength TEEN buffer than bulk chromatin or inner centromere chromatin stained for H3-P-T3 (red). Scale bar, 2 μm.
These results suggest that the centromere rest length is likely set by chromatin higher-order structure in vertebrates as it is in budding yeast (Bouck and Bloom, 2007) and is independent of condensin. Together, the data thus far suggest that depletion of condensin affects the mechanical properties of the inner centromeric chromatin rather than the structure of the kinetochore itself.
ATPase Activity of Condensin Is Essential for the Maintenance of the Rigidity of the Centromeric Chromatin
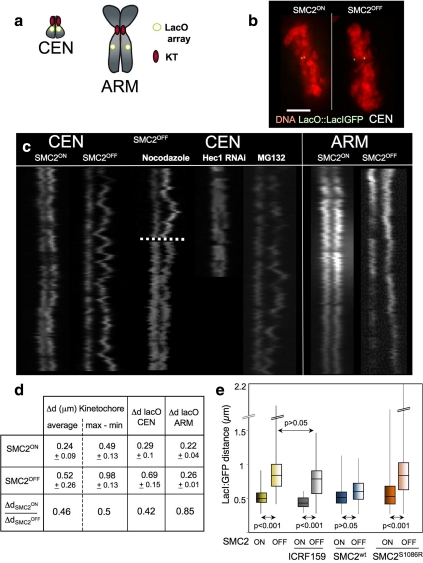
Efforts to determine the role of condensin at centromeres by following individual kinetochore movements were complicated by the fact that DT40 cells contain more than 150 kinetochores. To address the molecular mechanism for condensin stabilization of centromeric chromatin under tension, we generated a simplified in vivo model system for monitoring sister chromatid movements in metaphase. We isolated a SMC2ON/OFF cell line with a lacO array inserted in the pericentromeric region of a microchromosome (Figure 4a, CEN). This pericentric LacO array underwent movements resembling those of kinetochore pairs in living SMC2ON and SMC2OFF cells (Figure 4c; Supplementary Figure S2, a and c; Supplementary Movies 4 and 5), and on average, the LacI:GFP signals on sister chromatids were further apart in SMC2OFF:CEN cells fixed at metaphase (Figure 4b). Similar separation of pericentric regions was previously observed in wild-type budding yeast for reporter arrays adjacent to centromeres (Goshima and Yanagida, 2000; He et al., 2000; Pearson et al., 2001), but has not previously been observed in vertebrates.
Figure 4.
Pericentromeric movements require dynamic MTs and SMC2 ATPase activity. (a) Diagram showing LacO array integration sites in two different SMC2ON/OFF cell lines: CEN (integration in pericentromeric chromatin) and ARM (integration in the q arm of macro-chromosome). (b) Aligned metaphases with LacI-GFP signals in the same focal plane for SMC2ON and SMC2OFF cells. Scale bar, 5 μm. (c) Kymographs of the LacO array movements. In SMC2OFF cells pericentromeric chromatin undergoes excursions similar to those of sister kinetochores. These movements are abolished by nocodazole (dotted line shows time of addition) and Hec1 RNAi; they are unaffected by the proteasome inhibitor MG132. The ARM locus exhibits an increased spacing but no oscillations in SMC2OFF cells. (d) Summary of the average Δd and Δdmax values and their ratios for all loci determined from live cell imaging data. The Δdmax values for the kinetochores and pericentromeric integration are increased by 50 and 58%, respectively, in the absence of condensin, whereas no significant changes are obtained for the ARM integration. (e) Distribution of distances between the LacI:GFP signals in fixed metaphases of SMC2ON/SMC2OFF:CEN cells incubated with 20 μM MG132. Topo II inhibition (ICRF 159) does not restore the normal interlocus spacing. SMC2wt restores the normal interlocus spacing distribution but an ATPase-deficient SMC2 point mutant (SMC2S1086R) does not; n = 90. p < 0.001; Mann-Whitney U test.
Kymographs revealed the dynamic separation between the LacI:GFPCEN signals (Figure 4c), and their uncoordinated behavior was readily observed in movies of SMC2OFF:CEN cells (Supplementary Movies 4 and 5). Remarkably, the loss of condensin caused an approximately twofold increase in the maximum separation of the chromosomal region linking the two LacO arrays (Figure 4d). Thus, in this simplified system pericentric chromatin accurately reproduces the behavior of kinetochores. As with kinetochores, the excursions were reversible and MT-dependent. They were abolished by nocodazole (Figure 4c; Supplementary Figure 2f, and Supplementary Movie 6) or the RNAi knockdown of Hec1 (Figure 2b; Supplementary Figure 3g and Supplementary Movies 7–9). The oscillatory movements were unaffected by the addition of proteasome inhibitor MG132 (Figure 4c; Supplementary Figure 3e and Supplementary Movie 10). In controls, a LacO array integrated on the arm of a macro-chromosome (Figure 4a, ARM) maintained a constant relative distance between sister chromatids during metaphase in cells lacking condensin (Figure 4c; Supplementary Figure 3, b and d, and Supplementary Movies 11 and 12). This distance was larger than that in cells expressing SMC2 because, unlike inner centromeric chromatin, arm chromatin is 40% less compact in SMC2OFF cells (Hudson et al., 2003; Vagnarelli et al., 2006).
To begin to address the underlying mechanism by which condensin regulates centromere stiffness, we asked whether SMC2 ATPase activity is required for normal behavior of kinetochores under tension at metaphase. One strength of the DT40 cell conditional knockout system is that it is possible to perform complementation studies under conditions where the wild-type protein is essentially undetectable. Indeed, expression of wild-type SMC2 driven by a fragment of the endogenous SMC2 promoter restored the shorter distance between the two centromere-proximal sister loci in SMC2OFF:CEN cells (Figure 4e). However, SMC2S1086R, a mutant capable of assembling a condensin complex that targets to chromosomes (Supplementary Figure S1) but that lacks ATPase activity (Hudson et al., 2008) failed to rescue the spacing between sister kinetochores (Figure 4e). Therefore, normal stiffness of the centromeric chromatin requires SMC2 ATPase activity.
Loss of topoisomerase II (topo II) can reduce the interkinetochore distance in metaphase (Spence et al., 2007), and topo II can influence the longitudinal elasticity of mitotic chromosome arms (Kawamura and Marko, personal communication; Marko, 2008). Indeed, treatment with topo II inhibitor ICRF 159 slightly reduced the spacing between sister kinetochores, even in the presence of condensin (Figure 4e). However, addition of the drug failed to restore normal centromere stiffness to SMC2OFF cells lacking condensin. Thus, catenation regulated by topo II can have a minor effect on the compaction of inner centromeric chromatin, but is not a major factor regulating its compliance.
Proper Centromeric Stiffness Is Required for a Timely Silencing of the Spindle Assembly Checkpoint
A clue to the functional significance of the regulation of centromere stiffness by condensin was provided by the observation that in unperturbed cell cycles, the mitotic index in SMC2OFF cells is higher (7.7 ± 3.5%) than in SMC2ON cells (3.1 ± 1.2%), consistent with a mitotic delay (Figure 5, a and b, 0 time point). These cells accumulate in late prometaphase/metaphase before entering anaphase (Figure 5h). Time-lapse imaging cells revealed that condensin-depleted cells do not have problems in chromosome congression or in the maintenance of chromosome alignment (Figure 5k). Nonetheless, the mitotic delay appears to be due to activation of the spindle checkpoint.
Figure 5.
SMC2OFF cells exhibit persistent spindle checkpoint activation. (a and b) SMC2OFF cells can activate the spindle checkpoint normally in presence of colcemid (a) or taxol (b). n = 500 in each of three experiments. (c) Quantification of Mad2-positive kinetochores per metaphase. (d and f) Kinetochores of SMC2ON/OFF cells undergoing poleward excursions are negative for Mad2 and BubR1 (empty arrow and inset). Mad2:GFP-positive kinetochores are BubR1-positive in SMC2ON/OFF cells (filled arrows). (e and g) Staining SMC2ON/OFF kinetochores in aligned metaphases for Mad2. Scale bar, 5 μm. (h) Cells without condensin show an increase in late prometaphase and metaphase cells. n = 50 in each of three experiments. (i) SMC2OFF spindles are longer than SMC2ON spindles; n = 110. (j) Kinetochores of SMC2OFF cells engaged in poleward excursions are MCAK-negative (inset and empty arrow). Scale bar, 5 μm. (k and k′) Chromosomes of SMC2ON and SMC2OFF cells maintain a stable metaphase alignment in the presence of MG132.
SMC2OFF cells activate the spindle checkpoint normally in the presence of colcemid or taxol (Figure 5, a and b). The mitotic delay observed in otherwise unperturbed cell cycles appears to arise from sustained checkpoint activation, as we observed an increase in the number of SMC2OFF cells with aligned metaphases having two or more Mad2:GFP-positive kinetochores (Figure 5, c–g). However, kinetochores undergoing P excursions lacked detectable Mad2 or BubR1, or the MT de-polymerase MCAK (Figure 5, f and j), suggesting that KTs engaged in these extended movements exhibit a full MT occupancy. Instead, the Mad2-positive kinetochores were present on chromosomes apparently aligned on the metaphase plate.
Condensin Sets the Stiffness of the Centromeric Chromatin
If we model the inner centromeric chromatin as a spring using Hooke's law as a simplifying assumption (Gardner et al., 2005), then Fy = −kΔd, where F is the force applied, k is the spring constant or stiffness of the spring, and Δd is the distance stretched beyond the rest length. We postulate that FMAX is likely to be approximately equivalent in the presence and absence of condensin because the number of kMTs was the same in SMC2ON and SMC2OFF cells (Figure 2i) and because kinetochores, spindles, and the rest length of the centromeric chromatin between sister kinetochores were close to normal in SMC2OFF cells (Figure 2; Supplementary Figure 1, d and e). Recent studies in budding yeast found that the nucleosome architecture of chromatin regulates the rest length but not the stiffness of centromeres (Bouck and Bloom, 2007).
The average extent of P excursions (Δd) in SMC2OFF cells (0.52 ± 0.26 μm) was about two times that observed in SMC2ON cells (0.24 ± 0.09 μm; Figures 4d and 6a; Supplementary Figure S2a). Therefore, according to Hooke's law the spring constant of centromeres lacking condensin is 0.46 that of wild-type centromeres. Strikingly, a nearly identical value was obtained when we analyzed the movements of the centromere-linked LacO-CEN locus (Figure 4d). This is the first evidence for a protein complex affecting specifically the spring constant (stiffness) of centromere chromatin.
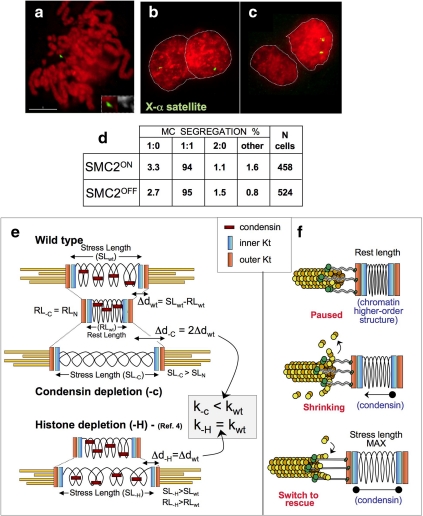
Figure 6.
Proposed role of condensin in regulating the compliance and MT dynamics at centromeres. (a–c) SMC2ON/OFF cell line carrying a human minichromosome was used to test the frequency of mis-segregation in the presence and absence of condensin. (a) Chromosome spread hybridized with a chromosome X-specific α-satellite probe; (b and c) binucleate cells showing two different types of segregation 1:1 (b) and 2:0 (c); (d) quantification of the experiment. (e) Without condensin, unstressed centromeric chromatin is as compact as wild type (rest length). On MT attachment, centromeres in SMC2OFF cells deform twice as much as SMC2ON centromeres (Δd). H3 or H4 depletion increased the rest length but the deformation due to spindle forces was unaltered (Bouck and Bloom, 2007). (f) Hypothetical model linking centromere chromatin elasticity and kinetochore MT dynamics.
DISCUSSION
Using a conditional knockout for the SMC2 subunit of the condensin complex coupled with live cell imaging, we have shown that the ATPase activity of SMC2 is required for the normal stiffness of inner centromeric chromatin when bioriented kinetochores are subjected to MT pulling forces in metaphase. We also found that kinetochores behave in DT40 cells as structurally independent domains that are linked by the elastic chromatin at the inner centromere. These dynamic properties of the centromeric chromatin are linked to spindle checkpoint activation and silencing. Using this system, we have shown that a weak centromeric spring causes delay in mitotic progression due to prolonged checkpoint activation.
The results obtained in our experimental model for observing kinetochore dynamics are in agreement with a previous study in which condensin depletion caused a reversible deformation of the centromeric chromatin accompanying uncoordinated sister kinetochore movements (Gerlich et al., 2006). We could not confirm results reported for Drosophila, in which similar kinetochore movements were observed, but where the distortions of the centromeric chromatin were irreversible (Oliveira et al., 2005).
Kinetochores Appear Structurally and Functionally Normal in Condensin-depleted DT40 Cells
Several studies have reported altered centromere/kinetochore structure in the absence of condensin. In yeast, condensin depletion causes loss of Cse4 from centromeres (Yong-Gonzalez et al., 2007). In human cells, RNAi for condensin caused aberrations in CENP-E and CREST signal geometry in one study (Ono et al., 2004) but no evident distortion of CENP-A signals in another (Gerlich et al., 2006). In Xenopus egg extracts, immunodepletion of condensin caused abnormal CENP-E localization (Wignall et al., 2003), and in Drosophila condensin RNAi, CID (the CENP-A homologue) was found to be distorted in metaphase (Jäger et al., 2005). Distortion of the kinetochore was also observed in the holocentric chromosomes of Caenorhabditis elegans after condensin RNAi (Hagstrom et al., 2002).
We show here that in DT40 cells, kinetochore overall structure is maintained after the depletion of condensin, as indicated by serial-sectioning EM and analysis of the distribution and/or copy number of several key kinetochore components. Likewise, kinetochore function is maintained, as indicated by spindle checkpoint activation and silencing when the kinetochores are engaged in poleward excursions at high degrees of centromere stretch. Furthermore, we showed previously that kinetochores in DT40 cells lacking detectable condensin segregate to opposite spindle poles during anaphase even when the chromatin trailing behind them becomes grossly distorted (Hudson et al., 2003; Vagnarelli et al., 2006). This is reminiscent of results in C. elegans showing that even though kinetochores in embryos lacking SMC2 were abnormal in metaphase, they adopted a normal morphology by anaphase (Kaitna et al., 2002). Furthermore, if the targeting subunit RepoMan is prevented from recruiting protein phosphatase 1 to the chromatids during anaphase, then the entire process of anaphase chromatid segregation appears to be completely normal in DT40 cells lacking detectible condensin (Vagnarelli et al., 2006).
The differences between the various studies are likely to have at least two explanations. First, DT40 kinetochores bind for four- to fivefold fewer MTs than the other metazoan kinetochores examined, and this could render them less susceptible to distortion in the absence of condensin. Second, the present study and that of Gerlich et al. (2006), which also failed to describe kinetochore structure abnormalities after condensin RNAi in human cells, were conducted mostly on live cells. Our analyses on fixed samples were performed using optimized fixation conditions that preserved the structure of both the chromatin and the kinetochore. This was particularly significant in this case because we have shown previously that condensin-depleted chromosomes are exquisitely sensitive to fixation conditions (Hudson et al., 2003), and disruption of kinetochore structure may have occurred in other studies during sample preparation.
An altered structure of the centromere might be expected to produce an increase in merotelic attachments and consequent chromosome mis-segregation. We did not observe this when monitoring the segregation of either lacI-GFP-tagged loci (Hudson et al., 2003; Vagnarelli et al., 2006) or a human mini-chromosome in cells with or without condensin (Figure 6, a–d), nor did we see any evidence of lagging kinetochores at anaphase in our earlier studies of the condensin knockout cells (Hudson et al., 2003; Vagnarelli et al., 2006). Because kMT attachment is believed to be a stochastic process, the probability of merotelic attachment may be less for kinetochores with fewer MT-binding sites. Regardless, our data show conclusively that condensin is not an obligate component of a system preventing merotelic attachments in vertebrate kinetochores.
Mitotic Delay in Condensin-depleted Cells Is Caused by Prolonged Activation of the Spindle Assembly Checkpoint
Our analysis suggests that centromere stretch has a biological function in regulating MT attachment to kinetochores. While this manuscript was under revision, two articles were published showing that intrakinetochore stretch during mitosis is important for silencing the spindle checkpoint (Maresca and Salmon, 2009; Uchida et al., 2009). These results could give the impression that centromere stretch between sister kinetochores is not required to silence the checkpoint. However, such an interpretation would be misleading. Indeed, many studies over the years have shown that tension can stabilize MT attachment to kinetochores and promote MT growth (Nicklas and Koch, 1969; Rieder and Salmon, 1994; Inoue and Salmon, 1995; Skibbens et al., 1995; Nicklas et al., 2001; Gardner et al., 2005; Figure 6, e and f).
Thus, although tension between sister kinetochores may not directly silence the spindle checkpoint signaling cascade (Maresca and Salmon, 2009; Uchida et al., 2009), this tension is likely to lessen the probability that kinetochores will release their MTs (Nicklas et al., 2001). Of course, kinetochores that release their MTs because of a lack of tension do activate the spindle checkpoint. Therefore, there is likely to be a critical, if indirect, link between the degree of interkinetochore centromere tension and activation/inactivation of the spindle checkpoint.
We did not analyze intrakinetochore stretch in the present study; however, our data are consistent with two possible models to explain the checkpoint activation and mitotic delay consequent upon condensin depletion. First, condensin depletion may affect tension within the kinetochore itself, thereby promoting checkpoint activation as suggested recently (Maresca and Salmon, 2009; Uchida et al., 2009). Second, the alteration in compliance of the centromeric chromatin that occurs upon condensin loss may create a gradient of tension within the spindle. This could cause kinetochores to release their MTs and activate the spindle checkpoint when tension is below a threshold, particularly in a sensitized system such as DT40 cells with only four MTs per kinetochore. Indeed, we observed an increase in Mad2-positive kinetochores located near the metaphase plate in the region where we would expect spindle tension to be lowest. Mad2 localization to kinetochores is widely accepted to be diagnostic of a lack of MT occupancy, and our result suggests that the kinetochores of chromosomes near the metaphase plate are more likely to release their MTs after condensin depletion.
In condensin-depleted cells the kinetochores that are engaging in the most obviously abnormal behavior—the poleward excursions—are not those that are signaling to the spindle checkpoint. We explain this by suggesting that even weakened springs can produce the same level of tension as stronger springs—this merely happens at a greater Δd (stretch) and is consistent with the observation that spindles are slightly longer in SMC2OFF cells (Figure 5i). Thus, the reduced stiffness of centromeric chromatin in condensin-depleted cells could create a gradient of tension: lowest near the surface of the centromere and progressively higher as the kinetochore is stretched further poleward. Because Aurora B activity is high on the chromosomes (Fuller et al., 2008), regions of lowered tension might be expected to favor kMT detachment and spindle checkpoint activation (King and Nicklas, 2000). Indeed, kinetochores undergoing excursions might be predicted to have particularly stable MT attachments, because they are located furthest from the Aurora B in the inner centromere (Cimini et al., 2006).
Decreased tension could also occur when uncoordinated sister kinetochores engage in simultaneous AP movements for longer than normal periods; however, this would not a priori show any preference for position on the spindle. Reproducibly, the Mad2-positive kinetochores were located close to the metaphase plate where tension would be expected to be lowest according to the gradient model.
SMC2 ATPase Activity Determines the Compliance of the Centromeric Chromatin
The loss of condensin causes an approximately twofold decrease in the stiffness of the centromeric “spring” when measured by two quite distinct reporter loci (Figure 4g). The underlying molecular mechanism is unknown, but SMC2 depletion causes changes in the localization and chromosomal association of DNA topo II alpha, KIF4A and a number of other chromosome scaffold components (Hudson et al., 2003; Gassmann et al., 2004). Thus, it is possible that condensin depletion alters the distribution or function of proteins involved in establishing or regulating sister chromatid cohesion, such as cohesin or Sgo1.
The twofold decrease in spring constant is consistent with a halving of the structural links between kinetochores. This suggests an obvious parallel with the ability of SMC-based cohesin rings to link pairs of chromatin fibers together (Nasmyth and Haering, 2005). Full understanding of the organization and dynamics of the centromere “spring” will therefore require a detailed elucidation of the mechanism of centromeric chromatin binding by condensin.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Robin Allshire, Alison Pidoux, and Jim Paulson for criticisms of the manuscript. This work was supported by the Wellcome Trust (P.V., W.C.E.), the Caledonian Research Foundation (D.H.), the Darwin Trust of Edinburgh (S.A.R.) and National Institutes of Health Grant GM06627 to B.M.E. W.C.E. is a Principal Research Fellow of The Wellcome Trust.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1127) on March 4, 2009.
REFERENCES
- Almagro S., Riveline D., Hirano T., Houchmandzadeh B., Dimitrov S. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J. Biol. Chem. 2004;279:5118–5126. doi: 10.1074/jbc.M307221200. [DOI] [PubMed] [Google Scholar]
- Bouck D. C., Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 2007;17:741–748. doi: 10.1016/j.cub.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C. B., Salmon E. D. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Coelho P. A., Queiroz-Machado J., Sunkel C. E. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 2003;116:4763–4776. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Dong Y., Hergert P., Strauss J., Hickey J. M., Salmon E. D., McEwen B. F. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., McDonald K. L., McIntosh J. R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Laemmli U. K. Architecture of metaphase chromosomes and chromosome scaffolds. J. Cell Biol. 1983;96:84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. Epigenetic control of centromere behavior. Annu. Rev. Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., Cleveland D. W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Fuller B. G., Lampson M. A., Foley E. A., Rosasco-Nitcher S., Le K. V., Tobelmann P., Brautigan D. L., Stukenberg P. T., Kapoor T. M. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. K., Pearson C. G., Sprague B. L., Zarzar T. R., Bloom K., Salmon E. D., Odde D. J. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Vagnarelli P., Hudson D., Earnshaw W. C. Mitotic chromosome formation and the condensin paradox. Exp. Cell Res. 2004;296:35–42. doi: 10.1016/j.yexcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J. M., Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Hagstrom K. A., Holmes V. F., Cozzarelli N. R., Meyer B. J. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J. M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B., Marko J. F., Chatenay D., Libchaber A. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol. 1997;139:1–12. doi: 10.1083/jcb.139.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D. F., Ohta S., Freisinger T., Macisaac F., Sennels L., Alves F., Lai F., Kerr A., Rappsilber J., Earnshaw W. C. Molecular and genetic analysis of condensin function in vertebrate cells. Mol. Biol. Cell. 2008;19:3070–3079. doi: 10.1091/mbc.E08-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D. F., Vagnarelli P., Gassmann R., Earnshaw W. C. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- Inoue S., Salmon E. D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger H., Rauch M., Heidmann S. The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma. 2005;113:350–361. doi: 10.1007/s00412-004-0322-4. [DOI] [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek P., Jantsch M., Loidl J., Glotzer M. The Aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- King J. M., Nicklas R. B. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 2000;113(Pt 21):3815–3823. doi: 10.1242/jcs.113.21.3815. [DOI] [PubMed] [Google Scholar]
- Loncarek J., Kisurina-Evgenieva O., Vinogradova T., Hergert P., La Terra S., Kapoor T. M., Khodjakov A. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T. J., Salmon E. D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko J. F. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008;16:469–497. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- Marshall O. J., Marshall A. T., Choo K. H. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J. Cell Biol. 2008;183:1193–1202. doi: 10.1083/jcb.200804078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B. F., Dong Y. Releasing the spindle assembly checkpoint without tension. J. Cell Biol. 2009;184:355–356. doi: 10.1083/jcb.200812016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., West R. R. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Koch C. A. Chromosome Micromanipulation III. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B., Waters J. C., Salmon E. D., Ward S. C. Checkpoint signals in grasshopper meiosis are sensitive to microtubule attachment, but tension is still essential. J. Cell Sci. 2001;114:4173–4183. doi: 10.1242/jcs.114.23.4173. [DOI] [PubMed] [Google Scholar]
- Oliveira R. A., Coelho P. A., Sunkel C. E. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol. 2005;25:8971–8984. doi: 10.1128/MCB.25.20.8971-8984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D. L., Hirano T. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Salmon E. D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A. L., Allshire R. C. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:569–579. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L., Salmon E. D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Korfali N., Villa P., Kottke T. J., Dingwall C., Kaufmann S. H., Earnshaw W. C. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 2002;21:1967–1977. doi: 10.1093/emboj/21.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N., Goldberg I., Wood E., Earnshaw W. C. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R. V., Rieder C. L., Salmon E. D. Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci. 1995;108:2537–2548. doi: 10.1242/jcs.108.7.2537. [DOI] [PubMed] [Google Scholar]
- Skibbens R. V., Skeen V. P., Salmon E. D. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J. M., Phua H. H., Mills W., Carpenter A. J., Porter A. C., Farr C. J. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 2007;120:3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Hudson D. F., Ribeiro S. A., Trinkle-Mulcahy L., Spence J. M., Lai F., Farr C. J., Lamond A. I., Earnshaw W. C. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 2006;8:1133–1142. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Ribeiro S. A., Earnshaw W. C. Centromeres: old tales and new tools. FEBS Lett. 2008;582:1950–1959. doi: 10.1016/j.febslet.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Warburton P. E., et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Waters J. C., Mitchison T. J., Rieder C. L., Salmon E. D. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall S. M., Deehan R., Maresca T. J., Heald R. The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J. Cell Biol. 2003;161:1041–1051. doi: 10.1083/jcb.200303185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Gonzalez V., Wang B. D., Butylin P., Ouspenski I., Strunnikov A. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 2007;12:1075–1090. doi: 10.1111/j.1365-2443.2007.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.