Abstract
The identification and characterization of proprotein convertase subtilisin-like/kexin type 9 (PCSK9) have provided new insights into LDL metabolism and the causal role of LDL in coronary heart disease (CHD). PCSK9 is a secreted protease that mediates degradation of the LDL receptor by interacting with the extracellular domain and targeting the receptor for degradation. Individuals with loss-of-function mutations in PCSK9 have reduced plasma levels of LDL cholesterol and are protected from CHD; these observations have validated PCSK9 as a therapeutic target and suggested new approaches for the treatment and prevention of CHD.
Keywords: low density lipoprotein receptor, proprotein convertase subtilisin-like/kexin type 9, low density lipoprotein, hypercholesterolemia
For the past four decades, characterization of patients with genetic disorders of lipid metabolism has revealed key components of the molecular machinery involved in biosynthetic and regulatory pathways that produce, transport, and eliminate cholesterol. These findings have led to the development of potent cholesterol-lowering drugs that reduce coronary heart disease (CHD). The identification of new forms of genetic hyperlipidemias continues to reveal novel participants, as exemplified by the discovery that mutations in proprotein convertase subtilisin-like/kexin type 9 (PCSK9) have profound effects on plasma levels of LDL cholesterol (LDL-C).
Here, we focus on advances in PCSK9 biology and structure during the last 2 years.
MISSENSE MUTATIONS IN PCSK9 REVEAL A KEY REGULATOR OF LDL METABOLISM
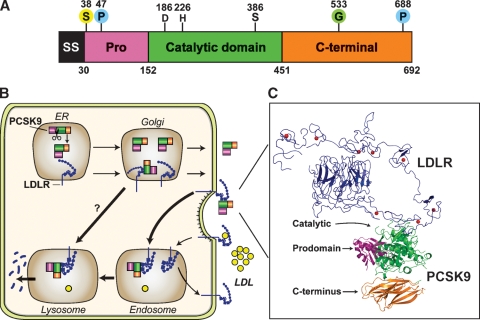
In 2003, Abifadel et al. (1) identified three families with autosomal dominant hypercholesterolemia and premature CHD caused by missense mutations in the ninth member of the proprotein convertase family, PCSK9. PCSK9 encodes a 692 amino acid protein that is expressed predominantly in liver, intestine, and kidney (2). The protein contains a signal sequence, a prodomain (amino acids 31–152), a catalytic domain (amino acids 153–451), and a C-terminal domain (amino acids 452–692) that are rich in cysteines and histidines (Fig. 1A). Overexpression of PCSK9 in livers of mice markedly reduces hepatic LDL receptor (LDLR) protein (but not mRNA) levels, causing hypercholesterolemia (3). This finding suggested that the missense mutations identified by Abifadel et al. (1) conferred a gain-of-function to the mutant protein. Subsequent studies revealed that inactivation of PCSK9 in humans and mice resulted in hypocholesterolemia (3).
Fig. 1.
PCSK9-mediated degradation of the LDLR. A: Schematic of the major domains of PCSK9. The location of the aspartate (D), histidine (H), and serine (S) that comprise the catalytic triad and the site of binding of the single N-linked sugar (N533) are shown. PCSK9 posttranslational modifications include glycosylation (G) (4), phosphorylations (P) at S47 and S688 (5), and sulfation (S) at Y38 and at another undefined tyrosine in the catalytic domain (6). B: Cellular trafficking and potential sites of PCSK9 action. After undergoing autocatalytic cleavage in the ER, the prodomain (purple) remains associated with the catalytic fragment (green) and the complex is secreted into the plasma. Secreted PCSK9 binds to LDLRs on the cell surface, and the LDLR/PCSK9 complex is internalized via the adaptor protein ARH (LDLRAP). PCSK9 may prevent the recycling of LDLRs from endosomes to the cell surface or direct LDLRs to lysosomes for degradation. A second intracellular pathway has been proposed (11) in which PCSK9 binds the LDLR in a post-ER compartment and targets LDLR for degradation in lysosomes. C: Model for full-length LDLR bound to PCSK9. The EGF-A domain of the LDLR (blue) at acidic pH and the PCSK9:EGF-A complex were superimposed. PCSK9 binds on the outside surface of LDLR. SS, signal sequence; Pro, prodomain.
PHYSIOLOGY OF PCSK9
Cellular itinerary of PCSK9
PCSK9 is synthesized as a 73 kDa zymogen in the endoplasmic reticulum (ER) and is modified en route to the cell surface (Fig. 1B). The protein undergoes autocatalytic cleavage between residues 152 and 153 (FAQ152↓SIP). The N-terminal prodomain remains tightly associated with the 63 kDa mature protein and acts as a chaperone to transport PCSK9 through the secretory pathway (4). A mutation in PCSK9 (C679×) that prevents folding of the C-terminal domain does not prevent autocatalytic cleavage, suggesting that cleavage is cotranslational (3). After cleavage, the last four amino acids of the prodomain blanket the catalytic triad, thereby restricting access to potential substrates. PCSK9 undergoes a series of posttranslational modifications, including glycosylation (4), phosphorylation (5), and tyrosine sulfation (6) (Fig. 1A). None of these modifications is required for secretion of PCSK9, and their role in PCSK9 function remains obscure.
Circulating levels of PCSK9
Mice in which PCSK9 has been selectively inactivated in liver have no detectable PCSK9 in the blood, suggesting that the liver is the major source of circulating PCSK9 (2). Recently, several laboratories have developed ELISAs to measure plasma PCSK9 levels in humans (3, 7–9). The mean concentration of PCSK9 varies widely between these assays (ranging from 500 ng/ml to 4 μg/ml) likely due to differences in antibody specificities and the standards used in the assays. PCSK9 levels correlate with LDL-C (r = 0.3–0.6) but not HDL-C (8, 9). Plasma levels of LDL-C and PCSK9 may be directly related because expression of PCSK9 promotes the degradation of hepatic LDLRs. The levels of these two proteins are not invariably coupled; treatment with high-dose statins reduces plasma levels of LDL-C but increases levels of circulating PCSK9 (8).
Kinetics of PCSK9 clearance
Recombinant PCSK9 has a half-life of ∼5 min in the blood of wild-type mice (10). Inactivating the LDLR increases the half-life of PCSK9 to ∼15 min, implicating LDLR as a major conduit for PCSK9 removal (10). The rapid clearance of PCSK9 from plasma, even in Ldlr−/− mice, indicates that the rate of PCSK9 synthesis must be high, given the low plasma concentrations of PCSK9.
Site of action of PCSK9
The effect of PCSK9 on LDL-C levels appears to be mediated solely through LDLRs since inactivation of Pcsk9 does not reduce plasma cholesterol levels in Ldlr−/− mice (2). Parabiosis experiments in wild-type and PCSK9 transgenic mice demonstrate that circulating PCSK9 can mediate degradation of LDLRs in the liver (3). Infusion of recombinant human PCSK9 into mice to levels comparable with those in human plasma caused a significant reduction of hepatic LDLRs (10). These experiments support PCSK9 acting primarily at the cell surface, although it remains possible that the protein interferes with the movement of the LDLR in the secretory pathway (Fig. 1B) (11).
It is not clear to what extent PCSK9 affects LDLRs in tissues other than the liver. Intravenous infusion of PCSK9 into mice at levels as high as 32 μg/h for 6 h abolished hepatic LDLR expression but failed to reduce LDLRs in adrenals (10). Moreover, LDLRs were not increased in adrenals of Pcsk9−/− mice (3). However, in another study, LDLRs were reduced in several tissues, particularly in lung, after the injection of 100 μg of PCSK9 (12).
Why PCSK9 preferentially degrades LDLRs in hepatocytes is unknown. A similar mystery surrounds ARH (LDLRAP), an adaptor protein required for LDLR internalization in hepatocytes but not in most other cell types (3). A physiological explanation might be that PCSK9 selectively suppresses LDLRs in the liver to prevent the immediate reuptake of nascent VLDL particles, thereby promoting their delivery to peripheral tissues.
Regulation of PCSK9
PCSK9 is regulated primarily at the level of transcription by sterol regulatory element binding proteins (SREBPs) (3), which regulate genes involved in fatty acid synthesis (SREBP-1c) and cholesterol metabolism (SREBP-2) (13). An SREBP binding site in the promoter of PCSK9 has been characterized (14). Although SREBP-1c has been implicated in the regulation of PCSK9 (15), SREBP-2 appears to play a more central role under physiological conditions (3). Manipulations that suppress SREBP-2, such as fasting or cholesterol feeding, are associated with lower PCSK9 mRNA levels in livers of mice (14). Mice that lack hepatic SREBP-2 have reduced PCSK9 mRNA levels, whereas mice that lack SREBP-1c do not (unpublished observations).
Activators of the nuclear receptors farnesoid X receptor or peroxisome proliferator-activated receptor α reduce PCSK9 mRNA in cultured human hepatocytes (16, 17). The physiological significance of these regulators requires further study.
PCSK9:LDLR BINDING AND STRUCTURE
Biochemical and crystallographic studies have provided new insights into how PCSK9 binds LDLRs and promotes degradation. The extracellular domain of the LDLR consists of an N-terminal ligand binding domain (R1-R7) that mediates binding to LDL, followed by an epidermal growth factor (EGF)-precursor homology domain that contains a pair of EGF-like repeats (EGF-A and EGF-B) separated from a third EGF-like repeat (EGF-C) by a β-propeller domain (18). After LDL binds to the LDLR, the receptor-ligand complex undergoes endocytosis and is delivered to the endosome (Fig. 1B) (13). The bound lipoproteins are released in the acidic environment of the endosome, and the LDLR recycles to the cell surface.
Structural requirements for PCSK9 binding and function
PCSK9 binds specifically to the EGF-A domain of the LDLR in a calcium-dependent manner (19). Other regions of the LDLR that do not contact PCSK9 are required for PCSK9-mediated degradation, including the β-propeller domain and at least three ligand binding repeats (20). The reason for these structural requirements is not known. The C-terminal domain of PCSK9 does not bind to the LDLR, but this region is required for LDLR degradation (20). Thus, the C terminus may bind another protein that directs LDLRs to lysosomes, or the domain may prevent the binding of a protein required for recycling of the LDLR from endosomes to the cell surface.
In the crystal structure of the LDLR at acidic pH, the ligand binding repeats 4 and 5 closely associate with the β-propeller domain (18). These findings suggested that the LDLR changes from an open to a closed conformation at acid pH, thereby facilitating the release of LDL particles (18). The pH-dependent conformational change in LDLR is not required for LDLR degradation by PCSK9 (20), but the affinity of PCSK9 binding to the LDLR is enhanced at acidic pH, suggesting that PCSK9 binds more avidly to LDLRs in the endosome/lysosomal compartments (19, 21, 22). An unresolved question is whether PCSK9 and LDL compete for binding to the LDLR.
PCSK9 crystal structure
The structure of PCSK9 was reported by three groups in 2007 (21, 23, 24), all of whom obtained similar structures despite using a wide range of pH conditions. The catalytic domain resembles that of other subtilisin-like serine proteases except for the high-affinity interactions with the prodomain that shields the active site from exogenous substrates. The C-terminal domain is a unique structure that contains three six-stranded β-sheet subdomains arranged with quasi-three-fold symmetry.
PCSK9:LDLR-AB cocrystal
The structure of PCSK9 in complex with the EGF-AB fragment from the LDLR revealed that PCSK9 primarily contacts the N-terminal region of the EGF-A repeat (25). The prodomain and the C-terminal domain of PCSK9 do not contact the EGF-A domain (Fig. 1C). The interface between PCSK9 and EGF-A is >20 Å from the catalytic site and is formed primarily by hydrophobic residues surrounded by several specific polar interactions. High levels of PCSK9 and LDLR expression in cells promotes associations of PCSK9 with regions of the LDLR outside of EGF-A, which may explain reports that PCSK9 binds VLDLR and apoER2 under certain conditions (27).
GAIN-OF-FUNCTION MUTATIONS IN PCSK9
Mutations in PCSK9 are responsible for a small fraction (<2%) of genetic hypercholesterolemias (28). In contrast with familial hypercholesterolemia (FH), which is caused by >1,000 different mutations in the LDLR, only a small number of missense mutations in PCSK9 cause hypercholesterolemia. Structural studies of PCSK9 provide insight into the mechanisms by which two gain-of-function mutations in PCSK9, D374Y and F216L, may function to increase LDLR degradation.
PCSK9(D374Y) is ∼10-fold more active than wild-type PCSK9 in mediating degradation of LDLRs, owing to an ∼5- to 30-fold increased affinity for the receptor (3, 21, 22). In the structure of the PCSK9:EGF-AB interface, PCSK9-D374 forms a salt bridge with EGF-A-H306. Changing D374 to Y places the hydroxyl group of tyrosine close enough to the carbonyl oxygen of EGF-A-C319 to permit formation of a new hydrogen bond (29). Surprisingly, replacing D374Y with other amino acids, including alanine and phenylalanine, increases the binding affinity between PCSK9 and the LDLR (30). Thus, the aspartate at residue 374 may ensure that PCSK9 binds with relatively low affinity to EGF-A at neutral pH, perhaps preventing the depletion of LDLRs on the sinusoidal membrane and limiting interactions with other proteins that contain EGF-A repeats.
Substitution of histidine for aspartate at the same position (D374H) is also associated with hypercholesterolemia (31). Amino acid substitutions at D374 are associated with higher plasma LDL-C levels and earlier CHD than is typically observed in heterozygous FH (28, 32), likely because the mutant PCSK9 causes a >50% reduction in LDLR. Controversy exists as to whether the patients with these mutations have altered responsiveness to HMG-CoA reductase inhibitors (statins) (28, 32).
The gain-of-function mutation F216L does not increase the affinity of PCSK9 for the LDLR (21, 25). The F216L mutation, and a mutation in an adjacent residue (R215H) (33), may alter a potential furin/PC5/6A-dependent cleavage site in PCSK9 (6). If this mechanism is correct, then these mutations would be predicted to prolong the half-life of PCSK9, causing these individuals to have higher circulating levels of the protein.
The least understood gain-of-function mutation in PCSK9 is S127R, which resides in the prodomain (Fig. 1A). The mutation reduces autocatalytic cleavage of PCSK9, resulting in decreased PCSK9 secretion. The mutant protein has only a modest increase in affinity for the LDLR (21, 22). In functional studies of a double mutant PCSK9 that contains the D374Y and S127R mutations, an additive affect is seen on the ability of the protein to reduce LDL uptake in cells, suggesting that the two mutations might work through independent mechanisms (30). The S127R mutation may interfere with intracellular trafficking of the LDLR to the cell surface, as has been suggested (11, 34).
LOSS-OF-FUNCTION MUTATIONS IN PCSK9 AND CHD
Several loss-of-function alleles in PCSK9 have been identified in individuals with hypocholesterolemia (3). Most of these mutations are rare, but three were sufficiently common to examine the relationship between LDL-C levels and CHD. In the Atherosclerosis Risk in Communities Study, a missense mutation in the prodomain (R46L), present in 3% of Caucasians, was associated with a 15% reduction in LDL-C and a 46% reduction in CHD (3). In the same study, two nonsense mutations (Y142× and C679×), present in 2% of African-Americans, caused a 28% reduction in LDL-C and an 88% reduction in CHD (3). The reduction in CHD associated with the PCSK9-R46L mutation has been replicated in two case-control studies (35, 36).
Could the greater than expected reduction in CHD observed in association with PCSK9 variants be due to an effect of PCSK9 on CHD that is independent of the effect on LDL-C levels? Recently, common sequence variations in other genes (e.g., APOE, HMGR, and APOB) have been linked to LDL-C levels and CHD risk (37). These findings are consistent with the loss-of-function variants in PCSK9 reducing CHD risk through lowering plasma levels of LDL-C.
To probe the association between hypocholesterolemia and cancer, Folsom, Peacock, and Boerwinkle (38) compared the frequency of PCSK9 loss-of-function alleles in subjects with and without various cancers (colon, lung, breast, and prostate). No differences in allele frequencies between the two groups were found. Although additional studies are required, these data suggest that low plasma levels of cholesterol are a consequence, not a causal contributor, to cancer.
Two young women with total PCSK9 deficiency have been identified (39). Both subjects had very low plasma levels of LDL-C (14 mg/dl and 16 mg/dl), providing perhaps the most compelling evidence of the importance of PCSK9 in LDL metabolism. No adverse clinical sequellae were reported in either individual. Determining the long-term consequences of PCSK9 deficiency will require careful clinical assessment of additional, older PCSK9-deficient individuals.
THERAPEUTIC APPROACHES TO INHIBITING PCSK9 ACTION
Despite the evidence from human studies that PCSK9 would be an excellent drug target for the treatment of hypercholesterolemia, enthusiasm for the development of small-molecule inhibitors of the protease has been tempered by the finding that the catalytic activity of PCSK9 is not required for LDLR degradation (26). However, an intracellular inhibitor of PCSK9 catalytic activity may be effective since autocatalytic processing of PCSK9 is required for secretion of the protein from the ER.
An alternative strategy to inhibit PCSK9 synthesis is to degrade the PCSK9 mRNA with antisense oligonucleotides or small interfering RNA. Antisense oligonucleotides administered to mice reduced PCSK9 expression by >90% and lowered plasma cholesterol levels by ∼53% (40). A single intravenous injection of an small interfering RNA delivered in lipidoid nanoparticles to cynomologous monkeys reduced plasma PCSK9 levels by ∼70% and plasma LDL-C levels by ∼56% (41). Plasma LDL-C levels remained significantly lower 3 weeks after injection.
A third option to inhibit PCSK9 activity is to prevent binding of PCSK9 to LDLRs at the cell surface with small molecules, peptides, or antibodies directed against PCSK9. Overexpressing soluble LDLR (42) or adding EGF-A fragments to cultured cells (43) inhibits the ability of exogenously added PCSK9 to mediate LDLR degradation. The success of this approach depends on PCSK9 functioning primarily at the cell surface.
New agents to treat hypercholesterolemia should work additively or synergistically with statins. Several lines of evidence suggest that an inhibitor of PCSK9 will augment the hypolipidemic effects of these powerful drugs. LDL clearance is accelerated and plasma cholesterol levels are reduced in statin-treated Pcsk9−/− mice compared with untreated Pcsk9−/− mice (3). Statins increase PCSK9 transcription, resulting in higher plasma PCSK9 levels (7, 8). The elevation in PCSK9 attenuates the increase in hepatic LDLR, which may explain why most LDL-C lowering occurs with the first dose of statin (>25%), and further doublings of the dose reduce plasma LDL levels by only ∼6% (44).
ARE THERE OTHER FUNCTIONS OF PCSK9?
Currently, there is little evidence that PCSK9 has functions other than promoting LDLR degradation. No other proteins that contains an EGF-A repeat are predicted to interact with PCSK9, based on the structure of the PCSK9:EGF-AB complex. The only phenotype of PCSK9 deficiency detected in humans is lower plasma cholesterol levels, but it remains possible that aging or exposure to environmental challenges, such as infectious agents or dietary toxins, may reveal other phenotypes associated with absence of PCSK9. For example, mice expressing no PCSK9 only in liver have impaired liver regeneration after a partial hepatectomy (2).
IMPLICATIONS FOR HUMAN HEALTH
The reduced CHD risk in individuals with loss-of-function mutations in PCSK9 indicates that LDL is necessary for the development of CHD and suggests that a modest, lifelong decrease in LDL-C levels would confer substantial protection against CHD, even in the presence of other risk factors (e.g., hypertension, diabetes, and smoking) (3). The reduction in CHD associated with defective PCSK9 alleles is greater than would be predicted from multiple statin trials. This observation highlights the importance of duration of exposure to LDL-C as well as the absolute level of LDL-C. Loss-of-function mutations in PCSK9 are associated with reduced plasma levels of LDL-C, starting in childhood (45). In contrast with a static measurement, such as a plasma level of LDL-C, genotypes provide an integrated measure of cholesterol exposure.
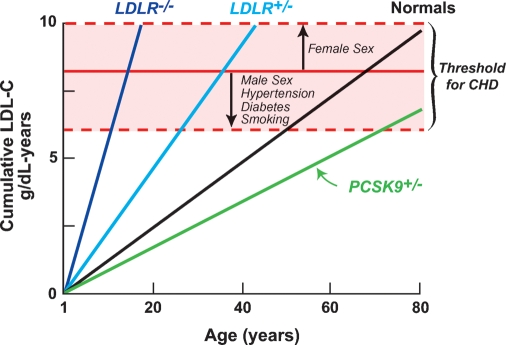
Figure 2 compares the cumulative LDL exposure (expressed as grams of cholesterol per year) over a lifetime in FH patients (46) and normal individuals (47). CHD occurs after a theoretical threshold of LDL exposure is exceeded (indicated by a red line). This threshold is reached in childhood in FH homozygotes and in early middle age in FH heterozygotes and is significantly delayed in individuals who are heterozygous for PCSK9 loss-of-function mutations. These findings raise new questions. Would modest reductions in plasma levels of LDL-C starting at an earlier age enhance the benefit of cholesterol-lowering therapy? Will new methods to estimate the lifetime exposure of blood vessels to LDL provide better indicators of absolute CHD risk than do current guidelines?
Fig. 2.
Relationship between cumulative LDL-C exposure and age. Cumulative plasma levels of LDL-C were estimated from mean plasma levels of LDL-C for FH homozygotes (46), FH heterozygotes (46), age-adjusted LDL-C levels in normal individuals [calculated from National Health and Nutrition Education Survey III (47, 48)], and PCSK9 heterozygotes (3). The horizontal red line represents a theoretical threshold of the cumulative LDL exposure required for development of CHD. The height of the red line will be lower in the presence of additional CHD risk factors (e.g., male sex, smoking, diabetes, and hypertension).
FINAL COMMENTS
The discovery of PCSK9, the elucidation of the mechanism by which it regulates plasma LDL cholesterol, and the validation of PCSK9 as a therapeutic target for the treatment of hypercholesterolemia occurred in just 5 years. The progress achieved in such a short period is a compelling testament to the value of careful clinical characterization and classical genetic studies of unusual patients (1), even in the postgenome era.
Abbreviations
CHD, coronary heart disease
EGF, epidermal growth factor
ER, endoplasmic reticulum
FH, familial hypercholesterolemia
LDL-C, LDL cholesterol
LDLR, LDL receptor
PCSK9, proprotein convertase subtilisin-like/kexin type 9
SREBP, sterol regulatory element binding protein
Published, JLR Papers in Press, November 19, 2008.
References
- 1.Abifadel M., M. Varret, J. P. Rabes, D. Allard, K. Ouguerram, M. Devillers, C. Cruaud, S. Benjannet, L. Wickham, D. Erlich, et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34 154–156. [DOI] [PubMed] [Google Scholar]
- 2.Zaid A., A. Roubtsova, R. Essalmani, J. Marcinkiewicz, A. Chamberland, J. Hamelin, M. Tremblay, H. Jacques, W. Jin, J. Davignon, et al. 2008. Proprotein convertase subtilisin/kexin type 9 (PCSK9): hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology. 48 646–654. [DOI] [PubMed] [Google Scholar]
- 3.Horton J. D., J. C. Cohen, and H. H. Hobbs. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seidah N. G., S. Benjannet, L. Wickham, J. Marcinkiewicz, S. B. Jasmin, S. Stifani, A. Basak, A. Prat, and M. Chretien. 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 100 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewpura T., A. Raymond, J. Hamelin, N. G. Seidah, M. Mbikay, M. Chretien, and J. Mayne. 2008. PCSK9 is phosphorylated by a Golgi casein kinase-like kinase ex vivo and circulates as a phosphoprotein in humans. FEBS J. 275 3480–3493. [DOI] [PubMed] [Google Scholar]
- 6.Benjannet S., D. Rhainds, J. Hamelin, N. Nassoury, and N. G. Seidah. 2006. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J. Biol. Chem. 281 30561–30572. [DOI] [PubMed] [Google Scholar]
- 7.Mayne J., T. Dewpura, A. Raymond, M. Cousins, A. Chaplin, K. A. Lahey, S. A. Lahaye, M. Mbikay, T. C. Ooi, and M. Chretien. 2008. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans. Lipids Health Dis. 7 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Careskey H. E., R. A. Davis, W. E. Alborn, J. S. Troutt, G. Cao, and R. J. Konrad. 2008. Atorvastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 49 394–398. [DOI] [PubMed] [Google Scholar]
- 9.Lambert G., N. Ancellin, F. Charlton, D. Comas, J. Pilot, A. Keech, S. Patel, D. R. Sullivan, J. S. Cohn, K. A. Rye, et al. 2008. Plasma PCSK9 concentrations correlate with LDL and total cholesterol in diabetic patients and are decreased by fenofibrate treatment. Clin. Chem. 54 1038–1045. [DOI] [PubMed] [Google Scholar]
- 10.Grefhorst A., M. C. McNutt, T. A. Lagace, and J. D. Horton. 2008. Plasma PCSK9 preferentially reduces liver LDL receptors in mice. J. Lipid Res. 49 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell K. N., E. A. Fisher, and J. L. Breslow. 2005. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA. 102 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt R. J., T. P. Beyer, W. R. Bensch, Y. W. Qian, A. Lin, M. Kowala, W. E. Alborn, R. J. Konrad, and G. Cao. 2008. Secreted proprotein convertase subtilisin/kexin type 9 reduces both hepatic and extrahepatic low-density lipoprotein receptors in vivo. Biochem. Biophys. Res. Commun. 370 634–640. [DOI] [PubMed] [Google Scholar]
- 13.Brown, M. S., and J. L. Goldstein. 2008. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J. Lipid Res. S15–S27. [DOI] [PMC free article] [PubMed]
- 14.Jeong H. J., H. S. Lee, K. S. Kim, Y. K. Kim, D. Yoon, and S. W. Park. 2008. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 49 399–409. [DOI] [PubMed] [Google Scholar]
- 15.Costet P., B. Cariou, G. Lambert, F. Lalanne, B. Lardeux, A. L. Jarnoux, A. Grefhorst, B. Staels, and M. Krempf. 2006. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281 6211–6218. [DOI] [PubMed] [Google Scholar]
- 16.Langhi C., C. Le May, S. Kourimate, S. Caron, B. Staels, M. Krempf, P. Costet, and B. Cariou. 2008. Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett. 582 949–955. [DOI] [PubMed] [Google Scholar]
- 17.Kourimate S., C. Le May, C. Langhi, A. L. Jarnoux, K. Ouguerram, Y. Zair, P. Nguyen, M. Krempf, B. Cariou, and P. Costet. 2008. Dual mechanisms for the fibrate-mediated repression of proprotein convertase subtilisin/kexin type 9. J. Biol. Chem. 283 9666–9673. [DOI] [PubMed] [Google Scholar]
- 18.Rudenko G., L. Henry, K. Henderson, K. Ichtchenko, M. S. Brown, J. L. Goldstein, and J. Deisenhofer. 2002. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 298 2353–2358. [DOI] [PubMed] [Google Scholar]
- 19.Zhang D. W., T. A. Lagace, R. Garuti, Z. Zhao, M. McDonald, J. D. Horton, J. C. Cohen, and H. H. Hobbs. 2007. Binding of proprotein convertase subtilisin/kexin yype 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282 18602–18612. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D. W., R. Garuti, W. J. Tang, J. C. Cohen, and H. H. Hobbs. 2008. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 105 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham D., D. E. Danley, K. F. Geoghegan, M. C. Griffor, J. L. Hawkins, T. A. Subashi, A. H. Varghese, M. J. Ammirati, J. S. Culp, L. R. Hoth, et al. 2007. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat. Struct. Mol. Biol. 14 413–419. [DOI] [PubMed] [Google Scholar]
- 22.Fisher T. S., P. Lo Surdo, S. Pandit, M. Mattu, J. C. Santoro, D. Wisniewski, R. T. Cummings, A. Calzetta, R. M. Cubbon, P. A. Fischer, et al. 2007. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 282 20502–20512. [DOI] [PubMed] [Google Scholar]
- 23.Hampton E. N., M. W. Knuth, J. Li, J. L. Harris, S. A. Lesley, and G. Spraggon. 2007. The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain. Proc. Natl. Acad. Sci. USA. 104 14604–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piper D. E., S. Jackson, Q. Liu, W. G. Romanow, S. Shetterly, S. T. Thibault, B. Shan, and N. P. Walker. 2007. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure. 15 545–552. [DOI] [PubMed] [Google Scholar]
- 25.Kwon H. J., T. A. Lagace, M. C. McNutt, J. D. Horton, and J. Deisenhofer. 2008. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA. 105 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNutt M. C., T. A. Lagace, and J. D. Horton. 2007. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J. Biol. Chem. 282 20799–20803. [DOI] [PubMed] [Google Scholar]
- 27.Poirier S., G. Mayer, S. Benjannet, E. Bergeron, J. Marcinkiewicz, N. Nassoury, H. Mayer, J. Nimpf, A. Prat, and N. G. Seidah. 2008. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J. Biol. Chem. 283 2363–2372. [DOI] [PubMed] [Google Scholar]
- 28.Humphries S. E., R. A. Whittall, C. S. Hubbart, S. Maplebeck, J. A. Cooper, A. K. Soutar, R. Naoumova, G. R. Thompson, M. Seed, P. N. Durrington, et al. 2006. Genetic causes of familial hypercholesterolaemia in patients in the UK: relation to plasma lipid levels and coronary heart disease risk. J. Med. Genet. 43 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottomley M. J., A. Cirillo, L. Orsatti, L. Ruggeri, T. S. Fisher, J. C. Santoro, R. T. Cummings, R. M. Cubbon, P. Lo Surdo, A. Calzetta, et al. 2009. Structural and biochemical characterization of the wild type PCSK9/EGF(AB) complex and natural familial hypercholesterolemia mutants. J. Biol. Chem. 284 1313–1323. [DOI] [PubMed] [Google Scholar]
- 30.Pandit S., D. Wisniewski, J. C. Santoro, S. Ha, V. Ramakrishnan, R. M. Cubbon, R. T. Cummings, S. D. Wright, C. P. Sparrow, A. Sitlani, et al. 2008. Functional analysis of sites within PCSK9 responsible for hypercholesterolemia. J. Lipid Res. 49 1333–1343. [DOI] [PubMed] [Google Scholar]
- 31.Bourbon M., A. C. Alves, A. M. Medeiros, S. Silva, and A. K. Soutar. 2008. Familial hypercholesterolaemia in Portugal. Atherosclerosis. 196 633–642. [DOI] [PubMed] [Google Scholar]
- 32.Naoumova R. P., I. Tosi, D. Patel, C. Neuwirth, S. D. Horswell, A. D. Marais, C. van Heyningen, and A. K. Soutar. 2005. Severe hypercholesterolemia in four British families with the D374Y mutation in the PCSK9 gene: long-term follow-up and treatment response. Arterioscler. Thromb. Vasc. Biol. 25 2654–2660. [DOI] [PubMed] [Google Scholar]
- 33.Cameron J., O. L. Holla, J. K. Laerdahl, M. A. Kulseth, T. Ranheim, T. Rognes, K. E. Berge, and T. P. Leren. 2008. Characterization of novel mutations in the catalytic domain of the PCSK9 gene. J. Intern. Med. 263 420–431. [DOI] [PubMed] [Google Scholar]
- 34.Homer V. M., A. D. Marais, F. Charlton, A. D. Laurie, N. Hurndell, R. Scott, F. Mangili, D. R. Sullivan, P. J. Barter, K. A. Rye, et al. 2008. Identification and characterization of two non-secreted PCSK9 mutants associated with familial hypercholesterolemia in cohorts from New Zealand and South Africa. Atherosclerosis. 196 659–666. [DOI] [PubMed] [Google Scholar]
- 35.McPherson R., and N. Kavaslar. 2007. Statins for primary prevention of coronary artery disease. Lancet. 369 1078. [DOI] [PubMed] [Google Scholar]
- 36.Kathiresan S. 2008. A PCSK9 missense variant associated with a reduced risk of early-onset myocardial infarction. N. Engl. J. Med. 358 2299–2300. [DOI] [PubMed] [Google Scholar]
- 37.Willer C. J., S. Sanna, A. U. Jackson, A. Scuteri, L. L. Bonnycastle, R. Clarke, S. C. Heath, N. J. Timpson, S. S. Najjar, H. M. Stringham, et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folsom A. R., J. M. Peacock, and E. Boerwinkle. 2007. Sequence variation in proprotein convertase subtilisin/kexin type 9 serine protease gene, low LDL cholesterol, and cancer incidence. Cancer Epidemiol. Biomarkers Prev. 16 2455–2458. [DOI] [PubMed] [Google Scholar]
- 39.Hooper A. J., A. D. Marais, D. M. Tanyanyiwa, and J. R. Burnett. 2007. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 193 445–448. [DOI] [PubMed] [Google Scholar]
- 40.Graham M. J., K. M. Lemonidis, C. P. Whipple, A. Subramaniam, B. P. Monia, S. T. Crooke, and R. M. Crooke. 2007. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J. Lipid Res. 48 763–767. [DOI] [PubMed] [Google Scholar]
- 41.Frank-Kamenetsky M., A. Grefhorst, N. N. Anderson, T. S. Racie, B. Bramlage, A. Akinc, D. Butler, K. Charisse, R. Dorkin, Y. Fan, et al. 2008. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. USA. 105 11915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian Y. W., R. J. Schmidt, Y. Zhang, S. Chu, A. Lin, H. Wang, X. Wang, T. P. Beyer, W. R. Bensch, W. Li, et al. 2007. Secreted PCSK9 downregulates low density lipoprotein receptor through receptor-mediated endocytosis. J. Lipid Res. 48 1488–1498. [DOI] [PubMed] [Google Scholar]
- 43.Shan L., L. Pang, R. Zhang, N. J. Murgolo, H. Lan, and J. A. Hedrick. 2008. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem. Biophys. Res. Commun. 375 69–73. [DOI] [PubMed] [Google Scholar]
- 44.Jones P. H., M. H. Davidson, E. A. Stein, H. E. Bays, J. M. McKenney, E. Miller, V. A. Cain, and J. W. Blasetto; STELLAR Study Group. 2003. Comparison of the efficacy and safety of rosuvastatin versus atorvastatin, simvastatin and pravastatin across doses (STELLER* Trial). Am. J. Cardiol. 92 152–160. [DOI] [PubMed] [Google Scholar]
- 45.Hallman D. M., S. R. Srinivasan, W. Chen, E. Boerwinkle, and G. S. Berenson. 2007. Relation of PCSK9 mutations to serum low-density lipoprotein cholesterol in childhood and adulthood (from The Bogalusa Heart Study). Am. J. Cardiol. 100 69–72. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein, J. L., H. H. Hobbs, and M. S. Brown. 2001. Familial hypercholesterolemia. In The Metabolic and Molecular Bases of Inherited Disease. C. Scriver, A. Beaudet, W. Sly, and D. Valle, editors. McGraw Hill, New York. 2863–2913.
- 47.Third report of the National Cholesterol Education Program (NCEP) expert panel on the detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) final report. Circulation. 2002. 106: 3143–3421. [PubMed]
- 48.Hickman T. B., R. R. Briefel, M. D. Carroll, B. M. Rifkind, J. I. Cleeman, K. R. Maurer, and C. L. Johnson. 1998. Distributions and trends of serum lipid levels among United States children and adolescents ages 4–19 years: data from the Third National Health and Nutrition Examination Survey. Prev. Med. 27 879–890. [DOI] [PubMed] [Google Scholar]