Abstract
Cells are devices whose structures delimit function. For example, in the nervous system, neuronal and glial shapes dictate paths of information flow. To understand how cells acquire their shapes, we examined the formation of a sense organ in C. elegans. Using time-lapse imaging, we found that sensory dendrites form by stationary anchoring of dendritic tips during cell-body migration. A genetic screen identified DEX-1 and DYF-7, extracellular proteins required for dendritic tip anchoring, which act cooperatively at the time and place of anchoring. DEX-1 and DYF-7 contain, respectively, zonadhesin and zona pellucida domains, and DYF-7 self-associates into multimers important for anchoring. Thus, unlike other dendrites, amphid dendritic tips are positioned by DEX-1 and DYF-7 without the need for long-range guidance cues. In sequence and function, DEX-1 and DYF-7 resemble tectorins, which anchor stereocilia in the inner ear, suggesting that a sensory dendrite anchor may have evolved into part of a mechanosensor.
INTRODUCTION
Some of the most remarkable architecture on the planet is found at the level of single cells. Neurons, in particular, adopt precisely patterned shapes that extend over millimeters (or, in some organisms, meters), and they elaborate complex dendrites with stereotyped lengths and branching patterns that occupy defined volumes of space (Parrish et al., 2007). The fidelity with which each neuron assumes and maintains its shape directly affects its function, with miswiring leading to a range of neurological disorders (Yaron and Zheng, 2007). The shapes of neurons are matched in complexity only, perhaps, by the shapes of the glia that surround them. Moreover, the shapes of neurons and glia must be closely coordinated for a functional nervous system to assemble. And, while the problem of generating cell shape takes on particularly astounding dimensions in the nervous system, the same problem – and, possibly, the same solutions – affect every tissue of the body.
The nematode C. elegans provides a useful model in which to study cell shape. Most of the 959 somatic cells of an adult hermaphrodite exhibit an essentially invariant shape from animal to animal (Sulston et al., 1983). Each of these cell shapes, including the complete wiring diagram of the nervous sytem, has been catalogued by three-dimensional reconstruction of electron micrographic serial sections (Ward et al., 1975; White et al., 1976; White et al., 1986). As most cells acquire their shapes in a defined time window from about 300-420 min post-fertilization, morphogenesis occurs on a time scale compatible with time-lapse imaging, a technique made even more powerful by the optical transparency of C. elegans. Finally, the facile genetics of this organism make it straightforward to isolate mutants affecting cell shape and thereby identify the relevant genes.
To examine how cell shape is determined in neurons and glia, we decided to use developmental imaging and genetics to study development of the amphids, a bilaterally symmetric pair of sense organs in the head of the animal. Each amphid consists of 12 neurons which exhibit specific responses to smells (AWA, AWB, and AWC neurons), temperature (AFD neuron), and taste, touch, osmotic conditions, and pheromones (ASE, ADF, ASG, ASH, ASI, ASJ, ASK, and ADL neurons) (Bargmann, 2006; Ward et al., 1975). Despite their diverse sensory functions, these 12 neurons share a similar overall shape, each extending an axon laterally into the nerve ring, or “brain,” and a single dendrite reaching to the tip of the nose. The axons do not arborize, but are unbranched or bifurcated, and make synapses en passant. The dendrites are also unbranched, each terminating at the nose in a sensory cilium bearing receptors responsible for that neuron’s specific sensory modalities (Bargmann, 2006). The 12 dendrites associate with two glia: the sheath glial cell, which extends a process to the nose where it ensheathes all 12 dendritic endings; and the socket glial cell, which extends a process to the nose where it forms a pore through which some neurons are directly exposed to the environment (Shaham, 2006; Ward et al., 1975). This sense organ, therefore, is shaped essentially as a linear bundle (Fig. 1A), allowing a single aspect of neuronal and glial shape to be analyzed without the added complications of dendritic field tiling or branching, features that have been carefully explored elsewhere (Gao, 2007). Thus, the amphid offers a simple system for asking a basic question about cell shape, namely, how is the length of a sensory dendrite, or a glial cell, established.
FIGURE 1. dex-1 and dyf-7 are required for dendrite extension.
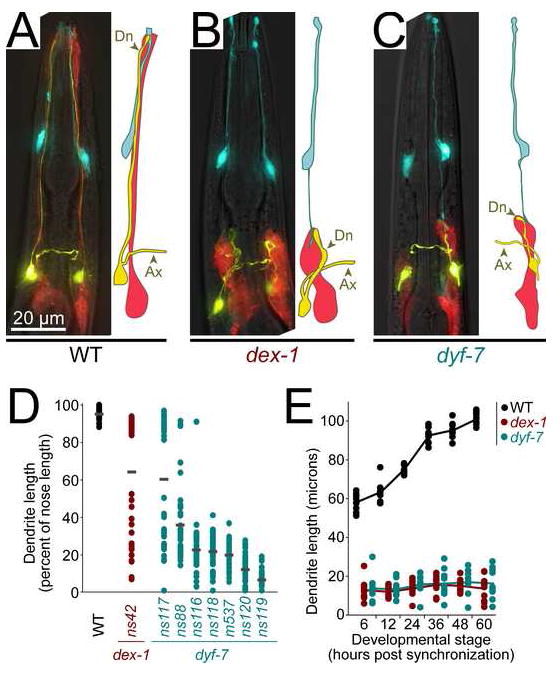
A, Wild-type; B, dex-1(ns42); and C, dyf-7(m537) animals expressing odr-1pro:YFP (AWC neurons, yellow), F16F9.3pro:mCherry (sheath glia, red), and itr-1pro:CFP (socket glia, blue). Ax, axon; Dn, dendrite. D, Animals expressing gcy-5pro:GFP (ASER neuron) with or without str-2pro:GFP (AWCL or AWCR), to mark individual dendrites, were synchronized at the first larval stage (L1) and dendrite and nose lengths measured. Each dot represents an individual (n=40 per genotype). Gray bars, means. E, Same as (D), but cohorts of L1s with short dendrites were selected by visual inspection, released from synchronization, and dendrite lengths were measured after 6 h (L1), 12 h (second larval stage, L2), 24 h (third larval stage, L3), 36 h (early fourth larval stage, L4), 48 h (late L4), and 60 h (adult). Lines connect means (n=10 per genotype at each stage).
RESULTS
DEX-1 and DYF-7 are required for dendrite and glial process extension
To identify factors that determine the shapes of neurons and glia, we generated a strain expressing fluorescent proteins in two amphid neurons (ASE and AWC) and the sheath glial cell, performed random chemical mutagenesis, and screened visually for animals with morphologically abnormal neurons and glia. We also screened a panel of candidate mutants known to affect sensory neuron function (Starich et al., 1995). We identified a novel class of mutants in which all 12 dendrites and the sheath glial process fail to extend to the tip of the nose, a phenotype we call Dex (Dendrite extension defective) (Fig 1A-C). The mutations define two genes, dex-1 and dyf-7 (see Methods).
The Dex phenotype is specific to the lengths of the dendrites and sheath glial process. Neuronal and glial cell bodies are properly positioned and appear healthy; both cell types express appropriate cell-specific markers, including a marker whose expression depends on normal axon-axon contacts; and overall axon morphology appears normal (Fig. 1A-C, Supp. Fig. 1). Dendrites are ensheathed by the sheath glial cell and possess sensory cilia to which odorant receptors properly localize (Supp. Fig. 1). The socket glial cell extends a grossly wild-type process to the tip of nose, but also extends an ectopic posterior process that contacts the shortened sheath glial cell (Fig. 1B and 1C), suggesting that all relevant cell-cell contacts in the organ – namely, between the sheath glial cell and each neuron, between the sheath and socket glia, and between the socket glial cell and hypodermal cells at the nose (Perkins et al., 1986; Ward et al., 1975) – remain intact. The specificity of the Dex phenotype to dendrite length indicates that dendrite and axon extension occur by different mechanisms, and that cell-cell contacts are probably not sufficient to confer normal cell shape.
Two additional features of the Dex phenotype struck us as potentially revealing. First, weakly penetrant alleles of dex-1 and dyf-7 produce bimodally distributed dendrite lengths, an effect seen most conspicuously in dyf-7(ns116) but also evident in dex-1(ns42), dyf-7(ns117) and, to a lesser extent, dyf-7(ns88) (Fig. 1D). These bimodal distributions suggest that dendrite growth in dex-1 and dyf-7 animals is limited by an initiation event that often fails, while downstream steps usually proceed to completion if the initiation event is successful.
Second, when dex-1 or dyf-7 larvae are born with short dendrites, those dendrites remain at constant length until adulthood, unlike wild-type dendrites, which scale almost two-fold in length to keep pace with overall larval growth (Fig. 1E, Supp. Fig 1D). However, dex-1 and dyf-7 neurons are not generally insensitive to larval growth, as their cell body size and axon length increase at rates similar to wild-type neurons (Supp. Fig. 1C-D). Thus, dex-1 and dyf-7 mutations cause defects at two steps: first, in establishing proper dendrite length during embryonic development and, second, in scaling dendrite length to keep pace with larval growth.
Dendrites extend as a coordinated bundle
Animals bearing the partially-penetrant alleles dex-1(ns42) and dyf-7(ns117) display dendrites and glial processes of variable lengths in a population, which allowed us to determine if each neuron and glial cell attains its length autonomously, or if lengths are coordinated within an amphid bundle. If each cell attains its length autonomously, then neurons in the same bundle would be of similar lengths no more often than one would expect based on the overall distribution of lengths in the population. However, a correlation between dendrite lengths would indicate coordination within each amphid bundle, suggesting a non-cell-autonomous mechanism of growth.
To distinguish between these possibilities, we generated a strain in which a single neuron, AWC, expresses green fluorescent protein (GFP) stochastically in either the left or right amphid (Troemel et al., 1999), and another neuron, ADF, expresses red fluorescent protein (RFP) in both amphids (Fig. 2A). We then compared the length of the AWC dendrite to the lengths of ipsilateral (Fig. 2A, closed arrowheads) and contralateral (Fig. 2A, open arrowheads) ADF dendrites in dex-1(ns42) and dyf-7(ns117) animals.
FIGURE 2. Amphid sensory dendrite extension is coordinated.
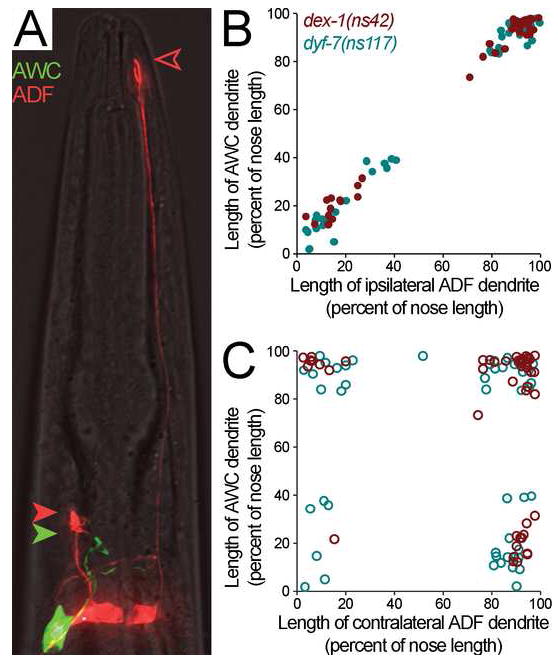
A, To compare lengths of ipsilateral and contralateral dendrites, animals bearing dex-1(ns42) or dyf-7(ns117) were made to express str-2pro:GFP (AWCL or AWCR neuron, green) and srh-142pro:RFP (ADFL and ADFR neurons, red). Closed green arrowhead, AWC dendritic tip; closed red arrowhead, ipsilateral ADF dendritic tip; open red arrowhead, contralateral ADF dendritic tip. B-C, Animals were collected as L4s and dendrite and nose lengths measured. AWC dendrite length was compared to the ipsilateral (B, closed circles) and contralateral (C, open circles) ADF dendrite lengths. Each dot represents an individual. dex-1(ns42), red symbols; dyf-7(ns117), blue symbols. Clustering of data points at corners reflects the bimodal distribution described in Fig. 1D.
In each mutant, dendrite lengths in the same amphid were perfectly correlated (Fig. 2B), while dendrite lengths in bilaterally apposed amphids were no more correlated than would be expected at random (Fig. 2C). Dendrite lengths were similarly well-correlated with the length of the sheath glial process (data not shown). Thus, at least two separable activities are involved in dendrite length establishment: a genetically uncharacterized activity that coordinates the relative lengths of dendrites and glia, and a dex-1 and dyf-7-dependent activity that sets the absolute length of the dendritic-glial bundle.
Dendrites form by dendritic tip anchoring during cell body migration
We considered two hypotheses for how dendrite length might be established. In one hypothesis, a dendritic growth cone emerges from the cell body and seeks a target at the nose. This mode of neurite outgrowth is well established for axon formation, and a similar mode of outgrowth has been observed using in vivo time-lapse imaging of highly branched dendrites forming in other systems (Williams and Truman, 2004; Wu and Cline, 2003). In an alternative hypothesis, the presumptive dendritic tip remains stationary while the cell body migrates away, a mode of outgrowth we call retrograde extension. Retrograde extension has not been directly observed in any system, but has been suggested by electron microscopic studies of developing sense organs in C. elegans embryos and male tails (Nguyen et al., 1999; Sulston et al., 1983). Time-lapse images of wild-type and dyf-7(m537) embryos expressing myristoylated GFP in most sensory neurons were consistent with retrograde extension (Movie 1), but due to the large number of cells labeled it was impossible to resolve individual dendrites. To overcome this limitation, we used optical cell marking (OCM) to observe the formation of single dendrites in living C. elegans embryos.
OCM entails cell-limited photoconversion of a broadly-expressed fluorescent protein – in this case, Kaede – to generate a marker with nearly exclusive single-cell specificity (Fig. 3A-C) (Ando et al., 2002). Using OCM we saw that in 4/4 wild-type embryos (Movies 2-5), amphid neurons were born near the presumptive nose and formed a projection towards it (Fig. 3D). The cell body then developed a leading edge antipodal to this projection and underwent a ~30-min, ~6-μm posterior migration while the dendritic tip remained anchored at its original position (Fig. 3E), forming a dendrite by retrograde extension (Fig. 3F).
FIGURE 3. Time-lapse imaging of dendrite extension.
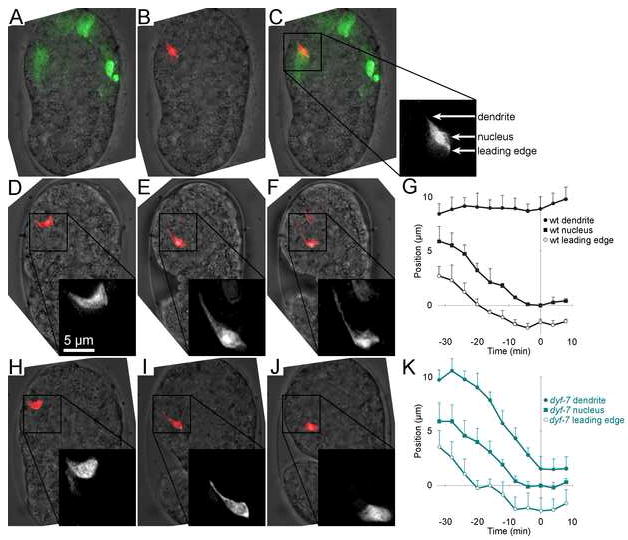
A-C, Dorsal view of a bean-stage embryo expressing the photoconvertible fluorescent protein Kaede in most sensory neurons. 20 50-msec pulses of a low-intensity 406 nm laser were used to convert Kaede from its native green-fluorescent state to its photoconverted red-fluorescent state in a single neuron. A, green, native Kaede; B, red, photoconverted Kaede; C, merged image. Box shows the vertical axis positions of the dendritic tip, nucleus, and leading edge of the photoconverted cell. D-F, Time-lapse sequence of the equivalent neuron in a different wild-type embryo. Inset is enlarged 2x. Scale bar applies to inset. D, 0 min; E, 51 min; F, 65 min. Optical stacks were collected every 4 min, and Kaede photoconversion was repeated every 16 min, as needed. G, Plot of dendrite, nucleus, and leading edge vertical axis positions over time in three wild-type embryos (Movies 1-3). Error bars, standard deviation (SD) among the individuals. A fourth wild-type embryo (Movie 4) began embryo elongation/rotation earlier and was excluded (see Supp. Fig. 2). H-J, Same as D-F, but with a dyf-7(m537) embryo. H, 0 min; I, 43 min; J, 52 min. K, Same as G, using four dyf-7(m537) embryos (Movies 4-8).
We did not attempt single-embryo analysis of dex-1 embryos because of the weak penetrance of the ns42 allele. However, in 4/4 dyf-7(m537) embryos (Movies 6-9), neurons were born in the proper position and formed projections towards the nose (Fig. 3H), but dendritic tips failed to remain anchored during cell body migration, were “dragged” along with the migrating cell body (Fig. 3I) and eventually disappeared (Fig. 3J). Thus, dyf-7 is required for anchoring dendritic tips during cell body migration.
We attempted to quantify the rate of movement of the dendritic tip, nucleus, and leading edge of each migrating cell by measuring their positions along the vertical image axis (y) at each time point (t) (see Supp. Methods). We aligned the movies in space and time using the endpoint of nuclear migration, defined by the abatement of nuclear velocity, as a registration mark ((y, t) = (0, 0); Supp. Fig. 2). Averaged trajectories of wild-type embryos from Movies 2-4 (Fig. 3G) and dyf-7 embryos from Movies 6-9 (Fig. 3K) showed that cell body migrations were virtually identical. However, in dyf-7 embryos the dendritic tip did not remain stationary but followed the migrating cell body with similar velocity, demonstrating that dyf-7 specifically affects dendritic tip anchoring but not cell body migration.
dex-1 and dyf-7 encode a tectorin-like protein pair
We used standard mapping and cloning techniques to identify the dex-1 and dyf-7 genes (see Methods). We found that dyf-7 encodes a predicted single-pass transmembrane protein with a small cytoplasmic domain and a large extracellular/lumenal domain similar to zona pellucida (ZP) proteins, best known for comprising the matrix surrounding vertebrate oocytes to which sperm bind (Fig. 4A) (Jovine et al., 2005). We mapped dex-1 to a 0.4 cM interval that included a gene predicted to encode a single-pass transmembrane protein with a domain similar to zonadhesin (Fig. 4A), a sperm protein that binds the ZP matrix (Hardy and Garbers, 1995), and thus a candidate DYF-7 interactor. Sequence analysis and genetic rescue showed that this gene is dex-1 (see Methods). In addition to its zonadhesin-like domain, DEX-1 is predicted to include two domains similar to nidogens, a family of proteins found in basement membranes, including neuronal basement membrane in C. elegans (Kim and Wadsworth, 2000).
FIGURE 4. dex-1 and dyf-7 encode transmembrane proteins and interact genetically.
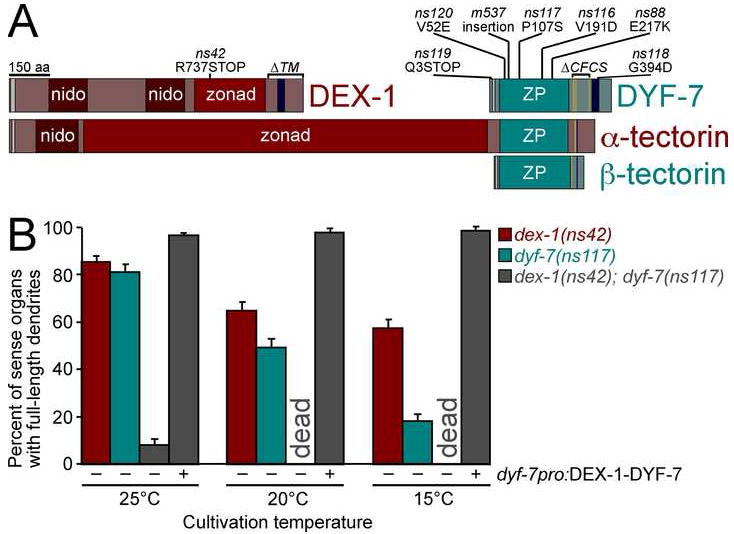
A, Schematic diagram of DEX-1, DYF-7, α-tectorin, and β-tectorin. Nidogen (nido), zonadhesin-like (zonad) and zona pellucida (ZP) domains; consensus furin cleavage sites (CFCS, gold bars); signal sequences (gray bars); and transmembrane segments/GPI anchors (dark blue bars) are indicated. B, Animals expressing gcy-5pro:GFP and str-2pro:GFP to mark individual dendrites, and bearing the cold-sensitive mutations dex-1(ns42), dyf-7(ns117), or both were cultivated at permissive (25°C), standard (20°C), or restrictive (15°C) temperatures and dendrite lengths scored. dex-1(ns42); dyf-7(ns117) animals were scored with (+) and without (−) a dyf-7pro:DEX-1-DYF-7 fusion transgene. In each case, n≥100. For transgene experiments, three lines were scored. Error bars, standard error of the mean (SEM) except, in transgene experiments, SD among lines.
The sole dex-1 mutation is predicted to truncate its zonadhesin domain (Fig. 4A). The most penetrant alleles of dyf-7 are predicted to prematurely terminate its coding sequence (ns119, m537), interfere with its transmembrane segment (ns118), or change a hydrophobic residue to a charged residue between the first and second conserved cysteines of the ZP domain (V52E in ns120). Less penetrant alleles correspond to other point mutations in the ZP domain (ns117, ns116, ns88) (Figs. 1D, 4A).
In addition to their similarity to proteins involved in sperm-egg binding, DEX-1 and DYF-7 also share domain composition with α- and β-tectorin (Fig. 4A), two vertebrate inner ear proteins found in a matrix (the tectorial membrane) that anchors the sensory endings of hair cells and is required for hearing (Legan et al., 1997; Petit et al., 2001). Mutations in α-tectorin are linked to instances of familial deafness and one such mutation, α-tectorin(G1824D) (Verhoeven et al., 1998), corresponds well to DYF-7(V52E), despite the low overall sequence similarity between their ZP domains (Supp. Fig. 3).
We reasoned that if DEX-1 and DYF-7 constitute a tectorin-like matrix, then mutations in each protein might interact genetically. Indeed, the weakly-penetrant, cold-sensitive alleles dex-1(ns42) and dyf-7(ns117) strongly synergize, such that the residual dendrite-anchoring activity of DYF-7(P107S) depends almost entirely on the presence of wild-type DEX-1 (Fig. 4B, 25°C). Cultivation under restrictive conditions revealed an even stronger synergy, in the form of a synthetic lethal phenotype (Fig. 4B, 20°C and 15°C) caused by defects in the excretory system (data not shown). These genetic interactions are consistent with the notion that the DEX-1 zonadhesin and DYF-7 ZP domains might physically interact.
In vertebrates, α-tectorin provides sufficient activity for hearing, while β-tectorin contributes more subtly (Russell et al., 2007). We noted that α-tectorin resembles a DEX-1-DYF-7 fusion (Fig. 4A), suggesting that α-tectorin may have arisen by duplication of an ancestral dyf-7-like gene downstream of, and in frame with, an ancestral dex-1-like gene. As a test of this hypothesis, we expressed a DEX-1-DYF-7 fusion and assayed its ability to rescue the dex-1(ns42); dyf-7(ns117) double mutant. The DEX-1-DYF-7 fusion restored dendrite length and organismal viability at 25°C, 20°C, and 15°C (Fig. 4B), while DEX-1 or DYF-7 individually did not (Supp. Fig. 4). These results support a split-gene origin for α-tectorin, and suggest a similarity in function between DEX-1 and DYF-7 and α-tectorin.
DEX-1 and DYF-7 act at the time and place of dendrite anchoring
To determine when dex-1 and dyf-7 act, we performed reciprocal temperature shifts of the cold-sensitive mutants at various developmental stages and scored dendrite lengths in adults. Shifting animals to the permissive temperature before, but not after, the time of dendrite formation was sufficient to promote dendrite extension (Fig. 5A-B, open circles). Conversely, shifting animals to the non-permissive temperature before, but not after, the time of dendrite formation inhibited dendrite extension (Fig. 5A-B, closed circles). Similarly, expression of dyf-7 cDNA, under control of a heat-shock inducible promoter, before the time of dendrite formation was necessary and sufficient for rescue of a completely penetrant dyf-7 mutation (Supp. Fig. 5A). Thus, dex-1 and dyf-7 activities are necessary and sufficient at the time of dendrite extension.
FIGURE 5. Timing and localization of DEX-1 and DYF-7 expression.
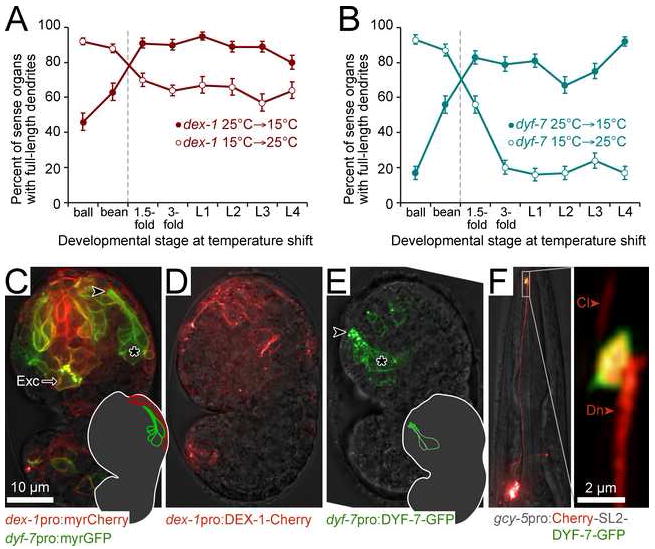
Animals expressing gcy-5pro:GFP and str-2pro:GFP and bearing the cold-sensitive mutations dex-1(ns42) (A) or dyf-7(ns117) (B) were subjected to temperature shifts at the indicated stages, and dendrite lengths scored in adults. Ball, bean, 1.5-fold, and 3-fold are morphologically-defined embryonic stages; L1, L2, L3 and L4 are larval stages. Late bean stage (dashed vertical line) corresponds to Fig. 3, when dendrites grow. Error bars, SEM. C, Ventral view of wild-type embryo at the stage of dendrite growth expressing dex-1pro:myristyl-mCherry and dyf-7pro:myristyl-GFP. *, amphid neuron cell bodies. , dendrite tip. Exc, excretory cell. Inset, schematic showing position of amphid neurons (green) and neighboring dex-1-expressing cells (red). D-E, Lateral views of wild-type embryos expressing dex-1pro:DEX-1-mCherry or dyf-7pro:DYF-7-GFP. Symbols and scale as in C. Inset, schematic showing position of amphid bundle and accumulation of DYF-7-GFP at dendritic tips. F, An animal expressing gcy-5pro:mCherry-SL2-DYF-7-GFP [SL2 (splice leader 2) permits expression of two transcripts as a single operon], to simultaneously mark the cytoplasm and DYF-7-GFP in the ASER neuron. Cl, cilium. Dn, dendrite. Inset is magnified 10x.
To determine when and where dex-1 and dyf-7 are expressed, we generated myristoylated fluorescent reporter transgenes using dex-1 or dyf-7 promoter sequences that are sufficient for rescue when used to express the corresponding cDNAs. These promoters drive reporter expression in embryos at the time of dendrite formation, with dyf-7pro expressed in most sensory neurons, including all amphid sensory neurons, and dex-1pro expressed in many non-neuronal adjacent cells, including hypodermal cells, seen as a row lateral to the amphid neuron bundle and, in a separate group, surrounding the sensory depression at the anterior pole of the embryo (Fig. 5C). The promoters also drive expression in the excretory cell, consistent with the lethality observed in the double mutant (Fig. 5C). The activity of each promoter was first apparent in bean-stage embryos, peaked in late embryogenesis, diminished in L1 larvae and was negligible in older larvae and adults (Movie 10, Supp. Fig. 5B).
We next used rescuing fluorescent fusion proteins to determine the subcellular localization of DEX-1 and DYF-7. DEX-1-mCherry localizes in spots and patches restricted to the head and tail, where sensory organs are forming (Fig. 5D). DYF-7-GFP accumulates in bright puncta at dendritic tips during retrograde extension, with weaker staining at the plasma membrane of the dendrite and cell body (Fig. 5E, Movie 11). Colocalization of DEX-1-mCherry and DYF-7-GFP at dendritic tips was occasionally observed (Supp. Fig. 5C). To further characterize DYF-7 localization, we forced expression of DYF-7-GFP in a single amphid neuron post-embryonically, when it is not normally expressed, to take advantage of the greater spatial resolution afforded by a mature neuron. In this system, DYF-7-GFP localizes in a novel domain at the dendritic tip, adjacent to the sensory cilium, indicating it contains a signal targeting it to the dendritic tip (Fig. 5F). The timing, expression, and localization of DEX-1 and DYF-7 position them to play a direct role in dendritic tip anchoring.
DEX-1 and DYF-7 are secreted
We imagined two models for dendrite anchoring. First, DEX-1 on neighboring cells could bind to DYF-7 at the dendritic tip, thus adhering the dendritic tip to cells at the nose. Alternatively, DEX-1 and DYF-7 could be proteolytically released from the cell surface and assembled into a local extracellular matrix to which the dendritic tip binds via a yet-unidentified receptor. Indeed, α- and β-tectorin are released from their membrane anchors by a furin-family protease and, together with collagens, assemble a matrix to which ciliated hair cells bind via an unidentified receptor (Legan et al., 1997; Petit et al., 2001). We reasoned that if DEX-1 and DYF-7 are similarly released, their activities might be independent of the cell type in which they are expressed, a prediction not compatible with the cell adhesion model.
We found that a dex-1 mutant was rescued by dex-1 cDNA expressed using its endogenous promoter, the dyf-7 promoter, or a glial enhancer element from the lin-26 gene (Fig. 6A) (Landmann et al., 2004), but not when using the pha-4 promoter expressed at the same developmental stage in foregut and midgut cells, anatomically separated from the amphid (Fig. 6A) (Mango et al., 1994). Mosaic analysis also indicated that dex-1 could act from multiple lineages (see Supp. Methods).
FIGURE 6. Secretion of DEX-1 and DYF-7.
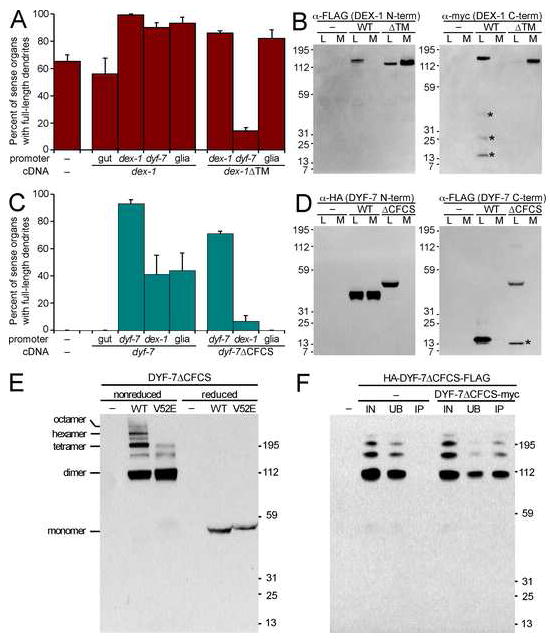
A, Animals expressing gcy-5pro:GFP and str-2pro:GFP, and bearing dex-1(ns42) and transgenes with the indicated promoters (gut, pha-4pro; dex-1pro; dyf-7pro; glia, lin-26 E1 enhancer:myo-2 minimal promoter) driving dex-1 or dex-1ΔTM cDNA were cultivated at 20°C and dendrite lengths scored. The enhanced defect in dyf-7pro:DEX-1ΔTM may reflect premature nonproductive interaction of DEX-1ΔTM and DYF-7 in the secretory pathway. Each bar is the mean of three transgenic lines, n=100 per line; error bars, SD among lines. B, S2 insect cells were transfected, or not (–), with FLAG-DEX-1-myc or FLAG-DEX-1ΔTM-myc and cultured at 25°C for 2 d. Equivalent samples of cell lysate (L) and medium (M) were collected and analyzed by immunoblot. Left, anti-FLAG; right, anti-myc. Due to increased reactivity of DEX-1ΔTM C-terminal myc, these samples were diluted 1:20. *, non-specific degradation. C, same as A except animals express only gcy-5pro:GFP and bear dyf-7(m537) and transgenes with the indicated promoters driving DYF-7 or DYF-7ΔCFCS. D, same as B except cells were transfected with HA-DYF-7-FLAG or HA-DYF-7ΔCFCS-FLAG. Left, anti-HA; right, anti-FLAG. *, non-specific degradation. E, S2 cells were transfected, or not (−), with HA-DYF-7ΔCFCS-FLAG (WT) or the same construct bearing the V52E mutation (V52E). Cell lysates were collected under nonreducing (no β-mercaptoethanol (β-me), 50°C) or reducing (5% β-me, boiling) conditions and analyzed by anti-HA immunoblot. Multimers are indicated. F, S2 cells were transfected, or not (−), with HA-DYF-7ΔCFCS-FLAG alone or in conjunction with DYF-7ΔCFCS-myc. Lysates were collected under nonreducing conditions, immunoprecipated using anti-myc antibody-conjugated agarose, and dilution-normalized volumes of starting material (input, IN), unbound (UB) and immunoprecipitated (IP) fractions were analyzed by anti-HA immunoblot.
To further examine proteolysis and secretion of DEX-1, we expressed an epitope-tagged construct in Drosophila S2 cells in vitro, a system that allows rapid production of large quantities of protein at a temperature compatible with C. elegans protein function. We observed no proteolytic cleavage or secretion of DEX-1 in this system (Fig. 6B), suggesting secretion in vivo would require a protease absent in S2 cells. DEX-1 lacks an optimal consensus furin cleavage site (CFCS) and α-tectorin and zonadhesin undergo proteolytic cleavages at non-CFCS sequences, suggesting the existence of a conserved protease that cleaves within zonadhesin domains (Bi et al., 2003; Legan et al., 1997). Consistent with this idea, a derivative of DEX-1 lacking its transmembrane anchor (DEX-1ΔTM) was efficiently secreted in vitro (Fig. 6B) and was competent for rescue when expressed using the dex-1 or glial promoters, although not the dyf-7 promoter (Fig. 6A). These results demonstrate that DEX-1 is capable of acting as a secreted factor.
Similarly, the dyf-7 cDNA was competent for rescue whether expressed in neurons, dex-1-expressing cells, or glia, but not in gut (Fig. 6C), and mosaic analysis indicated sufficiency in multiple amphid sensory neuron lineages (see Supp. Methods). In S2 cells in vitro, DYF-7 is proteolyzed into a ~45 kD N-terminal fragment, 50% of which is secreted into medium, and a ~15 kD C-terminal fragment, 100% of which is retained by cells (Fig. 6D), consistent with proteolytic cleavage at any of 3 CFCS sequences between the ZP domain and transmembrane segment (Fig. 4A). Deletion of the CFCS-bearing sequence (DYF-7ΔCFCS) abolished almost all proteolytic cleavage and resulted in a single ~50 kD product, 100% of which was retained by cells (Fig. 6D). Despite its inability to undergo proteolytic cleavage, DYF-7ΔCFCS was competent to anchor dendrites in vivo when expressed using its endogenous, neuron-specific promoter, but not when expressed in other cell types, suggesting that the ability of DYF-7 to act from multiple cell types depends on its proteolytic release from cell surfaces into the extracellular environment adjacent to neurons (Fig. 6C).
DYF-7 self-associates into multimers important for dendritic tip anchoring
ZP domains are disulfide-dependent polymerization modules thought to underlie the matrix-forming activity of proteins like tectorins (Jovine et al., 2006; Monne et al., 2008). To determine whether DYF-7 might similarly multimerize to form a matrix, we examined its behavior under non-reducing conditions. DYF-7ΔCFCS extracted from S2 cells displayed a “multimer of dimers” profile similar to that described for other ZP proteins (Fig. 6E) (Jovine et al., 2006). Less abundant species corresponding to trimers, pentamers, and heptamers were also present (Fig. 6E). No multimerization of wild-type DYF-7 secreted into medium was observed (data not shown), possibly because high local concentrations of the ZP domain are required for multimerization, as has been shown in other systems (Jovine et al., 2006).
To determine whether these high-molecular-weight complexes reflect self-association, we co-expressed DYF-7ΔCFCS constructs bearing HA-FLAG and myc epitope tags, performed anti-myc immunoprecipitation, and probed for the HA-FLAG-tagged protein in the unbound and immunoprecipitated fractions. As expected, about half of the dimer-associated HA-DYF-7ΔCFCS-FLAG co-precipitated with DYF-7ΔCFCS-myc, with the remaining half presumably representing HA-DYF-7ΔCFCS-FLAG homodimers (Fig. 6F). Recovery was more efficient with higher-order oligomers, and no immunoprecipitation was seen in the absence of DYF-7ΔCFCS-myc (Fig. 6F).
We wondered whether any of the DYF-7 point mutations we isolated due to their effect on dendrite anchoring might act by disrupting multimerization. We found that, when introduced into the ΔCFCS construct, the DYF-7(V52E) point mutant failed to multimerize normally despite dimerizing and thus, presumably, being at least partly folded (Fig. 6E). Using a semi-quantitative fluorescence-based detection system, we found that the relative abundance of the DYF-7(V52E) monomer, dimer, and trimer are increased while the tetramer and higher forms are severely diminished, an effect not observed with the chemically similar point mutant DYF-7(V191D) (Supp. Fig. 6). This result suggests that V52 may lie near the ZP multimerization interface, that this interface is structurally distinct from the dimerization interface and, most importantly, that the ability of DYF-7 to anchor dendritic tips is associated with multimerization and, by extension, matrix formation. By analogy, patients with the α-tectorin(G1824D) mutation may fail to multimerize α-tectorin.
DISCUSSION
Molecular mechanism of sensory dendrite extension
Our studies suggest the following model: Sensory neurons are born near the presumptive nose, polarize towards it, and express DYF-7. DYF-7 is trafficked towards this polarization; is released from the membrane, likely by a furin-family protease; and self-associates into dimers which further self-associate into higher-order multimers. Meanwhile, neighboring cells express transmembrane DEX-1, which likely undergoes processing by a non-furin protease and is released into the extracellular space. Then, acting together in the extracellular space, DEX-1 and DYF-7 anchor dendritic tips, possibly by co-assembling into a tectorial membrane-like matrix. At this point, the neuron cell body migrates posteriorly, probably guided by the repulsive cue SLT-1/Slit, which is highly expressed at the nose (Hao et al., 2001), and its receptor SAX-3/Robo, which is required for normal amphid cell body migration (Zallen et al., 1999). With the dendritic tip anchored at the nose, the dendrite stretches to span whatever distance the cell body may move, thus providing a simple yet robust system for attaining proper dendrite length and dendritic tip position.
There are at least two types of models for how DEX-1 and DYF-7 could generate an anchoring force. First, the DEX-1 nidogen domains could bind immobile basement membrane components while the DEX-1 zonadhesin domain could bind the DYF-7 ZP domain, which in turn could interact with the dendritic tip. Although a single such cable might be incapable of resisting the pull of cell migration, the multimerization of DYF-7 would allow the formation of myriad parallel cables, like Velcro fibers, over which the force could be distributed. Alternatively, DEX-1 and DYF-7 could co-assemble a large free-standing matrix attached to the dendritic tip, which by steric hindrance alone could act like a fluke anchor, restraining the dendritic tip from moving through the densely-packed environment at the nose tip. A key to distinguishing these models will be defining what physical interaction exists between DEX-1 and DYF-7, an important challenge as the physical interactions that bind the tectorins together, or that underlie zonadhesin-ZP sperm-egg interactions, have been difficult to ascertain.
Finally, it is critical to identify the receptor that connects DEX-1 and DYF-7 to dendritic tips. Its identity may provide insight into the functionally equivalent but so far elusive molecules in the hearing system that connect hair cell stereocilia to the tectorial membrane.
Retrograde extension is a novel mode of dendrite growth
In general, neurite development occurs in two phases: a “wiring” phase, during which connections are established, and a “scaling” phase, during which those connections are maintained in the face of mechanical distortion caused by organismal movement and growth. Wiring often occurs by anterograde extension (Fig. 7A), in which an axonal or dendritic projection emerges from a stationary cell body and is guided by extracellular cues towards its destination (Huber et al., 2003). These cues can be built into the cellular landscape or, in a phenomenon called axon towing, can be produced by migrating cells which escort the axon to its destination (Gilmour et al., 2004). A different mode of wiring occurs in cerebellar granule cell development, during which the neuron extends a pair of processes parallel to the surface of the external granule layer of the cerebellum before extending a third process in the radial direction (Fig. 7B) (Solecki et al., 2006). The cell soma then translocates along the radial process, reversing its polarity (Solecki et al., 2004). Here, we describe a third mode of wiring, retrograde extension, in which the tip of a sensory dendrite remains stationary while a neurite is generated de novo by cell body migration (Fig. 7C). Unlike cerebellar granule cell development, in which neurite formation precedes and is independent of cell soma translocation (Kerjan et al., 2005; Renaud et al., 2008), during retrograde extension it is the movement of the cell body itself that generates the neurite.
FIGURE 7. Three modes of neurite outgrowth.
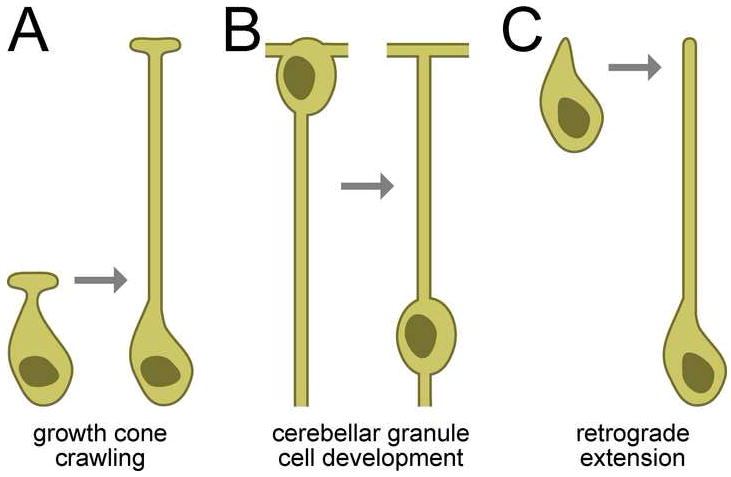
A, Many axons and dendrites form by a projection migrating away from a stationary cell body. B, During cerebellar granule cell development, a neurite forms in the same manner but the cell body then translocates along it. Neurite regions above the nucleus in this schematic are axons; below it are dendrites. C, Amphid sensory dendrites form by the stationary anchoring of dendritic tips during cell body migration, so the neurite is generated de novo by the migrating cell body.
Mechanisms of neurite scaling are less well understood, although extracellular matrix proteins seem to play a key role. In C. elegans, the extracellular matrix protein DIG-1 is required to maintain axon positioning during mechanical distortion caused by organismal movement: in dig-1 mutants, axons are misplaced, but the defect is suppressed in animals unable to move (Benard et al., 2006). In Drosophila mutants lacking the ZP-domain protein NompA, sensory dendrites attach normally to mechanosensitive bristles during embryogenesis, but become detached before adulthood. Although not known, detachment may be due to a scaling defect (Chung et al., 2001). Glial morphogenesis also depends critically on scaling; for example, radial glia contact both the ventricular zone (VZ) and pial surface when the cortex consists only of a primary neuroepithelium, and then maintain these contacts during cortical expansion, ultimately increasing to >100 μm in length in the mouse (Altman and Bayer, 1990; Noctor et al., 2002). Intriguingly, these glial contacts are lost in mutants lacking the nidogen-binding domain of laminin γ1 (Halfter et al., 2002; Haubst et al., 2006).
DEX-1 and DYF-7 are required for both retrograde extension and scaling. One interpretation of this result is that these phenomena may rely on a single physical mechanism – the anchoring of a dendritic tip relative to its extracellular environment – to resist the distortion caused by cell migration, in one case, and by organismal growth, in the other. As nidogen and zonadhesin proteins are expressed throughout the vertebrate nervous system, and ZP proteins are expressed in many sense organs and in the brain, a DEX-1-DYF-7-like anchoring activity may be conserved in the wiring and scaling of the vertebrate nervous system as well.
ZP proteins and the evolution of sense organs
There are 41 predicted ZP proteins in C. elegans, and 16 in humans (Finn et al., 2006). In addition to DYF-7, ZP proteins related to sense organ development include C. elegans RAM-5, required for morphogenesis of male-specific sense organs (Yu et al., 2000); Drosophila NompA, required for post-embryonic integrity of mechanosensitive sense organs (Chung et al., 2001); and, in mammals, the tectorins of the inner ear (Legan et al., 1997); olfactorin in the olfactory epithelium (Di Schiavi et al., 2005); and vomeroglandin and ebnerin, distinct products of the DMBT1 gene that are expressed in the pheromone-responsive vomeronasal organ (Matsushita et al., 2000) and in the taste-modulating secretions deposited by von Ebner’s gland onto taste buds (Li and Snyder, 1995), respectively. Thus, the association of ZP proteins with sense organs is conserved across species and across sensory modalities.
DEX-1 and DYF-7 resemble the tectorins in their overall domain structure, in being proteolytically released from membrane anchors and secreted to form extracellular matrices, and in anchoring the ciliated endings of sensory cells; however, DEX-1 and DYF-7 anchor dendritic tips to resist the force of cell migration while tectorins anchor stereocilia to resist the deflection caused by sound waves. Over two centuries ago, the theologian William Paley was fascinated by the anatomy of the ear, which he took as evidence of the hand of “the Author of nature” (Paley, 1803). However, the similarity between DEX-1-DYF-7 and the tectorins provides evidence that such a remarkable “instrument adapted to the reception of sound” as the tectorial membrane of the human ear may have evolved from an ancient module required for morphogenesis of a nematode sense organ.
METHODS
Strains and plasmids
Strains were constructed in the N2 background and cultured under standard conditions (Brenner, 1974; Stiernagle, 2006) unless otherwise noted. Transgenes and plasmids are listed in Supporting Tables I and II.
Isolation of mutants and genetic mapping
Animals of genotype nsIs53 IV; ntIs1 V; kyIs136 X were mutagenized using 70 mM ethyl methanesulfonate (EMS, Sigma) at 20°C for 4 h. Nonclonal F2 progeny were examined on an Axioplan 2 fluorescence microscope (Zeiss) with 63x/1.4 NA objective (Zeiss) and dual-band filter set (Chroma, Set 51019), and animals with aberrant amphid morphologies were recovered. In parallel, the markers were crossed to dyf-1 through dyf-13 mutants, for which amphid function was known to be defective but amphid structure had not been examined (Starich et al., 1995). dyf-7(m537) and dyf-8(m539) were identified as phenotypically Dex.
Linkage mapping and SNP analysis refined the dyf-7 map position to a 0.7 cM interval on LG X (Supp. Methods) that included C43C3.3, a gene previously identified as dyf-8 by Stephen Wicks (pers. comm.). We found that dyf-7(m537) contains a frameshift-insertion in C43C3.3 of eight imperfect 10-mer repeats (Supp. Table III); cross-progeny of dyf-7(m537) and dyf-8(m539) are phenotypically Dex; and dyf-7(m537) is rescued by the C43C3 cosmid (96 ± 4.6% of amphids with full-length dendrites; mean of three lines, n=100 per line, ± SD) and by a C43C3.3pro:C43C3.3 cDNA transgene (Fig. 6C).
Linkage mapping and SNP analysis mapped dex-1 to a 0.4 cM interval on LG III. Based on the identity of dyf-7, we scanned the dex-1 interval for candidate ZP or ZP-interacting proteins (Supp. Methods). Sequencing of D1044.2 from dex-1(ns42) revealed a C>T transition (Supp. Table III), and dex-1(ns42) was rescued by the D1044 cosmid (97 ± 4.7% of amphids with full-length dendrites; mean of three lines, n=100 per line, ± SD) and by a D1044.2pro:D1044.2 cDNA transgene (Fig. 6A).
Additional Dex mutants were isolated by screening nonclonal mutagenized F2 progeny using a fluorescence-equipped dissecting microscope (Leica) for the Dex phenotype; ns88 was isolated independently by Taulant Bacaj. These mutants mapped to LG X, and each bore a mutation in dyf-7 (Supp. Table III).
Dendrite length measurements
AxioVision 4.4 software (Zeiss) was used to measure the distance from the anterior limit of cell body fluorescence to the anterior limit of dendrite fluorescence and the nose tip. For temperature shifts, staged embryos were selected based on morphology. For population measurements, a fluorescence-equipped dissecting microscope was used to score dendrite length.
Microscopy and image processing
Images were collected on a Deltavision Core imaging system (Applied Precision) with a PlanApo 60x/1.42 NA or UPLSApo 100x/1.40 NA oil-immersion objective and a Photometrics CoolSnap HQ camera (Roper Scientific). For OCM, nsIs96 (dyf-7pro:Kaede) embryos at late ball stage were mounted (Supp. Methods) and photoconversion performed by a series of 20 50-msec pulses of a 406 nm laser at 10% power. At 4 min intervals, a stack of 12-16 optical sections at 0.4 μm spacing was acquired using white light and fluorescence (excitation 572/35 nm, emission 630/60 nm). To compensate for loss of signal by photobleaching, photoconversion of newly-synthesized Kaede was repeated every 12-24 min. Time-lapse images were acquired until the embryo began to twitch at 1.5-fold stage.
Deconvolution and analysis of images were performed with Softworx (Applied Precision) and IVE/Priism (Chen et al., 1996). Maximum brightness projections were obtained using contiguous optical sections; the upper and lower limit of the region of interest was not necessarily the same among time points or wavelengths. Figures and movies were assembled using Photoshop 7 (Adobe Software) and QuickTime Pro (Apple Computer). Quantitative analysis of cell migration is detailed in Supp. Methods.
Protein expression and analysis
Drosophila Schneider (S2) cells (Invitrogen) were cultured at 25°C and transfected with FuGene HD (Roche), incubated for 2 days, and lysed in sample buffer (60 mM Tris HCl pH 8.0, 2% sodium dodecylsulfate (SDS), 10% glycerol, 5% β-mercaptoethanol as appropriate, 0.01% bromophenol blue, 1x Complete protease inhibitor cocktail (Roche)) or IP buffer (60 mM Tris HCl pH 8.0, 1% Tergitol type NP-40 (Sigma), 10% glycerol, 1x Complete protease inhibitor cocktail). Immunoprecipitation was performed with goat anti-myc-conjugated agarose (Genetex) for 2 h at 4°C. Samples were analyzed on NuPage 4-12% Bis-Tris gels (Invitrogen) and immunoblotting was performed using the following antibodies: DEX-1 N-terminal FLAG, rabbit polyclonal anti-FLAG (Sigma) 1:5000; DEX-1 C-terminal myc, rabbit polyclonal anti-myc (AbCam) 1:10,000; DYF-7 N-terminal HA, rat monoclonal anti-HA 3F10 (Roche) coupled to horseradish peroxidase (HRP), 1:10,000; DYF-7 C-terminal FLAG, mouse monoclonal anti-FLAG M2 (Sigma) 1:10,000; goat polyclonal anti-rabbit (Pierce) and anti-mouse-IgG (Pierce) coupled to HRP, 1:10,000.
Supplementary Material
Acknowledgments
We thank Yun Lu for preparing the electron micrograph in Supp. Fig. 1B; Valeri Thomson, the Rockefeller University Bio-Imaging Resource Center and members of Ulrike Gaul’s laboratory for technical help; Cori Bargmann, Alison North and members of the Shaham laboratory for comments on the manuscript; and Myriam Heiman. M. G. H. was a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. S. S. is a Klingenstein Fellow in the Neurosciences and a Monique Weill-Caulier Scholar. This work was supported in part by a grant from the Patterson Trust and by NIH grant 1R01NS064273 to S. S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman J, Bayer SA. Vertical compartmentation and cellular transformations in the germinal matrices of the embryonic rat cerebral cortex. Exp Neurol. 1990;107:23–35. doi: 10.1016/0014-4886(90)90060-6. [DOI] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Chemosensation in C elegans. WormBook; 2006. pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard CY, Boyanov A, Hall DH, Hobert O. DIG-1, a novel giant protein, non-autonomously mediates maintenance of nervous system architecture. Development. 2006;133:3329–3340. doi: 10.1242/dev.02507. [DOI] [PubMed] [Google Scholar]
- Bi M, Hickox JR, Winfrey VP, Olson GE, Hardy DM. Processing, localization and binding activity of zonadhesin suggest a function in sperm adhesion to the zona pellucida during exocytosis of the acrosome. Biochem J. 2003;375:477–488. doi: 10.1042/BJ20030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Chung YD, Zhu J, Han Y, Kernan MJ. nompA encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron. 2001;29:415–428. doi: 10.1016/s0896-6273(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Di Schiavi E, Riano E, Heye B, Bazzicalupo P, Rugarli EI. UMODL1/Olfactorin is an extracellular membrane-bound molecule with a restricted spatial expression in olfactory and vomeronasal neurons. Eur J Neurosci. 2005;21:3291–3300. doi: 10.1111/j.1460-9568.2005.04164.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB. Molecular and cellular mechanisms of dendritic morphogenesis. Curr Opin Neurobiol. 2007;17:525–532. doi: 10.1016/j.conb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D, Knaut H, Maischein HM, Nusslein-Volhard C. Towing of sensory axons by their migrating target cells in vivo. Nat Neurosci. 2004;7:491–492. doi: 10.1038/nn1235. [DOI] [PubMed] [Google Scholar]
- Halfter W, Dong S, Yip YP, Willem M, Mayer U. A critical function of the pial basement membrane in cortical histogenesis. J Neurosci. 2002;22:6029–6040. doi: 10.1523/JNEUROSCI.22-14-06029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JC, Yu TW, Fujisawa K, Culotti JG, Gengyo-Ando K, Mitani S, Moulder G, Barstead R, Tessier-Lavigne M, Bargmann CI. C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron. 2001;32:25–38. doi: 10.1016/s0896-6273(01)00448-2. [DOI] [PubMed] [Google Scholar]
- Hardy DM, Garbers DL. A sperm membrane protein that binds in a species-specific manner to the egg extracellular matrix is homologous to von Willebrand factor. J Biol Chem. 1995;270:26025–26028. doi: 10.1074/jbc.270.44.26025. [DOI] [PubMed] [Google Scholar]
- Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, Gotz M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development. 2006;133:3245–3254. doi: 10.1242/dev.02486. [DOI] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- Jovine L, Darie CC, Litscher ES, Wassarman PM. Zona pellucida domain proteins. Annu Rev Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- Jovine L, Janssen WG, Litscher ES, Wassarman PM. The PLAC1-homology region of the ZP domain is sufficient for protein polymerisation. BMC Biochem. 2006;7:11. doi: 10.1186/1471-2091-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjan G, Dolan J, Haumaitre C, Schneider-Maunoury S, Fujisawa H, Mitchell KJ, Chedotal A. The transmembrane semaphorin Sema6A controls cerebellar granule cell migration. Nat Neurosci. 2005;8:1516–1524. doi: 10.1038/nn1555. [DOI] [PubMed] [Google Scholar]
- Kim S, Wadsworth WG. Positioning of longitudinal nerves in C. elegans by nidogen. Science. 2000;288:150–154. doi: 10.1126/science.288.5463.150. [DOI] [PubMed] [Google Scholar]
- Landmann F, Quintin S, Labouesse M. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev Biol. 2004;265:478–490. doi: 10.1016/j.ydbio.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Legan PK, Rau A, Keen JN, Richardson GP. The mouse tectorins. Modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J Biol Chem. 1997;272:8791–8801. doi: 10.1074/jbc.272.13.8791. [DOI] [PubMed] [Google Scholar]
- Li XJ, Snyder SH. Molecular cloning of Ebnerin, a von Ebner’s gland protein associated with taste buds. J Biol Chem. 1995;270:17674–17679. doi: 10.1074/jbc.270.30.17674. [DOI] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Matsushita F, Miyawaki A, Mikoshiba K. Vomeroglandin/CRP-Ductin is strongly expressed in the glands associated with the mouse vomeronasal organ: identification and characterization of mouse vomeroglandin. Biochem Biophys Res Commun. 2000;268:275–281. doi: 10.1006/bbrc.2000.2104. [DOI] [PubMed] [Google Scholar]
- Monne M, Han L, Schwend T, Burendahl S, Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456:653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- Nguyen CQ, Hall DH, Yang Y, Fitch DH. Morphogenesis of the Caenorhabditis elegans male tail tip. Dev Biol. 1999;207:86–106. doi: 10.1006/dbio.1998.9173. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley W. Natural theology: Evidence of the existence and attributes of the deity, collected from the appearances of nature) 1803 [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Petit C, Levilliers J, Hardelin JP. Molecular genetics of hearing loss. Annu Rev Genet. 2001;35:589–646. doi: 10.1146/annurev.genet.35.102401.091224. [DOI] [PubMed] [Google Scholar]
- Renaud J, Kerjan G, Sumita I, Zagar Y, Georget V, Kim D, Fouquet C, Suda K, Sanbo M, Suto F, et al. Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nat Neurosci. 2008;11:440–449. doi: 10.1038/nn2064. [DOI] [PubMed] [Google Scholar]
- Russell IJ, Legan PK, Lukashkina VA, Lukashkin AN, Goodyear RJ, Richardson GP. Sharpened cochlear tuning in a mouse with a genetically modified tectorial membrane. Nat Neurosci. 2007;10:215–223. doi: 10.1038/nn1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. Glia-neuron interactions in the nervous system of Caenorhabditis elegans. Curr Opin Neurobiol. 2006;16:522–528. doi: 10.1016/j.conb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Govek EE, Tomoda T, Hatten ME. Neuronal polarity in CNS development. Genes Dev. 2006;20:2639–2647. doi: 10.1101/gad.1462506. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C elegans. WormBook; 2006. pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I, Verstreken M, Van Hauwe P, Coucke P, Chen A, et al. Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet. 1998;19:60–62. doi: 10.1038/ng0598-60. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Williams DW, Truman JW. Mechanisms of dendritic elaboration of sensory neurons in Drosophila: insights from in vivo time lapse. J Neurosci. 2004;24:1541–1550. doi: 10.1523/JNEUROSCI.4521-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Time-lapse in vivo imaging of the morphological development of Xenopus optic tectal interneurons. J Comp Neurol. 2003;459:392–406. doi: 10.1002/cne.10618. [DOI] [PubMed] [Google Scholar]
- Yu RY, Nguyen CQ, Hall DH, Chow KL. Expression of ram-5 in the structural cell is required for sensory ray morphogenesis in Caenorhabditis elegans male tail. Embo J. 2000;19:3542–3555. doi: 10.1093/emboj/19.14.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Kirch SA, Bargmann CI. Genes required for axon pathfinding and extension in the C. elegans nerve ring. Development. 1999;126:3679–3692. doi: 10.1242/dev.126.16.3679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


