Abstract
Cholesterol biosynthesis is among the most intensely regulated processes in biology. Synthetic rates vary over hundreds of fold depending on the availability of an external source of cholesterol. Studies of this feedback regulatory process have a rich history. The field began 75 years ago when Rudolf Schoenheimer measured cholesterol balance in mice in a bottle. He found that cholesterol feeding led to decreased cholesterol synthesis, thereby introducing the general phenomenon by which end products of biosynthetic pathways inhibit their own synthesis. Recently, cholesterol feedback has been explained at a molecular level with the discovery of membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs), and an appreciation of the sterol-sensing role of their partner, an escort protein called Scap. The key element in Scap is a hexapeptide sequence designated MELADL (rhymes with bottle). Thus, over 75 years, Schoenheimer's bottle led to Scap's MELADL. In addition to their basic importance in membrane biology, these studies have implications for the regulation of plasma cholesterol levels and consequently for the development of atherosclerotic plaques, myocardial infarctions, and strokes. In this article we review the major milestones in the cholesterol feedback story.
Keywords: cholesterol biosynthesis, LDL receptor, SREBP pathway, Insig, oxysterols, HMG-CoA reductase, CopII-coated vesicles, membrane proteins, historical aspects
CLASSIC EXPERIMENTS
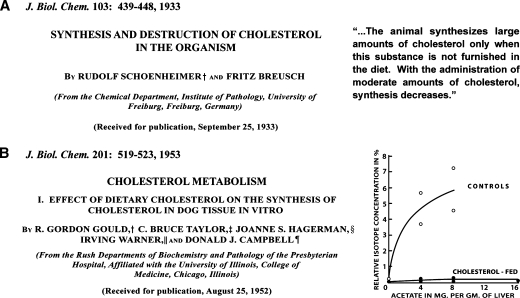
In 1933, Rudolf Schoenheimer, then a physician in Germany, published his classic studies of cholesterol balance in mice. Schoenheimer placed mice in ventilated bottles and fed them diets high and low in cholesterol. After a month, he measured the total content of cholesterol in the bottle. He demonstrated that mice produce cholesterol when the diet is cholesterol-poor, and they exhibit a net destruction of cholesterol when they consume cholesterol (1). Figure 1A shows the title page of Schoenheimer's classic 1933 paper on cholesterol feedback in living mice and the conclusions that he drew. These early experiments predated by many years the classic studies of Jacob and Monod, who elucidated the mechanism of feedback inhibition in bacteria (2).
Fig. 1.
Two classic papers on cholesterol homeostasis in animals. (A) In 1933, Schoenheimer and Breusch (1) published the first example of a biological feedback system. They used balance techniques to study cholesterol synthesis and degradation in mice on low and high cholesterol diets. Their major conclusion is shown in quotation marks. (B) In 1953, Gould et al. (4) performed one of the first studies to use radioisotopes to measure the synthesis of cholesterol. They showed a reduction in the incorporation of [14C]acetate into [14C]cholesterol in liver slices from dogs fed a high cholesterol diet (1% cholesterol). The animals were fed the diets for 7 days, and the incubation time was 3 hours.
Table 1 lists key experimental milestones, from the physiological to the molecular, that delineate the feedback control of cholesterol homeostasis. In the 1950s and 1960s, scientists in several laboratories elucidated the intricate pathway by which 2-carbon acetate is converted to 27-carbon cholesterol with its four conjoined rings. Central to this effort were discoveries made in the laboratories of Bloch, Lynen, Folkers, and Cornforth and Popjak (reviewed in Ref. 3). Those discoveries laid the groundwork for the identification of 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) reductase as the first committed enzyme in the cholesterol biosynthetic pathway and the primary site of feedback regulation. During this same period, the emergence of radioisotopes permitted the demonstration that cholesterol biosynthesis occurs in the liver and that the liver is subject to feedback repression when animals are fed cholesterol (Table 1).
TABLE 1.
Feedback control of cholesterol homeostasis: experimental milestones from 1933 to 2007
| Year | Experimental System | Major Finding | References |
|---|---|---|---|
| 1933 | Balance studies in mice | Dietary cholesterol suppresses whole body synthesis of cholesterol | (1) |
| 1951–54 | Tissue slices from dogs and rats | Dietary cholesterol inhibits [14C]acetate incorporation into cholesterol; major site of inhibition localized to liver | (4, 105–108) |
| 1951–60 | Extracts of yeast and rat liver | Pathway of cholesterol synthesis, involving 25 enzymatic steps, delineated | (3) |
| 1959–71 | Liver slices and microsomes from rats | HMG-CoA reductase identified as key enzyme in pathway; inhibited by dietary cholesterol | (109–112) |
| 1973–85 | Cultured mammalian cells | Cells obtain cholesterol from 2 sources: endogenous synthesis and receptor-mediated uptake of LDL – both subject to feedback control | (6) |
| 1984–85 | Cultured hamster UT-1 cells | cDNA and gene for HMG-CoA reductase cloned; feedback mediated by inhibition of gene transcription and acceleration of degradation | (18, 22, 27) |
| 1990 | Mutational analysis and transfection | Sterol regulatory element (SRE) in promoter/enhancer of cholesterol-regulated genes identified as 10-bp sequence | (30) |
| 1993–94 | Biochemical purification and molecular biology | cDNA for SREBP family of membrane-bound transcription factors cloned | (32, 35) |
| 1995–03 | Cultured mammalian cells and livers of mice | SREBP pathway delineated; feedback mediated by two sterol-sensing membrane proteins, Scap and Insig | (60, 76, 92, 98) |
| 2005–07 | Reconstituted ER-to-Golgi transport assays | Molecular switch mediating feedback localized to MELADL sequence in Scap, which governs transport of Scap/SREBPs into CopII vesicles | (81, 82) |
Figure 1B shows the title page and key experiment from a 1953 paper by Gordon Gould and colleagues who were the first to use radioactive isotopes to demonstrate feedback inhibition in dog liver (4). In this seminal 1953 paper, Gould et al. measured the incorporation of [14C]acetate into [14C]cholesterol in liver slices of dogs that had been fed a control diet (0% cholesterol) and a high cholesterol diet (1% cholesterol) for 7 days. The results showed a striking inhibition of cholesterol synthesis in the livers from the cholesterol-fed dogs. In the 1980s, quantitative studies in the laboratory of John Dietschy showed that the liver is the major site of total body cholesterol synthesis in rats, monkeys, and several other animal species (5).
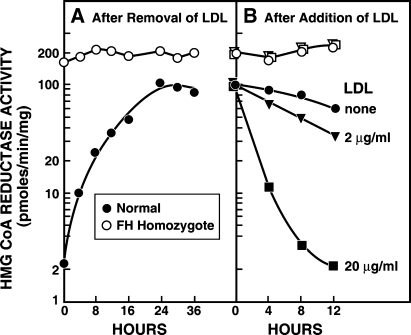
Our own collaborative studies in Dallas were initiated in 1972 when we began to investigate the feedback regulation of cholesterol synthesis in human fibroblasts grown in tissue culture (reviewed in Ref. 6). Our motivation was to understand familial hypercholesterolemia (FH), a genetic disease characterized by elevated levels of cholesterol-rich LDL in plasma and premature myocardial infarctions (7). Measuring HMG-CoA reductase activity in cell-free extracts from normal fibroblasts, we showed that enzyme activity was low when the cells were grown in the presence of LDL (Fig. 2A). Reductase activity rose by 50-fold when LDL was removed from the culture medium, and the activity rapidly declined when LDL was added back (Fig. 2B). We observed a striking difference in cells from a subject with the homozygous form of FH. When grown in LDL-containing medium, these cells had reductase activity that was 75-fold above normal (Fig. 2A). There was no further increase when LDL was removed from the medium and no decline when LDL was restored (Fig. 2B).
Fig. 2.
Feedback regulation of HMG-CoA reductase activity in fibroblasts from a normal subject (•) and from an FH homozygote (○). A: Monolayers of cells were grown in dishes containing 10% fetal calf serum. At zero time, the medium was replaced with fresh medium containing 5% human serum from which the lipoproteins had been removed. At the indicated time, extracts were prepared, and HMG-CoA reductase activity was measured. B: After a 24-h incubation with 5% human lipoprotein-deficient serum, human LDL was added to give the indicated cholesterol concentration. HMG-CoA reductase activity was measured in cell-free extracts at the indicated time. [Reprinted with permission from Goldstein and Brown (7)].
Those striking findings led us on a 10-year quest that resulted in the discovery of the LDL receptor pathway and the appreciation of the general phenomenon of receptor-mediated endocytosis (reviewed in Refs. 6 and 8). We found that normal fibroblasts have a cell surface receptor that binds LDL and internalizes the lipoprotein in coated pits. The internalized LDL is degraded in lysosomes and the cholesterol is made available for membrane synthesis. The liberated cholesterol leads to a major decrease in HMG-CoA reductase activity. This decrease is attributable to a reduction in the amount of enzyme protein rather than inhibition of its enzymatic activity. FH homozygote cells lack LDL receptors, and therefore they are unable to take up LDL. This finding explained their constitutively high levels of HMG-CoA reductase activity.
HMG-COA REDUCTASE: CDNA CLONING AND REGULATION
After we worked out the steps in the LDL receptor pathway, we turned to an analysis of the mechanisms by which HMG-CoA reductase protein levels are reduced by lipoprotein-derived cholesterol. We were aided by the observation that cholesterol accumulation also reduces the number of LDL receptors, thus shutting off the external supply of cholesterol as well as endogenous synthesis (9, 10). Indirect evidence indicated that the major mechanism for the reduction in HMG-CoA reductase and the LDL receptor is through a reduction in the mRNAs for these two proteins. This finding suggested that the transcription of both genes was regulated by a common factor. T. Y. Chang and colleagues reached a similar conclusion from the analysis of mutant CHO cells auxotrophic for cholesterol (11). By this time, the molecular biologists had provided us with the tools necessary to clone cDNAs and genes. Together with Wolfgang Schneider, we purified the LDL receptor from the richest natural source, namely the adrenal glands of cows (12), We then obtained a partial protein sequence, and together with Tokuo Yamamoto and David Russell, we cloned the cDNA (13). The gene structure was worked out by Thomas Südhof, then a postdoctoral fellow (14).
Cloning of HMG-CoA reductase defied a biochemical approach. The enzyme is bound to membranes of the endoplasmic reticulum (ER), and it resisted all attempts at purification. Instead, we used a genetic trick. Following a precedent established by Robert Schimke with inhibitors of dihydrofolate reductase (15), we treated cultured chinese hamster ovary (CHO) cells with compactin, then a recently discovered competitive inhibitor of HMG-CoA reductase (16). When CHO cells were grown in the absence of LDL, they were forced to rely on endogenous cholesterol synthesis, and hence they were killed by the reductase inhibitor. However, a small number of cells survived because they produced sufficient reductase to overcome the inhibition. These resistant cells were grown up, and the concentration of compactin was increased, again killing the vast majority of cells. The rare survivors were again grown up, and the dose of compactin was again ratcheted up. After many months of ratcheting and selection, we obtained a line of CHO cells that resisted high concentrations of compactin. We estimated that HMG-CoA reductase constituted about 2% of the total protein in these cells, which we designated UT-1 cells in honor of our parent university (17).
At this time in the early 1980s, few if any “housekeeping” genes had been cloned. The cloning methods were so primitive that one could only clone a cDNA if the mRNA constituted a high percentage of the total mRNA in the cell. Because of the compactin selection, the UT-1 cells fit these criteria. Two courageous postdoctoral fellows, Dan Chin and Ken Luskey, set out to clone reductase. The cDNA library from UT-1 cells contained only 360 partial cDNAs, but from it we were able to isolate the cDNA for HMG-CoA reductase (18) and also for HMG-CoA synthase, the enzyme that precedes reductase in the cholesterol biosynthetic pathway (19). Sequencing of the reductase cDNA revealed that the protein contains 887 amino acids. The N-terminal 339 amino acids form eight membrane-spanning helices, thereby anchoring the protein to ER membranes (20, 21). The C-terminal 548 amino acids contain all of the catalytic activity. Using the newly invented techniques of mutagenesis, we produced a cDNA that encoded only the catalytic portion of reductase. This truncated protein was completely soluble, and it was able to restore cholesterol-independent growth to UT-2 cells, a line of reductase-deficient cells that we had isolated (22). If the soluble catalytic domain contains all of the activity, why is it attached to the N-terminal membrane-bound domain? The answer lies in regulated degradation. When attached to the membrane, reductase is stable only in sterol-depleted cells. The enzyme is rapidly degraded when cellular sterol levels rise. Gregorio Gil showed that this rapid degradation does not occur when cells express only the catalytic domain of reductase as a soluble protein (22). Thus, sterols control HMG-CoA reductase activity in part by accelerating the degradation of the membrane-bound enzyme (22, 23).
With cDNAs for the LDL receptor and HMG-CoA reductase in hand, we were soon able to show directly that the mRNAs for both proteins decline rapidly when sterols are added to cultured cells (24). Our experiments were aided by the observation that oxygenated derivatives of cholesterol, such as 25-hydroxycholesterol (25-HC), are even more effective than cholesterol when added to the culture medium in solvents (25, 26). A variety of experiments demonstrated that the sterol-mediated reduction in mRNA levels is caused by a decline in the rate of transcription. Indirect evidence indicated the presence of a transcription factor that enhanced the transcription (27) of both genes and that was rendered inactive when cellular sterol levels rose (reviewed in Ref. 28).
SREBPS: MEMBRANE-BOUND TRANSCRIPTION FACTORS
The search for the postulated sterol-regulated transcription factor took several years and required the efforts of many postdoctoral fellows. The breakthrough came when Südhof identified a 42-bp sequence in the enhancer region of the LDL receptor promoter that conferred sterol-regulated transcription when inserted into a heterologous promoter (29). Subsequent studies of the 42-bp element by Jeffrey Smith and Timothy Osborne further localized the key component to a 10-bp sequence that we named the sterol regulatory element (SRE) (30). Navigating skillfully through a minefield of potential artifacts, Xiaodong Wang and Michael Briggs used 500 l of cultured HeLa cells as a source to purify a nuclear protein of ∼60 kDa that bound specifically to the SRE (31). We named this protein SRE-binding protein (SREBP). A partial amino acid sequence was obtained, and this was used to clone a cDNA for SREBP. The DNA sequence revealed that SREBP was not a 60 kDa protein, but rather a 125 kDa protein (32). The 60 kDa peptide purified from nuclear extracts was only the N-terminal portion of the 125 kDa protein. Sequence analysis revealed that the 60 kDa nuclear fragment belongs to a large family of transcription factors designated basic-helix-loop-helix-leucine zipper (bHLH-Zip) proteins. All other members of this family recognize palindromic DNA sequences called E-boxes. SREBP differs because the SRE is not a palindromic E-box. Moreover, SREBP has a tyrosine in the DNA binding domain that differs from a conserved arginine residue found in all other bHLH-Zip proteins (33). Crystallography studies showed that this tyrosine is the key to the ability of SREBP to recognize the nonpalindromic SRE (34).
Initially, we believed that there was only one SREBP. However, there was an issue of concern. The cloned cDNA for SREBP encoded five of the six peptides that we had identified by protein sequencing in our purified SREBP (32). The cDNA did not encode the sixth peptide. We were tempted to pass off this anomaly as a contaminant in our purified SREBP. However, Xianxin Hua decided to try to clone the cDNA encoding the sixth peptide. He used nested PCR reactions to clone this cDNA, which was found to encode a closely related protein (35). We named the first protein SREBP-1 and the second one SREBP-2. Further studies showed that SREBP-1 existed in two major forms, owing to alternate promoters giving rise to alternate first exons. We named these two versions SREBP-1a and SREBP-1c (32, 36). At about the same time, the laboratory of Bruce Spiegelman embarked on studies to identify novel bHLH-Zip transcription factors that promote adipogenesis. When they probed an adipocyte cDNA library with an E-box sequence, they isolated a cDNA encoding an apparent 95 kDa protein that they named ADD1 (37). This cDNA turned out to be a truncated form of SREBP-1c, the truncation occurring because of an apparent sequence anomaly in the original ADD1 cDNA (36). SREBP-1c/ADD1 was subsequently shown by Spiegelman's laboratory (38) and our laboratory (39) to activate genes for fatty acid synthesis.
THE SREBP PATHWAY: A SURPRISE
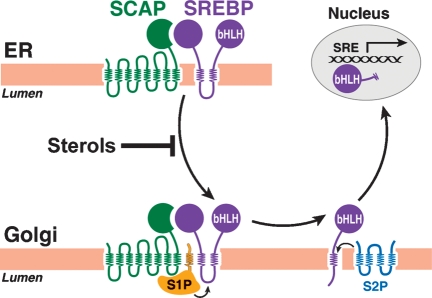
The biggest surprise in the SREBP story came when Roy Sato, a postdoctoral fellow, prepared an antibody against the nuclear fragment of SREBP-1a and used the antibody to stain cultured fibroblasts that had been incubated with and without an exogenous source of cholesterol. In sterol-depleted cells, the SREBP was in the nucleus, as expected. Remarkably, in sterol-overloaded cells, the SREBP was not in the nucleus. Rather, it exhibited a lacy distribution consistent with localization on ER membranes (40). At this point, we examined the predicted protein sequence more carefully. A hydrophobicity plot revealed that the N-terminal fragment of ∼480 amino acids containing the bHLH-Zip domain was followed by a sequence of 80 amino acids that included two classic membrane-spanning helices separated by a 30 amino acid hydrophilic sequence (41). This putative membrane attachment domain is followed by a segment of ∼550 amino acids that we called the regulatory domain. Protease protection experiments showed that SREBPs are oriented in the ER membranes in a hairpin fashion. The N-terminal bHLH-Zip domain and the C-terminal regulatory domain project into the cytosol, and the 30-amino acid hydrophilic loop projects into the lumen (Fig. 3) (42).
Fig. 3.
The SREBP pathway. When cells are depleted of sterols, Scap transports SREBPs from the ER to the Golgi apparatus. Release of SREBPs from the membrane is initiated by Site-1 Protease (S1P), a Golgi-located protease that cleaves SREBPs in the luminal loop between the two membrane-spanning sequences. Once the two halves of the SREBP are separated, a second Golgi protease, Site-2 Protease (S2P), cleaves the NH2-terminal bHLH-Zip domain of SREBP at a site located within the membrane-spanning region. After the second cleavage, the NH2-terminal bHLH-Zip domain leaves the membrane, carrying three hydrophobic residues at its COOH-terminus. The cleaved SREBP enters the nucleus, where it activates genes controlling lipid synthesis and uptake.
In sterol-depleted cells, the bHLH-Zip domain of SREBP is released from the membrane by proteolysis so that it can enter the nucleus. Biochemical studies conducted by Juro Sakai revealed that the release process was much more complicated than we expected. Two proteolytic cleavages are required (Fig. 3) (43). The first protease cuts SREBPs at a conserved leucine residue within the 30-amino acid luminal loop. This protease, designated site-1 protease (S1P), separates the SREBPs into two halves, but the bHLH-Zip domain remains associated with the membrane by virtue of its single membrane-spanning helix. After cleavage by S1P, a second protease, designated site-2 protease (S2P), clips the N-terminal fragment, releasing the bHLH-Zip domain from the membrane. Even more remarkable, Elizabeth Duncan showed that S2P cleaves the protein within a transmembrane helix (44). When initially described, this type of cleavage reaction was considered unique, but later studies by others showed that several other proteins are also cleaved within membrane-spanning regions. We designated this general process as regulated intramembrane proteolysis (45, 46).
To identify the two postulated proteases, we turned to the use of somatic cell genetics in collaboration with Robert Rawson. We were aided by the availability of mutant CHO cells that are cholesterol auxotrophs. These cells, originally isolated in the laboratory of T. Y. Chang and subsequently in our laboratory, fail to increase the mRNAs for HMG-CoA reductase and the LDL receptor when deprived of cholesterol, and hence the cells die. Biochemical studies demonstrated that the defect in one of these cell lines, designated M19, lies in S2P (43). These cells cleave SREBPs at site-1, but the bHLH-Zip domain remains attached to the membrane because of the lack of S2P. In collaboration with T. Y. Chang's laboratory, we corrected this defect in M19 cells by transfecting human genomic DNA (47). After repeated rounds of transfection and selection, we eventually identified the human gene. The encoded protein contained the sequence HEXXH, which is part of the catalytic site in a family of zinc metalloproteases. Mutation of these residues destroyed the activity of S2P, supporting the notion that the protein is a zinc metalloprotease (47). However, S2P differs dramatically from previously characterized zinc metalloproteases in that it is extremely hydrophobic. Indeed, the HEXXH sequence is embedded in a highly hydrophobic region of the protein (48). Remarkably, hydrophobic proteases resembling S2P are found in all mammalian species, in Eubacteria, and even in Archaea. The laboratory of Richard Losick showed that a relative of S2P in Bacillus subtilis senses nutrient deprivation by proteolytically processing a membrane-bound transcription factor, which then initiates sporulation (49). Recent structural studies of another bacterial relative of S2P confirmed that the protein contains zinc and provide a potential gating mechanism for cleavage of transmembrane helices (50).
The reason for the ready isolation of S2P-deficient fibroblasts soon became apparent. CHO cells express only a single copy of the S2P gene on the X chromosome. In order to isolate mutant cells with a defect in S1P, we had to first transfect CHO cells with a cDNA encoding S2P so that the cells contained multiple copies of the gene. Using the amphotericin selection technique of Chang and Chang (51), we isolated CHO/pS2P cells that were auxotrophic for cholesterol, owing to a defect in site-1 cleavage (52). Juro Sakai used a clever expression cloning strategy to correct the defect in these cells by expressing cDNAs from a human cDNA library. He was quickly able to isolate the cDNA encoding S1P. S1P turned out to be a novel serine protease that is attached to cell membranes by a single hydrophobic sequence at its C terminus (53).
THE SREBP PATHWAY: SCAP AS A TRANSPORTER AND STEROL SENSOR
Throughout this work, we were puzzled by two questions: why do cells need two proteases to release SREBPs from membranes, and how do sterols regulate this process? The latter question began to be answered when we isolated the cDNA for Scap, whose letters initially stood for SREBP cleavage-activating protein. We isolated Scap through use of 25-RA cells, a sterol-resistant mutant CHO cell line isolated by T. Y. Chang. These cells process SREBPs normally in a sterol-depleted state, but they fail to shut off this processing when overloaded with sterols. Cell fusion studies showed that this phenotype was dominant. Therefore, Xianxin Hua prepared a cDNA library from the 25-RA cells and transfected pools of cDNAs into wild-type CHO cells. Using a reporter assay, he screened for cells that had become resistant to sterol-mediated suppression of SRE-dependent transcription. Through many reiterations, he isolated a cDNA encoding Scap (54). The N-terminal segment of ∼725 amino acids is organized into eight membrane-spanning helices (42) (Fig. 3). The C-terminal segment of ∼550 amino acids is hydrophilic and projects into the cytosol. This segment contains multiple copies of a sequence termed WD40, which is known to form β-propellers that mediate protein-protein interactions. When the Scap cDNA was isolated from wild-type CHO cells, we learned the reason for the dominant sterol-resistant phenotype. The Scap cDNA in 25-RA cells contained a point mutation. An aspartic acid at residue 443 was replaced by an asparagine. When cells expressed this mutant version of Scap (D443N), cleavage of SREBP was no longer suppressed by sterols (54).
The D443N mutation in Scap, which confers sterol resistance, is contained in a segment of the membrane attachment domain of Scap that is conserved in several other proteins whose activities are related to sterols. This segment includes five membrane-spanning helices. The segment is found in the membrane domain of HMG-CoA reductase where it participates in the sterol-triggered degradation of the protein (see below). This conserved sequence is also found in Patched and Dispatched, which interact with Hedgehog, a protein morphogen that contains a covalently attached cholesterol molecule. For these reasons, we named this segment the sterol-sensing domain (54, 55).
Biochemical experiments, primarily by Juro Sakai, demonstrated that the C-terminal WD40 domain of Scap binds to the C-terminal regulatory domain of SREBPs (Fig. 3) (56), and this binding is necessary for proteolytic activation of the SREBPs (57). The question remained as to how Scap facilitates proteolysis. This question was answered by Axel Nohturfft, who analyzed the glycosylation pattern of Scap and found that its N-linked sugars become resistant to digestion by endoglycosidase H when cells were deprived of sterols, but they remain in an endoglycosidase H-sensitive form when sterols are present (55). Inasmuch as endoglycosidase H resistance requires processing of carbohydrates in the Golgi complex, these experiments provided the first evidence that the Scap/SREBP complex moves from ER to Golgi in a sterol-sensitive fashion. Nohturfft, together with Peter Espenshade, confirmed these findings by transfecting cells with a plasmid encoding the membrane portion of Scap fused to green fluorescent protein (58). Fluorescence microscopy showed that this protein was localized to the ER in sterol-loaded cells, and it moved rapidly to the Golgi complex when sterols were depleted. This movement delivers Scap to S1P whose active form resides in the Golgi complex, as shown by Espenshade (59).
Figure 3 summarizes in diagrammatic form the steps in the SREBP pathway and illustrates the site at which the pathway is negatively controlled by sterols (60).
THE SREBP PATHWAY: IDENTIFICATION OF INSIGS
At this point, the cellular mechanism for sterol regulation was clear, but we still did not know the molecular mechanism. A clue came from the observation that sterol regulation was lost when we overexpressed Scap in cells by transfection. We postulated the existence of an ER retention protein that holds Scap in the ER when sterols are present (61). When Scap is overproduced, this retention protein becomes saturated, and the excess Scap moves to the Golgi complex, even when sterols are present. Tong Yang used sterol-regulated coimmunoprecipitation assays to identify the postulated sterol-dependent ER retention protein (62). She was aided by Rudi Aebersold's group in Seattle, who were able to determine protein sequences from the trace amount of material that coprecipitated with Scap only in the presence of sterols. The mRNA encoding the retention protein had been identified earlier as an mRNA that was induced by insulin treatment of rat livers. Its function was unknown, but it had been given the name Insig, which stands for insulin-induced gene (63). Even before Yang's discovery, Amber Luong, a graduate student in our laboratory, had identified Insig-1 as an SREBP-induced target gene in CHO cells (64). The convergence of Yang's and Luong's work gave us early confidence that Insig function was related to SREBP activation.
Further cloning experiments by Daisuke Yabe revealed the existence of two closely related Insigs, which we designated Insig-1 and Insig-2 (65). Gene knockdown experiments using RNA interference confirmed that Insig-1 and Insig-2 are both required in order for sterols to retain the Scap/SREBP complex in the ER (66). Later this conclusion was verified by workers in Russell DeBose-Boyd's laboratory, who isolated sterol-resistant mutant CHO cells whose defect was traced to mutations in Insig-1 and Insig-2 (67).
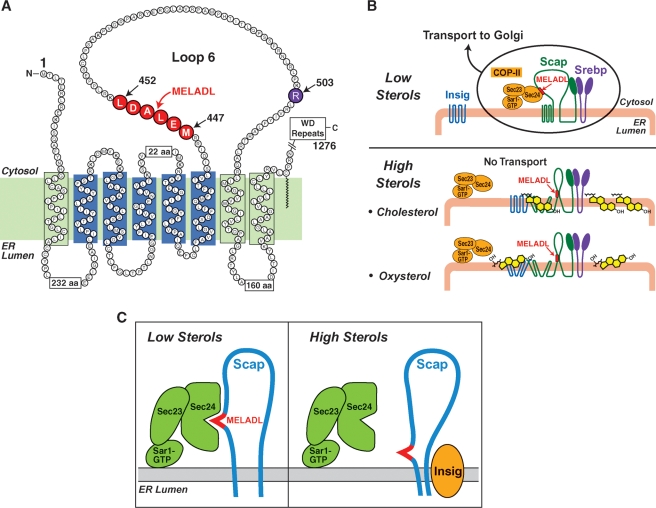
Co-immunoprecipitation and native gel electrophoresis experiments demonstrated that Scap binds to Insigs only in the presence of sterols, either cholesterol or 25-HC (62, 66). An important clue to the regulatory mechanism came from protease protection studies designed to reveal the conformation of Scap in sealed ER membrane vesicles. Andrew Brown showed that Scap has a different conformation when the vesicles are isolated from sterol-treated cells as opposed to sterol-deprived cells (68). The conformational change involves arginine-503, which resides in loop 6 between membrane-spanning helices 6 and 7 (Fig. 4A). Loop 6 is oriented toward the cytoplasm, and the protein should have been cleaved at arginine-503 when the sealed membranes were treated with trypsin. However, when ER membranes were isolated from sterol-depleted cells, trypsin failed to cleave arginine-503. Cleavage occurred only when the membranes were isolated from cholesterol-treated cells. The sterol-resistant mutant Scap(D443N) was resistant to this conformational change as were sterol-resistant Scaps with other point mutations (Y298C or L315F) (66). We concluded that sterol treatment changes the conformation of loop 6 and that this change is required for sterols to block ER-to-Golgi movement of the Scap/SREBP complex. The simplest explanation was that sterols bind directly to Scap and alter the conformation of loop 6, thereby causing Scap to bind to Insig.
Fig. 4.
Mechanisms by which sterols alter Scap's conformation and inhibit transport of SREBPs from ER to Golgi. A: Amino acid sequence and topology of the membrane domain of hamster Scap. The sterol-sensing domain of Scap (transmembranes 2-5) is shown in blue. Amino acids 447-452 (shown in red) denote the hexapeptide MELADL sequence that targets Scap to CopII coat proteins. Arginine-503 (shown in purple) denotes the amino acid residue that is subject to trypsin-induced proteolysis in ER membrane from cholesterol-treated cells but not sterol-depleted cells. B: Differential binding of cholesterol and oxysterols to Scap and Insig, respectively. When the concentration of cholesterol in the ER membrane is low, the hexapeptide MELADL sorting signal in loop 6 of Scap binds to Sec24, a component of the Sar1/Sec23/Sec24 complex of CopII coat proteins. This binding mediates the sequestration of Scap/SREBP complexes in CopII-coated vesicles that leave the ER. Cholesterol and oxysterols promote Scap binding to Insigs, but by different membranes. Cholesterol binds directly to Scap, triggering Scap to bind to Insig, an ER retention protein. Oxysterols bind directly to Insig, triggering Insig to bind to Scap. In both cases, the end result is a conformational change in the cytoplasmic loop 6 of Scap that prevents CopII proteins from gaining access to the MELADL sequence (C). The diagrams in A–C are likely to be oversimplifications because the protein complexes can potentially form large oligomeric complexes.
Postdoctoral fellow Chris Adams exposed a problem with the simple sterol-binding hypothesis. For 30 years, we had known that oxysterols such as 25-HC are more potent than cholesterol in suppressing cholesterol synthesis when added to cultured media in ethanol solution (26, 69). Similar results had been obtained by Kandutsch and Chen (25). We had made extensive use of this potency by using mixtures of cholesterol and 25-HC in most of our experiments on sterol regulation. Adams made the surprising observation that 25-HC did not cause the same conformational change in Scap that was caused by cholesterol. Moreover, when added to cultured cells, a photoactivated derivative of cholesterol could be cross-linked to Scap. No crosslinking was observed with a photoactivated derivative of 25-HC. Yet, 25-HC, like cholesterol, stimulated the formation of a Scap/Insig complex as detected on native gels, and knockdown of Insig-1 and Insig-2 abrogated 25-HC's ability to inhibit SREBP processing (66). Thus, 25-HC and cholesterol promote the interaction of Scap with Insigs, but they must do so by different mechanisms (66).
CHOLESTEROL VS. OXYSTEROLS: SOLVING THE PUZZLE
The aforementioned puzzle was solved by Arun Radhakrishnan, who established the first in vitro sterol-binding assay using the purified recombinant membrane domain of Scap (70). This was by no means a simple feat because the ligand ([3H]cholesterol) and the putative receptor (the membrane domain of Scap) are both insoluble and can only be maintained in detergent micelles. Binding requires that the [3H]cholesterol jump from a donor micelle to a micelle containing Scap. The hydrophobic nature of these interactions creates many opportunities for artifacts. Nevertheless, Radhakrishnan was able to establish conditions to demonstrate highly specific saturable binding of [3H]cholesterol to Scap. He showed that binding was absolutely dependent on the configuration of the hydroxyl group at the 3-position of the A ring of cholesterol (70). This hydroxyl group must be in the physiologic β-configuration in order for sterols to bind. On the other hand, neither the iso-octyl side chain nor the Δ5 double bond are required. Most important, the addition of a hydroxyl group on the side chain at position 25, 24, or 27 abrogated binding.
If 25-HC could not bind to Scap, how did it cause Scap to bind to Insigs? Initially, we postulated that ER membranes must contain another protein that binds 25-HC and serves as an oxysterol sensor. Several years earlier, Paul Dawson in our laboratory purified and cloned the cDNA encoding a soluble oxysterol-binding protein (OSBP) from cytosol (71, 72), but we were never able to show that this protein participated in sterol-mediated regulation of SREBP processing. We could find no evidence that sterol regulation required any of the related OSBPs that were subsequently isolated in other laboratories. Moreover, it appeared that Scap and Insig were the only mammalian proteins that were required for oxysterol-mediated regulation of SREBP processing. This conclusion emerged from the experiments of Irina Dobrosotskaya, who reconstituted oxysterol regulation in transfected insect cells in culture (73).
Insect cells do not synthesize sterols because they lack the enzymes necessary to form sterol rings. Although they have an SREBP, a Scap, and the two proteases, they use these proteins to regulate fatty acid biosynthesis, not sterol biosynthesis (74, 75). Insect cells lack Insigs. In these cells, cleavage of SREBP is not blocked by oxysterols or by cholesterol. Dobrosotskaya transfected insect cells with plasmids encoding mammalian SREBP, Scap, and Insig. Remarkably, processing of mammalian SREBP in these cells was blocked by oxysterols (73). Blocking did not occur when she expressed the sterol-resistant mutant Scap(D443N). These results argued against the requirement for a separate oxysterol-binding protein. It seemed highly unlikely that insect cells would have such a protein and that it would be able to block the processing of mammalian Scap.
The riddle was solved by Radhakrishnan who established a sterol-binding assay using purified recombinant Insig-2 (76). The challenge was even more difficult than that of the Scap binding assay because Insig proteins are even more hydrophobic than Scap and because they are easily denatured in vitro. Indeed, Insig-1 resisted purification, and, hence, Radhakrishnan used Insig-2. With his assay, Radhakrishnan showed that Insig-2 is an oxysterol binding protein. Insig-2 bound 25-HC and other oxysterols with high affinity and specificity. Although we have not been able to perform similar binding studies with Insig-1, the extensive sequence identity of the two proteins (85% identical within their membrane domains) (65) and their functional redundancy (66) strongly indicate that the two proteins bind oxysterols in a similar fashion. Remarkably, Insig-2 did not bind cholesterol. These experiments established that oxysterols and cholesterol trigger Scap binding to Insigs using two separate mechanisms. Oxysterols bind to the Insig partner, and cholesterol binds to Scap. Both of these binding reactions produce the same result: prevention of movement of the Scap/SREBP complex to the Golgi (Fig. 4B).
IN VITRO RECONSTITUTION OF ER EXPORT OF SCAP/SREBP IN COPII-COATED VESICLES
At this point, a crucial unresolved question was, what is the mechanism by which the Scap/SREBP complex is selectively moved from ER to Golgi, and how does the Scap/Insig interaction block this movement? In solving this problem, we took advantage of two decades of work by others, most notably the laboratories of Randy Schekman working in yeast (77) and William Balch working in animals cells (78). Studies by Rothman and Orci had shown that proteins move from ER to Golgi by being incorporated selectively into coated vesicles that bud from ER membranes (79). Later studies showed that these vesicles form as the result of an ordered series of reactions. The process is initiated when Sar1, a cytosolic protein, exchanges GTP for GDP and binds to ER membranes. Sar1 then recruits a heterodimer called Sec23/24 to the membrane. The Sec24 component selectively binds to cytoplasmically exposed sequences on ER membrane proteins that are selected for ER export. The bound proteins are incorporated into vesicles that bud from the ER membrane and subsequently move to the Golgi (Fig. 4B). These vesicles are known as CopII-coated vesicles.
Schekman and Balch had established assays in which ER membranes could be incubated in vitro and the incorporation of proteins into budding CopII-coated vesicles could be followed. Using these assays, Axel Nohturfft and Peter Espenshade showed that Scap was incorporated into these vesicles in vitro, but only when the ER membranes were isolated from sterol-depleted cells (58, 80). When they isolated membranes from sterol-loaded cells, the CopII-coated vesicles formed normally, but Scap was not incorporated even though the vesicles contained other proteins. Moreover, addition of 25-HC to sterol-depleted membranes in vitro selectively blocked the incorporation of Scap into budding vesicles. Thus, the in vitro assay mimicked faithfully the regulatory process that was seen in intact cells (81, 82).
Liping Sun went on to use purified recombinant Sar1 and Sec23/24 complex to conduct a series of Scap-binding assays with ER membranes from sterol-deprived or sterol-loaded cells (81, 82). He added the recombinant proteins to the membranes, and then he precipitated the Sar1 component using classic pulldown techniques. When the ER membranes were isolated from sterol-deprived cells, the Sar1/Sec23/24 complex bound to Scap. No such binding occurred when the membranes were isolated from sterol-loaded cells.
Using techniques of in vitro mutagenesis, Sun identified the region of Scap that is critical for binding the Sec23/24 complex (81). Remarkably, this region turned out to be loop 6, the same loop that was previously shown to undergo a conformational change upon cholesterol binding. Sun narrowed the critical region to a six-amino acid sequence designated MELADL (methionine-glutamic acid-leucine-alanine-aspartic acid-leucine) (Figs. 4A, B). When ER membranes are deficient in sterols, the MELADL sequence is accessible for Sec23/24 binding, and hence Scap is incorporated into CopII vesicles (Figs. 4B, C). When cholesterol binds to Scap or when oxysterols bind to Insig, the sterol alters the conformation of loop 6 so that Scap binds to Insig, the MELADL sequence is no longer recognized by the CopII proteins, and transport is abrogated (Figs. 4B, C). Although cholesterol or oxysterols block the binding of CopII proteins to Scap, they do not prevent the binding of a Fab fragment of an antibody directed against the MELADL sequence. Moreover, insertion or deletion of single amino acids between MELADL and the membrane prevent CopII binding and Scap/SREBP transport. From this and other evidence, we suggested that the conformational change does not cover up the MELADL sequence completely, but rather it alters its position in relation to the membrane (82). According to this hypothesis, the CopII proteins are located at a fixed distance from the ER membrane surface (Fig. 4C). When Scap changes its conformation, the CopII proteins are no longer able to reach the MELADL sequence, whereas the smaller Fab fragment can still have access. Of course, many other types of conformation change could prevent CopII binding. The issue will not be solved until the structure of the Scap/Insig complex is determined; a daunting task.
In the most recent studies of this regulatory system, Radhakrishnan developed an effective method to isolate pure ER membranes from cultured CHO cells (83). This method allowed a demonstration of the switch-like nature of the process by which cholesterol blocks Scap/SREBP transport. When ER cholesterol is less than 5% of total ER lipids, the Scap/SREBP complex moves to the Golgi. When cholesterol rises above 5%, transport is abruptly halted. This switch-like nature of this regulatory response is similar to that observed when ligands bind cooperatively to multimeric proteins. The classic example is the binding of oxygen to tetrameric hemoglobin. This type of binding reaction can be quantified by Hill plots. Interestingly, the Hill plot for the relation of ER cholesterol to Scap/SREBP transport yielded a Hill coefficient of approximately 4. Our previous studies showed that the membrane domain of Scap forms tetramers (70), and it is tempting to speculate that the switch-like response relates to the cooperative binding of 4 cholesterol molecules to each Scap tetramer. Thus far, we have been unable to test this cooperative binding model in our in vitro cholesterol-binding reactions because the existence of detergent micelles complicates the kinetics of the binding reaction. Other models might also account for the switch-like nature of the regulatory response, including cooperative binding of Scap to Insig. Further in vitro binding studies may clarify this mechanism.
A major unanswered question in the sterol sensing mechanism relates to the stoichiometry of the protein complexes that are formed. When purified in detergents, the membrane domain of Scap is a tetramer (70) and Insigs are dimers (76). SREBPs are dimers. If these same oligomers exist in the membrane, then there is the potential to build up long chains of Insig/Scap/SREBP complexes when sterols are added. It has not been possible to study these potential polymers definitively in cell extracts because the complexes tend to come apart when solubilized with detergents. Nevertheless, further attempts to solve this sticky problem are required.
CONVERGENT FEEDBACK: ANOTHER LEVEL OF CONTROL
Recent studies by Jin Ye have revealed another level of control in this pathway. We had earlier observed that the gene encoding Insig-1, one of the two Insigs, is activated by SREBPs, thus providing a feedback mechanism by which nuclear SREBPs trigger the buildup of their own inhibitor, namely Insig-1 (65). Ye and Yi Gong showed that the degradation rate of Insig-1 protein is also regulated. When cells are incubated in the absence of sterols, Insig-1 is rapidly ubiquitinated and degraded. When sterols accumulate, Insig binds to Scap, and this stabilizes Insig-1 (84, 85). These two methods of Insig-1 regulation led us to propose a model for convergent feedback inhibition of SREBP processing (84).
When SREBPs reach the nucleus, they activate transcription, causing increased cholesterol synthesis. Fresh SREBPs will continue to be sent to the nucleus until two criteria have been met: 1) the synthesis of Insig-1 mRNA has been increased sufficiently to allow new Insig-1 molcules to be made; and 2) sufficient cholesterol has been produced in order to stabilize Insig-1 and permit it to hold back the Scap/SREBP complex in the ER. The requirement for this convergence may serve to even out oscillations that might occur as cholesterol synthesis is turned on and off. Importantly, Insig-2 does not show either of the characteristics necessary for convergent feedback inhibition; it is not a transcriptional target of SREBP (65) and it does not have a rapid turnover (86). In cultured cells like CHO cells, the amount of Insig-2 is so low as not to interfere with the convergent regulatory scheme. However, in liver the nonconvergent properties of Insig-2 may be relevant, especially during fasting and refeeding (87, 88).
INSIGS AS MEDIATORS OF STEROL-TRIGGERED DEGRADATION OF HMG-COA REDUCTASE
The discovery of Insigs allowed us to solve a paradox that had plagued us ever since the cloning of HMG-CoA reductase nearly 20 years earlier. As noted above, we and others had noted that HMG-CoA reductase is rapidly degraded when oxysterols are added to cells. Our laboratory had shown that rapid degradation depends upon the polytopic membrane attachment domain of reductase (22), and this conclusion was confirmed and amplified by the Simoni laboratory (89). However, the mechanism for this degradation was difficult to study because the rapid degradation was abolished when HMG-CoA reductase was overexpressed by transfection. As with Scap transport, the rapid degradation of reductase appeared to require the stoichiometric activity of another protein, and this protein was saturated and overwhelmed at high levels of reductase expression.
Inasmuch as Insigs were rate-limiting for Scap transport and inasmuch as Scap and HMG-CoA reductase share a sterol-sensing domain, it seemed reasonable to propose that Insigs were also rate-limiting for reductase degradation. This hypothesis was tested by Navdar Sever and Russell DeBose-Boyd. The results were dramatic. Concomitant overexpression of Insig-1 or Insig-2 restored sterol-accelerated degradation when reductase was overexpressed (90, 91). Moreover, they showed that 25-HC triggered the binding of HMG-CoA reductase to Insig. Thus, sterols cause the binding of either of two proteins to Insig;, namely, Scap and HMG-CoA reductase (92). The results of these binding reactions are different. Whereas Scap binding displaces CopII proteins and causes ER retention, reductase binding triggers ubiquitination and degradation.
DeBose-Boyd has gone on to expose several of the proteins necessary for ubiquitination and degradation of reductase. The pathway is closely related to the general process of ER-associated protein degradation (ERAD). Ubiquitination requires an E3 ubiquitin ligase designated gp78 and a proteasome-associated protein called VCP (93). The mammalian process differs in detail from the process described for yeast HMG-CoA reductase, which is thought to be triggered by the denaturation of the reductase caused by the buildup of nonsterol intermediates in the sterol biosynthetic pathway (94). In mammalian cells, the process is triggered primarily by sterols, either oxysterols or dihydrolanosterol, an intermediate in cholesterol synthesis (95, 96). Geranylgeraniol, a nonsterol intermediate, plays a contributory role at a postubiquitination step (97). A crucial question in both systems is the physical means by which a polytopic protein such as HMG-CoA reductase is extracted from ER membranes in a form that can be degraded by proteasomes. The problem is especially acute with reductase because the process must avoid any cleavage between the membrane attachment domain and the soluble catalytic domain. If the catalytic domain is released into the cytoplasm, it will be catalytically active and long-lived, defeating the purpose of the rapid degradation machinery.
SREBP PATHWAY IN THE LIVER
As described in the introduction, feedback inhibition of cholesterol synthesis by dietary cholesterol was first described in mice and the site of control was later shown to be the liver. To determine whether the SREBP pathway mediates this feedback, we carried out a series of gene manipulation experiments in mouse liver. These studies were performed in collaboration with Guosheng Liang, Jin Shimano, Jay Horton, Luke Engelking, Iichiro Shimomura and Robert Hammer (reviewed in Ref. 98). In addition to the effects on cholesterol synthesis, these studies demonstrated an important regulatory role for SREBP-1c in mediating the insulin stimulation of fatty acid and triglyceride synthesis in liver. This topic is beyond the scope of this review (see Ref. 99).
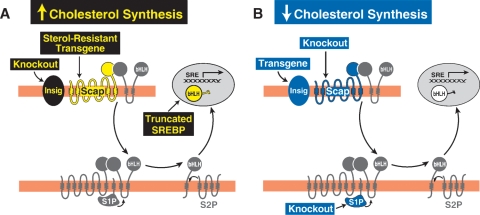
The results of the hepatic cholesterol synthesis studies are summarized in Fig. 5. As predicted from the cell culture studies, three manipulations led to increases in hepatic cholesterol synthesis (Fig. 5A). These were: 1) production of a truncated form of SREBP-2 that lacks the membrane attachment domain and reaches the nucleus without a requirement for proteolysis (100); 2) insertion of a transgene encoding a sterol-resistant mutant Scap(D443N), which does not bind Insig in the presence of sterols (101); and 3) knockout of the genes encoding both forms of Insig (Insig-1 and Insig-2) (102). In addition to overproduction of cholesterol, all of these gene-altered mice resisted suppression of cholesterol synthesis by cholesterol feeding. As a result, they continued to produce cholesterol even though the livers were packed with cholesterol stored as cholesteryl esters.
Fig. 5.
Alterations in hepatic cholesterol synthesis in gene-manipulated mice. A: Increased cholesterol synthesis occurs in livers of the following gene-manipulated mice: transgenic mice expressing a truncated form of SREBP-2 (100) that lacks the transmembrane and C-terminal regulatory domains; transgenic mice expressing a mutant version of Scap (D443N) that does not bind Insig in the presence of sterols and thus undergoes constitutive ER-to-Golgi transport (62, 101); and knockout mice lacking both Insig-1 and Insig-2 (102). B: Decreased cholesterol synthesis occurs in livers of the following gene-manipulated mice: transgenic mice that overexpress Insig-1 (87); knockout mice lacking Scap (103); and knockout mice lacking site-1 protease (104).
Three gene alterations decreased cholesterol synthesis in liver (Fig. 5B). These were: 1) knockout of Scap (103); 2) knockout of site-1 protease (104); and 3) insertion of a transgene encoding Insig-1 (87). Earlier studies in cultured cells had shown that overproduction of Insig led to a block in Scap/SREBP transport even when cholesterol levels were very low (62).
Considered together, the findings in Fig. 5 confirm the essential role of the SREBP pathway in cholesterol synthesis in mammalian liver, and they provide a molecular explanation for the feedback inhibition that was first observed by Schoenheimer in bottled mice 75 years ago. Thus, Schoenheimer's bottle led to Scap's MELADL.
Acknowledgments
This review is dedicated to the memory of four of our scientific heroes who sadly passed away within the last year: Arthur Kornberg, Daniel E. Koshland, Jr., George E. Palade, and Earl R. Stadtman. Their love for research and their emphasis on the highest standards of experimentation have inspired our scientific lives. We thank Russell DeBose-Boyd, Jin Ye, and Guosheng Liang for critical review of the manuscript.
Abbreviations
bHLH-Zip, basic-helix-loop-helix-leucine zipper
CHO, Chinese hamster ovary
ER, endoplasmic reticulum
FH, familial hypercholesterolemia
HMG-CoA, 3-hydroxy-3-methylglutaryl CoA
S1P, site-1 protease
S2P, site-2 protease
Scap, SREBP cleavage-activating protein
SRE, sterol regulatory element
SREBP, SRE-binding protein
The original research described in this review was supported by grants from the National Institutes of Health (HL20948), the Perot Family Foundation, and the Moss Heart Foundation.
Published, JLR Papers in Press, October 29, 2008.
References
- 1.Schoenheimer R., and F. Breusch. 1933. Synthesis and destruction of cholesterol in the organism. J. Biol. Chem. 103 439–448. [Google Scholar]
- 2.Jacob F., and J. Monod. 1961. On the regulation of gene activity. Cold Spring Harb. Symp. Quant. Biol. 26 193–211. [DOI] [PubMed] [Google Scholar]
- 3.Bloch K. 1965. The biological synthesis of cholesterol. Science. 50 19–128. [DOI] [PubMed] [Google Scholar]
- 4.Gould R. G., C. B. Taylor, J. S. Hagerman, I. Warner, and D. J. Campbell. 1953. Cholesterol metabolism: I. Effect of dietary cholesterol on the synthesis of cholesterol in dog tissue in vitro. J. Biol. Chem. 201 519–523. [PubMed] [Google Scholar]
- 5.Spady D. K., and J. M. Dietschy. 1983. Sterol synthesis in vivo in 18 tissues of the squirrel monkey, guinea pig, rabbit, hamster, and rat. J. Lipid Res. 24 303–315. [PubMed] [Google Scholar]
- 6.Brown M. S., and J. L. Goldstein. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232 34–47. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein J. L., and M. S. Brown. 1973. Familial hypercholesterolemia: identification of a defect in the regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity associated with overproduction of cholesterol. Proc. Natl. Acad. Sci. USA. 70 2804–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein J. L., R. G. W. Anderson, and M. S. Brown. 1979. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature. 279 679–685. [DOI] [PubMed] [Google Scholar]
- 9.Brown M. S., and J. L. Goldstein. 1975. Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell. 6 307–316. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein J. L., and M. S. Brown. 1984. Progress in understanding the LDL receptor and HMG CoA reductase, two membrane proteins that regulate the plasma cholesterol. J. Lipid Res. 25 1450–1461. [PubMed] [Google Scholar]
- 11.Chin J., and T-Y. Chang. 1981. Evidence for coordinate expression of 3-hydroxy-3-methylglutaryl coenzyme A reductase and low density lipoprotein binding activity. J. Biol. Chem. 256 6304–6310. [PubMed] [Google Scholar]
- 12.Schneider W. J., U. Beisiegel, J. L. Goldstein, and M. S. Brown. 1982. Purification of the low density lipoprotein receptor, an acidic glycoprotein of 164,000 molecular weight. J. Biol. Chem. 257 2664–2673. [PubMed] [Google Scholar]
- 13.Yamamoto T., C. G. Davis, M. S. Brown, W. J. Schneider, M. L. Casey, J. L. Goldstein, and D. W. Russell. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 39 27–38. [DOI] [PubMed] [Google Scholar]
- 14.Sudhof T. C., J. L. Goldstein, M. S. Brown, and D. W. Russell. 1985. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 228 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schimke R. T., R. J. Kaufman, F. W. Alt, and R. F. Kellems. 1978. Gene amplification and drug resistance in cultured murine cells. Science. 202 1051–1056. [DOI] [PubMed] [Google Scholar]
- 16.Endo A., M. Kuroda, and K. Tanzawa. 1976. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 72 323–326. [DOI] [PubMed] [Google Scholar]
- 17.Chin D. J., K. L. Luskey, R. G. W. Anderson, J. R. Faust, J. L. Goldstein, and M. S. Brown. 1982. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold elevation in 3-hydroxy-3-methylglutaryl CoA reductase. Proc. Natl. Acad. Sci. USA. 79 1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin D. J., K. L. Luskey, J. R. Faust, R. J. MacDonald, M. S. Brown, and J. L. Goldstein. 1982. Molecular cloning of 3-hydroxy-3-methylglutaryl coenzyme A reductase and evidence for regulation of its mRNA in UT-1 cells. Proc. Natl. Acad. Sci. USA. 79 7704–7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil G., J. L. Goldstein, C. A. Slaughter, and M. S. Brown. 1986. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster: I. Isolation and sequencing of a full-length cDNA. J. Biol. Chem. 261 3710–3716. [PubMed] [Google Scholar]
- 20.Liscum L., J. Finer-Moore, R. M. Stroud, K. L. Luskey, M. S. Brown, and J. L. Goldstein. 1985. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 260 522–530. [PubMed] [Google Scholar]
- 21.Roitelman J., E. H. Olender, S. Bar-Nun, W. A. Dunn, Jr., and R. D. Simoni. 1992. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl Coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 117 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil G., J. R. Faust, D. J. Chin, J. L. Goldstein, and M. S. Brown. 1985. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 41 249–258. [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi M., J. L. Goldstein, and M. S. Brown. 1988. Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme A reductase: mevalonate-derived product inhibits translation of mRNA and accelerates degradation of enzyme. J. Biol. Chem. 263 8929–8937. [PubMed] [Google Scholar]
- 24.Metherall J. E., J. L. Goldstein, K. L. Luskey, and M. S. Brown. 1989. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 264 15634–15641. [PubMed] [Google Scholar]
- 25.Kandutsch A. A., and H. W. Chen. 1974. Inhibition of sterol synthesis in cultured mouse cells by cholesterol derivatives oxygenated in the side chain. J. Biol. Chem. 249 6057–6061. [PubMed] [Google Scholar]
- 26.Brown M. S., and J. L. Goldstein. 1974. Suppression of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and inhibition of growth of human fibroblasts by 7-ketocholesterol. J. Biol. Chem. 249 7306–7314. [PubMed] [Google Scholar]
- 27.Reynolds G. A., S. K. Basu, T. F. Osborne, D. J. Chin, G. Gil, M. S. Brown, J. L. Goldstein, and K. L. Luskey. 1984. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5′ untranslated regions. Cell. 38 275–286. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein J. L., and M. S. Brown. 1990. Regulation of the mevalonate pathway. Nature. 343 425–430. [DOI] [PubMed] [Google Scholar]
- 29.Sudhof T. C., D. W. Russell, M. S. Brown, and J. L. Goldstein. 1987. 42-bp element from LDL receptor gene confers end-product repression by sterols when inserted into viral TK promoter. Cell. 48 1061–1069. [DOI] [PubMed] [Google Scholar]
- 30.Smith J. R., T. F. Osborne, J. L. Goldstein, and M. S. Brown. 1990. Identification of nucleotides responsible for enhancer activity of sterol regulatory element in low density lipoprotein receptor gene. J. Biol. Chem. 265 2306–2310. [PubMed] [Google Scholar]
- 31.Wang X., M. R. Briggs, X. Hua, C. Yokoyama, J. L. Goldstein, and M. S. Brown. 1993. Nuclear protein that binds sterol regulatory element of LDL receptor promoter: II. Purification and characterization. J. Biol. Chem. 268 14497–14504. [PubMed] [Google Scholar]
- 32.Yokoyama C., X. Wang, M. R. Briggs, A. Admon, J. Wu, X. Hua, J. L. Goldstein, and M. S. Brown. 1993. SREBP-1, a basic helix-loop-helix leucine zipper protein that controls transcription of the LDL receptor gene. Cell. 75 187–197. [PubMed] [Google Scholar]
- 33.Kim J. B., G. D. Spotts, Y-D. Halvorsen, H-M. Shih, T. Ellenberger, H. C. Towle, and B. M. Spiegelman. 1995. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 15 2582–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parraga A., L. Bellsolell, A. R. Ferre-D'Amare, and S. K. Burley. 1998. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 6 661–672. [DOI] [PubMed] [Google Scholar]
- 35.Hua X., C. Yokoyama, J. Wu, M. R. Briggs, M. S. Brown, J. L. Goldstein, and X. Wang. 1993. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA. 90 11603–11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimomura I., H. Shimano, J. D. Horton, J. L. Goldstein, and M. S. Brown. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tontonoz P., J. B. Kim, R. A. Graves, and B. M. Spiegelman. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J. B., and B. M. Spiegelman. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10 1096–1107. [DOI] [PubMed] [Google Scholar]
- 39.Shimano H., J. D. Horton, I. Shimomura, R. E. Hammer, M. S. Brown, and J. L. Goldstein. 1997. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 99 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 77 53–62. [DOI] [PubMed] [Google Scholar]
- 41.Hua X., J. Sakai, M. S. Brown, and J. L. Goldstein. 1996. Regulated cleavage of sterol regulatory element binding proteins (SREBPs) requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 271 10379–10384. [DOI] [PubMed] [Google Scholar]
- 42.Nohturfft A., M. S. Brown, and J. L. Goldstein. 1998. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J. Biol. Chem. 273 17243–17250. [DOI] [PubMed] [Google Scholar]
- 43.Sakai J., E. A. Duncan, R. B. Rawson, X. Hua, M. S. Brown, and J. L. Goldstein. 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 85 1037–1046. [DOI] [PubMed] [Google Scholar]
- 44.Duncan E. A., U. P. Davé, J. Sakai, J. L. Goldstein, and M. S. Brown. 1998. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273 17801–17809. [DOI] [PubMed] [Google Scholar]
- 45.Brown M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 100 391–398. [DOI] [PubMed] [Google Scholar]
- 46.Ye, J., M. S. Brown, and J. L. Goldstein. 2004. Regulated intramembrane proteolysis (Rip). In Encyclopedia of Biological Chemistry. W. J. Lennarz and M. D. Lane, editors. Elsevier, Oxford. 665–670.
- 47.Rawson R. B., N. G. Zelenski, D. Nijhawan, J. Ye, J. Sakai, M. T. Hasan, T-Y. Chang, M. S. Brown, and J. L. Goldstein. 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1 47–57. [DOI] [PubMed] [Google Scholar]
- 48.Zelenski N. G., R. B. Rawson, M. S. Brown, and J. L. Goldstein. 1999. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274 21973–21980. [DOI] [PubMed] [Google Scholar]
- 49.Rudner D. Z., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA. 96 14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng L., H. Yan, Z. Wu, N. Yan, Z. Wang, P. D. Jeffrey, and Y. Shi. 2007. Structure of a Site-2 protease family intramembrane metalloprotease. Science. 318 1608–1612. [DOI] [PubMed] [Google Scholar]
- 51.Chang T-Y., and C. C. Y. Chang. 1982. Revertants of a Chinese hamster ovary cell mutant resistant to suppression by an analogue of cholesterol: isolation and partial biochemical characterization. Biochemistry. 21 5316–5323. [DOI] [PubMed] [Google Scholar]
- 52.Rawson R. B., D. Cheng, M. S. Brown, and J. L. Goldstein. 1998. Isolation of cholesterol-requiring mutant CHO cells with defects in cleavage of sterol regulatory element binding proteins at Site-1. J. Biol. Chem. 273 28261–28269. [DOI] [PubMed] [Google Scholar]
- 53.Sakai J., R. B. Rawson, P. J. Espenshade, D. Cheng, A. C. Seegmiller, J. L. Goldstein, and M. S. Brown. 1998. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell. 2 505–514. [DOI] [PubMed] [Google Scholar]
- 54.Hua X., A. Nohturfft, J. L. Goldstein, and M. S. Brown. 1996. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage activating protein (SCAP). Cell. 87 415–426. [DOI] [PubMed] [Google Scholar]
- 55.Nohturfft A., M. S. Brown, and J. L. Goldstein. 1998. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc. Natl. Acad. Sci. USA. 95 12848–12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakai J., A. Nohturfft, D. Cheng, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1997. Identification of complexes between the COOH-terminal domains of sterol regulatory element binding proteins (SREBPs) and SREBP cleavage-activating protein (SCAP). J. Biol. Chem. 272 20213–20221. [DOI] [PubMed] [Google Scholar]
- 57.Sakai J., A. Nohturfft, J. L. Goldstein, and M. S. Brown. 1998. Cleavage of sterol regulatory element binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J. Biol. Chem. 273 5785–5793. [DOI] [PubMed] [Google Scholar]
- 58.Nohturfft A., D. Yabe, J. L. Goldstein, M. S. Brown, and P. J. Espenshade. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 102 315–323. [DOI] [PubMed] [Google Scholar]
- 59.Espenshade P. J., D. Cheng, J. L. Goldstein, and M. S. Brown. 1999. Autocatalytic processing of Site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274 22795–22804. [DOI] [PubMed] [Google Scholar]
- 60.Brown M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89 331–340. [DOI] [PubMed] [Google Scholar]
- 61.Yang T., J. L. Goldstein, and M. S. Brown. 2000. Overexpression of membrane domain of SCAP prevents sterols from inhibiting SCAP/SREBP exit from endoplasmic reticulum. J. Biol. Chem. 275 29881–29886. [DOI] [PubMed] [Google Scholar]
- 62.Yang T., P. J. Espenshade, M. E. Wright, D. Yabe, Y. Gong, R. Aebersold, J. L. Goldstein, and M. S. Brown. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110 489–500. [DOI] [PubMed] [Google Scholar]
- 63.Diamond R. H., K. Du, V. M. Lee, K. L. Mohn, B. A. Haber, D. S. Tewari, and R. Taub. 1993. Novel delayed-early and highly insulin-induced growth response genes: identification of HRS, a potential regulator of alternative pre-mRNA splicing. J. Biol. Chem. 268 15185–15192. [PubMed] [Google Scholar]
- 64.Luong, A. 2000. Identification of three novel SREBP-activated target genes: acetyl CoA synthetase, 3-β-hydroxysterol dehydrogenase, and CL-6/INSIG1. Ph.D. Dissertation. University of Texas Southwestern Medical Center, Dallas. TX.
- 65.Yabe D., M. S. Brown, and J. L. Goldstein. 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA. 99 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams C. M., J. Reitz, J. K. DeBrabander, J. D. Feramisco, M. S. Brown, and J. L. Goldstein. 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 279 52772–52780. [DOI] [PubMed] [Google Scholar]
- 67.Lee P. C. W., N. Sever, and R. A. DeBose-Boyd. 2005. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J. Biol. Chem. 280 25242–25249. [DOI] [PubMed] [Google Scholar]
- 68.Brown A. J., L. Sun, J. D. Feramisco, M. S. Brown, and J. L. Goldstein. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 10 237–245. [DOI] [PubMed] [Google Scholar]
- 69.Goldstein, J. L., J. R. Faust, G. Y. Brunschede, and M. S. Brown. 1975. Steroid requirements for suppression of HMG CoA reductase activity in cultured human fibroblasts. In Encyclopedia of Biological Chemistry. D. Kritchevsky, R. Paoletti, and W. L. Holmes, editors. Plenum Publishing Corp., New York. 77–84. [DOI] [PubMed]
- 70.Radhakrishnan A., L-P. Sun, H. J. Kwon, M. S. Brown, and J. L. Goldstein. 2004. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell. 15 259–268. [DOI] [PubMed] [Google Scholar]
- 71.Dawson P. A., D. R. van der Westhuyzen, J. L. Goldstein, and M. S. Brown. 1989. Purification of oxysterol binding protein from hamster liver cytosol. J. Biol. Chem. 264 9046–9052. [PubMed] [Google Scholar]
- 72.Dawson P. A., N. D. Ridgway, C. A. Slaughter, M. S. Brown, and J. L. Goldstein. 1989. cDNA cloning and expression of oxysterol binding protein, an oligomer with a potential leucine zipper. J. Biol. Chem. 264 16798–16803. [PubMed] [Google Scholar]
- 73.Dobrosotskaya I., J. L. Goldstein, M. S. Brown, and R. B. Rawson. 2003. Reconstitution of sterol-regulated endoplasmic reticulum-to-Golgi transport of SREBP-2 in insect cells by co-expression of mammalian SCAP and insigs. J. Biol. Chem. 278 35837–35843. [DOI] [PubMed] [Google Scholar]
- 74.Seegmiller A. C., I. Dobrosotskaya, J. L. Goldstein, Y. K. Ho, M. S. Brown, and R. B. Rawson. 2002. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell. 2 229–238. [DOI] [PubMed] [Google Scholar]
- 75.Kunte A. S., K. A. Matthews, and R. B. Rawson. 2006. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 3 439–448. [DOI] [PubMed] [Google Scholar]
- 76.Radhakrishnan A., Y. Ikeda, H. J. Kwon, M. S. Brown, and J. L. Goldstein. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA. 104 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antonny B., and R. Schekman. 2001. ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13 438–443. [DOI] [PubMed] [Google Scholar]
- 78.Bannykh S. I., N. Nishimura, and W. E. Balch. 1998. Getting into the Golgi. Trends Cell Biol. 8 21–25. [DOI] [PubMed] [Google Scholar]
- 79.Rothman J. E., and L. Orci. 1992. Molecular dissection of the secretory pathway. Nature. 355 409–415. [DOI] [PubMed] [Google Scholar]
- 80.Espenshade P. J., W-P. Li, and D. Yabe. 2002. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA. 99 11694–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun L-P., L. Li, J. L. Goldstein, and M. S. Brown. 2005. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 280 26483–26490. [DOI] [PubMed] [Google Scholar]
- 82.Sun L-P., J. Seemann, M. S. Brown, and J. L. Goldstein. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. USA. 104 6519–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Radhakrishnan A., J. L. Goldstein, J. G. McDonald, and M. S. Brown. 2008. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gong Y., J. N. Lee, P. C. W. Lee, J. L. Goldstein, M. S. Brown, and J. Ye. 2006. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 3 15–24. [DOI] [PubMed] [Google Scholar]
- 85.Lee J. N., B-L. Song, R. A. DeBose-Boyd, and J. Ye. 2006. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 281 39308–39315. [DOI] [PubMed] [Google Scholar]
- 86.Lee J. N., Y. Gong, X. Zhang, and J. Ye. 2006. Proteasomal degradation of ubiquitinated Insig proteins is determined by serine residues flanking ubiquitinated lysines. Proc. Natl. Acad. Sci. USA. 103 4958–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Engelking L. J., H. Kuriyama, R. E. Hammer, J. D. Horton, M. S. Brown, J. L. Goldstein, and G. Liang. 2004. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 113 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yabe D., R. Komuro, G. Liang, J. L. Goldstein, and M. S. Brown. 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 100 3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skalnik D. G., H. Narita, C. Kent, and R. D. Simoni. 1988. The membrane domain of 3-hydroxy-3-methylglutaryl-coenzyme A reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto β-galactosidase. J. Biol. Chem. 263 6836–6841. [PubMed] [Google Scholar]
- 90.Sever N., P. C. W. Lee, B-L. Song, R. B. Rawson, and R. A. DeBose-Boyd. 2004. Isolation of mutant cells lacking Insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 279 43136–43147. [DOI] [PubMed] [Google Scholar]
- 91.Sever N., T. Yang, M. S. Brown, J. L. Goldstein, and R. A. DeBose-Boyd. 2003. Accelerated degradation of HMG CoA reductase mediated by binding of Insig-1 to its sterol-sensing domain. Mol. Cell. 11 25–33. [DOI] [PubMed] [Google Scholar]
- 92.Goldstein J. L., R. A. DeBose-Boyd, and M. S. Brown. 2006. Protein sensors for membrane sterols. Cell. 124 35–46. [DOI] [PubMed] [Google Scholar]
- 93.Song B-L., and R. A. DeBose-Boyd. 2004. Ubiquitination of 3-hydroxy-3-methylglutaryl-CoA reductase in permeabilized cells mediated by cytosolic E1 and a putative membrane-bound ubiquitin ligase. J. Biol. Chem. 279 28798–28806. [DOI] [PubMed] [Google Scholar]
- 94.Shearer A. G., and R. Y. Hampton. 2005. Lipid-mediated, reversible misfolding of a sterol-sensing domain protein. EMBO J. 24 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song B-L., N. B. Javitt, and R. A. DeBose-Boyd. 2005. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1 179–189. [DOI] [PubMed] [Google Scholar]
- 96.Lange Y., D. S. Ory, J. Ye, M. H. Lanier, F-F. Hsu, and T. L. Steck. 2008. Effectors of rapid homeostatic responses of endoplasmic reticulum cholesterol and 3-hydroxy-3-methlglutaryl-CoA reductase. J. Biol. Chem. 283 1445–1455. [DOI] [PubMed] [Google Scholar]
- 97.Sever N., B-L. Song, D. Yabe, J. L. Goldstein, M. S. Brown, and R. A. DeBose-Boyd. 2003. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278 52479–52490. [DOI] [PubMed] [Google Scholar]
- 98.Horton J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown M. S., and J. L. Goldstein. 2008. Selective vs. total insulin resistance: a pathogenic paradox. Cell Metab. 7 95–96. [DOI] [PubMed] [Google Scholar]
- 100.Horton J. D., I. Shimomura, M. S. Brown, R. E. Hammer, J. L. Goldstein, and H. Shimano. 1998. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Invest. 101 2331–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Korn B. S., I. Shimomura, Y. Bashmakov, R. E. Hammer, J. D. Horton, J. L. Goldstein, and M. S. Brown. 1998. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N). J. Clin. Invest. 102 2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Engelking L. J., G. Liang, R. E. Hammer, K. Takaishi, H. Kuriyama, B. M. Evers, W-P. Li, J. D. Horton, J. L. Goldstein, and M. S. Brown. 2005. Schoenheimer effect explained - Feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 115 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsuda M., B. S. Korn, R. E. Hammer, Y-A. Moon, R. Komuro, J. D. Horton, J. L. Goldstein, M. S. Brown, and I. Shimomura. 2001. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 15 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang J., J. L. Goldstein, R. E. Hammer, Y-A. Moon, M. S. Brown, and J. D. Horton. 2001. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc. Natl. Acad. Sci. USA. 98 13607–13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gould R. G. 1951. Lipid metabolism and atherosclerosis. Am. J. Med. 11 209–227. [DOI] [PubMed] [Google Scholar]
- 106.Tomkins G. M., H. Sheppard, and I. L. Chaikoff. 1953. Cholesterol synthesis by liver: III. Its regulation by ingested cholesterol. J. Biol. Chem. 201 137–141. [PubMed] [Google Scholar]
- 107.Langdon R. G., and K. Bloch. 1953. The effect of some dietary additions on the synthesis of cholesterol from acetate in vitro. J. Biol. Chem. 202 77–81. [PubMed] [Google Scholar]
- 108.Frantz I. D., Jr., H. S. Schneider, and B. T. Hinkelman. 1954. Suppression of hepatic cholesterol synthesis in the rat by cholesterol feeding. J. Biol. Chem. 206 465–469. [PubMed] [Google Scholar]
- 109.Bucher N. L. R., K. McGarrahan, E. Gould, and A. V. Loud. 1959. Cholesterol biosynthesis in preparations of liver from normal, fasting, x–irradiated, cholesterol-fed, Triton, or 44-cholesten-3-one-treated rats. J. Biol. Chem. 234 262–267. [PubMed] [Google Scholar]
- 110.Siperstein M. D., and M. J. Guest. 1960. Studies on the site of the feedback control of cholesterol synthesis. J. Clin. Invest. 39 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Linn T. C. 1967. The effect of cholesterol feeding and fasting upon β-hydroxy-β-methylglutaryl coenzyme A reductase. J. Biol. Chem. 242 990–993. [PubMed] [Google Scholar]
- 112.Shapiro D. J., and V. W. Rodwell. 1971. Regulation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and cholesterol synthesis. J. Biol. Chem. 246 3210–3216. [PubMed] [Google Scholar]