Abstract
Chromosome translocation to generate the TEL-AML1 (also known as ETV6-RUNX1) chimeric fusion gene is a frequent and early or initiating event in childhood acute lymphoblastic leukemia (ALL). Our starting hypothesis was that the TEL-AML1 protein generates and maintains preleukemic clones and that conversion to overt disease requires secondary genetic changes, possibly in the context of abnormal immune responses. Here, we show that a murine B cell progenitor cell line expressing inducible TEL-AML1 proliferates at a slower rate than parent cells but is more resistant to further inhibition of proliferation by TGF-β. This facilitates the competitive expansion of TEL-AML1–expressing cells in the presence of TGF-β. Further analysis indicated that TEL-AML1 binds to a principal TGF-β signaling target, Smad3, and compromises its ability to activate target promoters. In mice expressing a TEL-AML1 transgene, early, pre-pro-B cells were increased in number and also showed reduced sensitivity to TGF-β–mediated inhibition of proliferation. Moreover, expression of TEL-AML1 in human cord blood progenitor cells led to the expansion of a candidate preleukemic stem cell population that had an early B lineage phenotype (CD34+CD38–CD19+) and a marked growth advantage in the presence of TGF-β. Collectively, these data suggest a plausible mechanism by which dysregulated immune responses to infection might promote the malignant evolution of TEL-AML1–expressing preleukemic clones.
Introduction
TEL(ETV6)-AML1(RUNX1), generated by the t(12;21) chromosome translocation, is the most common chimeric fusion gene in childhood cancer, selectively associated with B cell precursor acute lymphoblastic leukemia (ALL) (1–3). Observations on clinical samples — normal cord blood (4) and specimens from monozygotic twins (5) — plus animal modeling (6–9) indicate that this oncogene can induce a preleukemic phenotype but is insufficient for overt leukemogenesis. TEL-AML1 fusion generated during fetal hemopoiesis produces a clinically covert preleukemic clone that can persist postnatally for at least 15 years (10). A functional TEL-AML1 fusion gene arises in a B lineage progenitor/stem cell and induces clonal expansion with a relatively high frequency during normal prenatal hemopoiesis (~1% of individuals) but only converts to frank ALL in around 1% of cases (4). Epidemiological evidence now supports the view that a plausible mechanism for this critical conversion step could involve an abnormal immune response to infection (11). An unproven assumption here is that TEL-AML1 protein not only initiates the preleukemic phase but is sufficient in itself to sustain this covert condition for up to 15 years. Additional genetic abnormalities, including multiple gene deletions (12) observed at diagnosis of TEL-AML1–positive ALL, are generally considered to be secondary events associated with the transition of silent preleukemic stem cells (pre-LSCs) to overt ALL (10).
This pattern of natural history raises important functional questions including: (a) What transformed stem cell population generates and sustains the preleukemic (and ALL) clone? We have addressed this question of stem cell identity in a separate article, providing evidence that TEL-AML1 can generate a population of self-renewing human cord blood cells with a unique phenotype (CD34+CD38–CD19+), compatible with a very early stage of B cell lineage commitment (13). (b) What cellular signaling pathways are corrupted by TEL-AML1 to sustain such a persistent preleukemic state? (c) What phenotypic characteristics of a TEL-AML1–driven preleukemic clone might help explain the vulnerability of its constituent stem cells to secondary genetic changes possibly occurring in the context of an abnormal immune response to infection.
The t(12;21) translocation fuses protein dimerization domains of TEL with essentially all of the DNA binding and activating regions of AML1 (2). The latter is a key component of the core binding factor (CBF) complex (i.e., CBFα/RUNX1) that is critical for normal hematopoiesis; genes encoding protein in this complex are highly selected as targets for mutational change initiating acute leukemias (14). TEL-AML1 fusion converts AML1 to a predominantly negative transcriptional regulator impeding differentiation and involving the recruitment of corepressor molecules such as NCOR and Sin3A (15, 16). Examining how such transcriptional deregulation might lead to a sustained preleukemic clone that is additionally vulnerable — possibly under proliferative stress — to further mutation is difficult with clinical samples, as analysis of TEL-AML1 function will be confounded by additional genetic abnormalities (12). We have therefore developed 3 experimental or model systems wherein the impact of TEL-AML1 protein itself can be assessed: we have used both an in vitro murine pro-B progenitor cell line with hormone-inducible TEL-AML1 and a transgenic Eμ TEL-AML1 murine model. To provide a system that more closely approximates leukemogenesis in patients, we also exploited a recently developed system for expressing TEL-AML1 in human cord blood progenitor cells (13). The latter model allows us to assess the impact of TEL-AML1 expression on candidate pre-LSCs.
Our working hypothesis is that pre-LSCs with TEL-AML1 fusion genes transit to overtly leukemic stem cells with additional genetic abnormalities under the promotional impact of a dysregulated immune response to infection (11). We therefore wished to test the prediction that selective, proliferative advantage might be evident in the context of cytokine products of activated T cells. TGF-β and IFN-γ are key immune system network modulators released by regulatory T cells (17). We therefore focused on these molecules. An additional reason for a focus on TGF-β signaling was its well-recognized role as a locus for mutational disruption in cancer progression (18) and in early B cell progenitor regulation (19).
In all 3 model systems, we found evidence that TEL-AML1 compromises the TGF-β signaling pathway, which may contribute to both the persistence or maintenance of covert preleukemic clones in patients and their competitive positive selection in an inflammatory context.
Results
Induced expression of TEL-AML1 in BaF3 cells.
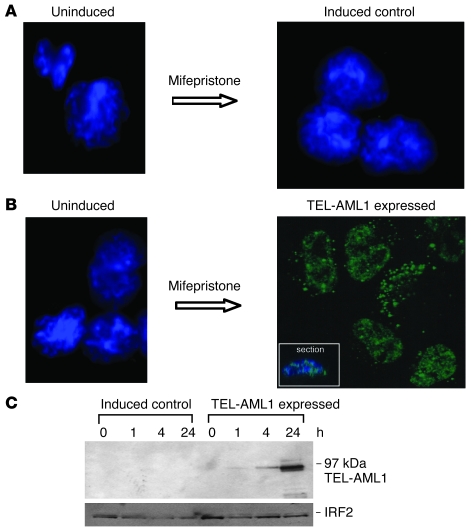
The GeneSwitch system (20) is based on an autoregulatory feedback loop that involves the binding of a GAL4 regulatory fusion protein, pSwitch, to GAL4 upstream activating sequences in both the promoter controlling expression of the GAL4 regulatory protein and that controlling expression of TEL-AML1. The expression of TEL-AML1 is itself controlled by the presence or absence of the agonist mifepristone, which brings about a conformational change in the pSwitch regulatory protein and its subsequent activation (Supplemental Methods and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI36428DS1). We established inducible TEL-AML1 in murine BaF3 cells, a putative pro-B cell line (21). Thirty double-positive stable clones expressing the regulatory protein that also showed inducible expression of TEL-AML1 by Western blot and staining via the V5 terminal tag were selected and expanded in liquid culture. Since we observed some degree of cell death in both inducible control and TEL-AML1–expressing clones at the recommended concentrations of mifepristone, we further titrated positive clones against decreasing concentrations of agonist to determine the lowest optimal conditions for protein expression without pleiotropic effects (data not shown). All subsequent experiments were performed with 12.5 pM mifepristone and 1.5 × 104 cells/ml for 3 days (unless otherwise stated). After incubation with mifepristone, predominant nuclear speckled staining was observed by confocal microscopy using an antibody against the V5 tag in all TEL-AML1–expressing cells but not control cells (Figure 1, A and B, and Supplemental Figure 2). The results from one representative clone (i.e., 1/27) are shown in Figure 1B and from another, in Supplemental Figure 3. In a number of different clones, expression of TEL-AML1 was observed by Western blot analysis in as little as 4 hours (Figure 1C and data not shown).
Figure 1. Induction of TEL-AML1 protein expression in BaF3 cells.
(A) Uninduced (left) and mifepristone-induced control cells (right) stained with both DAPI and an antibody against the TEL-AML1 V5 tag (original magnification, ×100). (B) Uninduced (left, DAPI and anti–V5 tag stained) and mifepristone-induced expression of TEL-AML1 (right, anti–V5 tag alone) (original magnification, ×100). The inset (×100) shows a confocal microscopy cross section. For DAPI staining, see Supplemental Figure 2E. (C) Western blot analysis over time of mifepristone-induced control and TEL-AML1–expressing BaF3 cell whole-cell protein extracts, blotted with anti-AML1 antibody. A loading control (IFN regulatory factor 2 [IRF2]) is shown below.
BaF3 cells expressing TEL-AML1 retain IL-3 dependence, grow more slowly than controls, but are less sensitive to the antiproliferative effects of TGF-β.
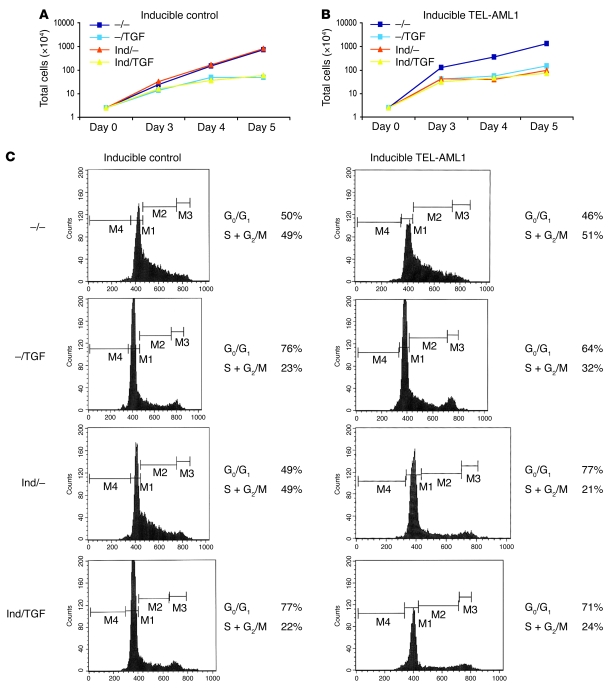
We next evaluated the growth profile of TEL-AML1–negative and –positive BaF3 cells in standard culture conditions by determining viable cell numbers (Figure 2). For this purpose, 2 TEL-AML1–expressing clones were used (with essentially identical results), and here we show the data for one of them. No difference in growth was detected with inducible control clones grown in the presence of mifepristone (Figure 2A), while expression of the fusion gene caused a slowing of cell growth (Figure 2B). We have also observed, consistently, a slower proliferation rate in several murine B cell progenitor cell lines following transfection with retroviral TEL-AML1 constructs (C. Champseix, S. Tsuzuki, T. Enver, and M. Greaves, unpublished observations). BaF3 cells expressing TEL-AML1 retained dependence on IL-3 for viability and proliferation (Supplemental Figure 4B).
Figure 2. Effects of TEL-AML1 expression and TGF-β on growth rates and cell cycle of BaF3 cells.
(A) Typical growth curve of control cells grown in the presence or absence of mifepristone inducer (Ind) and/or TGF-β (TGF) at 10 ng/ml. (B) Typical growth curve of clone 1/27 cells (inducible for TEL-AML1) grown in the presence or absence of inducer and/or TGF-β at 10 ng/ml. (C) Cell cycle analysis (BrdU) of inducible control and inducible TEL-AML1 cells grown in the presence or absence of inducer and/or TGF-β.
Next, we investigated whether TEL-AML1 expression, despite its negative impact on cell proliferation rate, could, nevertheless, provide some advantage in the context of growth inhibitors or apoptotic stimuli. We first tested the response of the cells to the addition of IFN-γ or TNF, IL-3 deprivation, or serum starvation, but saw no differences due to TEL-AML1 expression (Supplemental Figure 4B and data not shown). There was, however, a consistently different response of TEL-AML1–positive cells to the growth inhibitor TGF-β1 (TGF-β) at 10 ng/ml. As in previous studies (22), parental BaF3 cells were sensitive to the antiproliferative effects of TGF-β over a dose range of 1–100 ng/ml (see Methods and Supplemental Figure 4A), as were inducible control cells (Figure 2A), but TEL-AML1–positive BaF3 cells showed marked resistance to its effects, as evidenced by the finding that TGF-β did not further reduce their slower rate of proliferation (Figure 2B). The net result is that in the presence of TGF-β, TEL-AML1–positive cells proliferate at approximately the same rate as normal, equivalent cells.
In further examination of the inhibitory effects of TGF-β, we pulsed cells with BrdU for 30 minutes and analyzed the cell cycle of this synchronized population of cells after 10 hours of culture in the presence or absence of TGF-β (Figure 2C). TGF-β affected the cell cycle of TEL-AML1–negative cells, reducing the proportion of cells in S plus G2 relative to M phase (from 49% to 22%). In the absence of the mifepristone inducer, cycling of the TEL-AML1 clone was also reduced by TGF-β (51% to 32%). However, when TEL-AML1 expression was induced, the more slowly cycling cells were again resistant to further proliferative inhibition by TGF-β; i.e., there was no reduction in the proportion of cells in S plus G2 relative to M phase (Figure 2C; 21% vs. 24%). These data suggested that in the presence of TGF-β, cells expressing TEL-AML1, although proliferating slowly, might acquire at least proliferative parity if not a selective advantage.
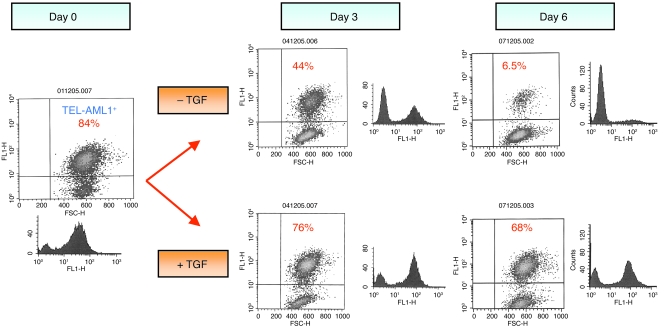
We next performed a coculture experiment with a range of percentages of TEL-AML1–negative and –positive cells in the presence or absence of TGF-β and monitored the percentage of TEL-AML1–positive cells. Figure 3 provides data from one of 5 independent experiments that gave consistent results. After 6 days in the absence of TGF-β, there was a large relative increase in the percentage of TEL-AML1–negative cells, in accordance with their faster growth. However, in the same in vitro culture conditions in the presence of TGF-β, 10-fold more TEL-AML1–positive cells were consistently present, maintaining proliferative parity (Figure 3).
Figure 3. Competition assay of a mix of TEL-AML1–expressing and –nonexpressing BaF3 cells grown in the presence or absence of TGF-β.
A mixture of TEL-AML1–expressing and –nonexpressing cells (84%:16%) was grown in the presence or absence of TGF-β, and the cell growth and TEL-AML1 expression profile were analyzed over time. Growth of TEL-AML1–expressing cells but not control cells is sustained in the presence of TGF-β. Flow cytometry analysis: the vertical axis represents fluorescence activity (for TEL-AML1 expression using an antibody against the V5 tag); the horizontal axis represents light scatter (cell size). In 4 repeat experiments, the starting percentage of TEL-AML1–positive cells was around 80%, and their persistence in the presence of TGF-β was consistent.
The increase in endogenous p27KIP1 transcription in response to TGF-β is blocked in the presence of TEL-AML1.
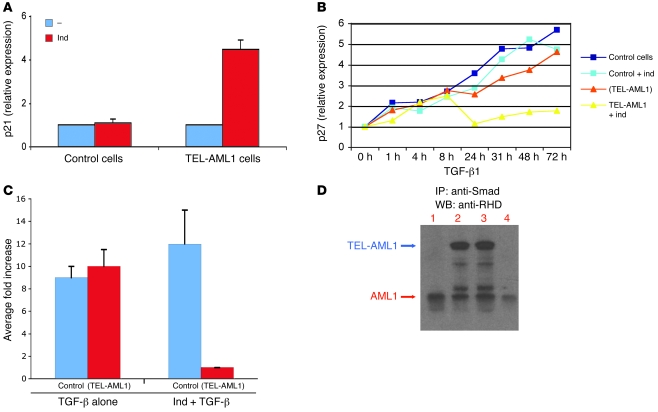
In the presence of TEL-AML1 alone, we saw a relative increase in transcript levels of the cyclin-dependent kinase (Cdk) inhibitor proteins p27KIP1 (CDKN1B) and p21 (CDKN1A) (Figure 4A and Supplemental Figure 5), which control G1 phase progression and S phase entry (23). This upregulation is concomitant with the change in cell cycle. However, it has also been shown that TGF-β induces G1 arrest in B cells via upregulation of p27KIP1 protein (23). We therefore investigated next whether the resistance of TEL-AML1–positive cells to TGF-β–induced growth inhibition was associated with an impact on p27KIP1 expression. Real-time quantitative PCR (Q-PCR) analysis of p27KIP1 expression levels was performed in TEL-AML1–positive and –negative cells treated with TGF-β for different periods of time (Figure 4B) and in the absence of TGF-β (Supplemental Figure 5). As anticipated, inducible control cells in the presence of TGF-β showed a progressive increase in p27KIP1 expression. Conversely, in the presence of TGF-β, TEL-AML1–positive cells exhibited minimal further increase in p27KIP1 expression. In some experiments (as in Figure 4B), there was a transient increase in p27KIP1 in the presence of TGF-β and TEL-AML1, but this was not consistently seen.
Figure 4. TEL-AML1 activates expression of p21 but blocks the TGF-β–mediated activation of p27 and inhibits the TGF-β response of a target gene promoter.
(A) Q-PCR analysis showing activation of p21 (CDKN1A) by TEL-AML1 in the absence of TGF-β. Control cells and cells inducible for TEL-AML1 were incubated for 3 days in the absence (–) or presence of inducer. cDNA was subjected to Q-PCR and normalized to GAPDH, to which relative expression of p21 is shown. Error bars represent the SD of an experiment performed in triplicate and repeated 3 times. (B) Q-PCR analysis showing a block in expression of TGF-β–induced p27KIP1 in the presence of TEL-AML1. cDNA was prepared from a TGF-β time-course analysis of both control and TEL-AML1–inducible cells in the presence or absence of the TEL-AML1–inducing agent (ind). Cells inducible for TEL-AML1 but not actually induced are indicated by parentheses, i.e., (TEL-AML1). Experiments were repeated 3 times. (C) Inhibition of the TGF-β–responsive IgA promoter by TEL-AML1 in a luciferase reporter assay. Activation of the IgA promoter was assayed by its transient transfection into control cells and cells inducible for TEL-AML1. Cells inducible for TEL-AML1 but not actually induced are indicated by parentheses. Growth was continued in the presence of TGF-β, either alone or after addition of the TEL-AML1–inducing agent. Error bars represent SD from 3 independent experiments. (D) TEL-AML1 associates with Smad3. Cell lysates from control and TEL-AML1–expressing cells were immunoprecipitated with anti-Smad3 antibody and half the IP subjected to Western blot analysis with an antibody against the runt homology domain (RHD) of AML1. Lane 1, uninduced cells; lane 2, TEL-AML1–induced; lane 3, TEL-AML1–induced + TGF; lane 4, control cells + TGF.
The expression of TEL-AML1 markedly represses the response of the mouse Igα promoter to TGF-β.
To determine whether TEL-AML1 expression interfered only with the antiproliferative activity of TGF-β or whether its overall signaling was affected, we tested the response to TGF-β of one of the known target gene promoters. Using transient transduction reporter assays with a construct that contained the mouse Igα promoter (positively regulated by TGF-β; ref. 24), we observed that expression of TEL-AML1 markedly repressed the response of this promoter to TGF-β (Figure 4C).
Smad2/3 signaling in the presence of TEL-AML1.
To gain some insight into the possible mechanism underlying the lack of response of TEL-AML1–positive cells to TGF-β, we analyzed the TGF-β pathway of these cells. First we investigated the early steps of the pathway: TGF-β binding to TGF-β II receptor and activation of the receptor complex by Smad2/3 phosphorylation in the presence or absence of a potent and specific inhibitor of TGF-β (SB-431542) (25, 26). By Western blot analysis with anti–p-Smad2/3, we showed that the phosphorylation of endogenous Smad2/3 protein, in response to TGF-β, was unaffected in TEL-AML1–positive cells, inferring that disruption to the TGF-β pathway by TEL-AML1 was subsequent to Smad2/3 activation (Supplemental Figure 6 and data not shown). Furthermore, using Q-PCR array analysis, we also analyzed the expression of a focused panel of genes known to be related to bone morphogenetic protein–mediated TGF-β signal transduction and assessed their status in the presence or absence of TEL-AML1. No changes in the levels of TGF-β II receptor gene expression were observed (data not shown), again consistent with downstream disruption of the pathway.
It has been reported that other leukemic fusion proteins that incorporate AML1, such as AML1/EVI-1 and AML1/ETO, associate with Smad3 (27, 28) and do so via the DNA binding Runt domain (27), or EVI1 in the case of AML1-EVI1 (28). Since this domain is retained in the TEL-AML1 fusion, we investigated whether TEL-AML1 was able to associate with Smad3. Smad3 immunoprecipitation from lysates of TEL-AML1–positive and –negative cells, grown for 48 hours in the presence or absence of TGF-β, confirmed association of TEL-AML1 with Smad3 (Figure 4D). We next used the CAGA12 promoter reporter gene to assess the response to TGF-β in the absence of a binding site for (TEL-)AML1. The CAGA12 promoter contains 12 tandem copies of the DNA binding site for Smad3 alone linked to luciferase (25) but no binding site for AML1. The results indicated reduced activation of the CAGA12 promoter by TGF-β in the presence of TEL-AML1, which cannot bind to this promoter (Supplemental Figure 7A). Furthermore, using EMSA with protein extracts isolated from REH t(12;21) cells and a DNA probe that contains the AMLU and AMLD binding sites as well as SMADA and SMADB binding sites (29), we show that TEL-AML1 is able to bind to its cognate binding site in the presence of Smad3 (Supplemental Figure 7B). The combined data suggest that the repressive effect seen at this TGF-β–responsive promoter in the presence of TEL-AML1 operates through interaction with Smad3 but may not require full DNA binding of the fusion protein itself. Similarly, the corepressor mSin3A is known to regulate diverse signaling pathways in both normal and malignant cell growth by forming large multiprotein repressor complexes (15, 16). In cells induced to express TEL-AML1, immunoprecipitation with antibody against mSin3a confirmed its interaction with the fusion protein (data not shown).
Functional impact of TEL-AML1 expression in progenitor cells from transgenic mice.
As the lineage affiliation of BaF3 is not unambiguous, we analyzed the effect of TEL-AML1 protein expression in an alternative murine model system: BM-derived B lineage progenitor cells from mice transgenic for TEL-AML1. As expression of the fusion gene was regulated by the IgH enhancer, we anticipated that TEL-AML1 protein would be selectively expressed in the B cell lineage and, possibly, at lower levels in earlier progenitors, in which the enhancer element might be accessible to transcription factors and therefore active (30). Two transgenic lines expressing Eμ TEL-AML1 were generated. In more than 100 mice followed for 18 months, no leukemias developed, commensurate with the view that TEL-AML1 is in itself insufficient to cause overt leukemia (9, 31).
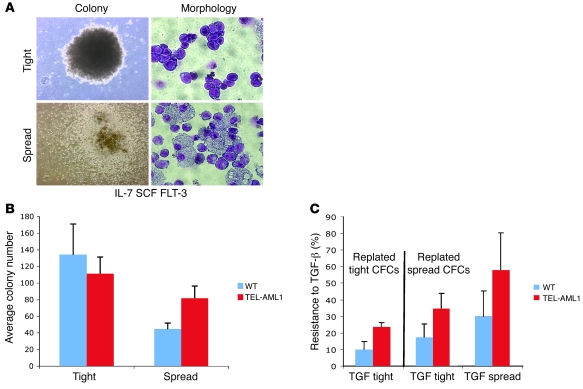
We cloned BM colony-forming cells from mice in the presence of Flt-3 ligand, IL-7, and SCF. Two morphological types of colonies were generated in both wild-type and TEL-AML1 mice (Figure 5A). These distinctive colonies were qualitatively distinguished, and we refer to them as “spread” (or diffuse) and “tight” (or dense). TEL-AML1 mice consistently showed higher numbers of spread colonies and fewer tight colonies compared with wild-type mice (Figure 5B). Replating assays revealed that spread colonies could form both spread and tight secondary colonies, but replated tight colonies only produced tight colonies. This suggests that spread CFCs are developmentally less differentiated than tight colonies, which is compatible with their phenotypes (see below). We next assessed the impact of TGF-β on progenitor CFCs with and without TEL-AML1 expression by selectively picking spread versus tight colonies and replating them in the presence of TGF-β. We observed that both spread and tight TEL-AML1–positive colonies were consistently less sensitive to the inhibitory effects of TGF-β than were wild-type colonies. Reduced sensitivity to TGF-β was reflected in colony numbers (Figure 5C and Supplemental Figure 8; data from 5 experiments). Additionally, residual colonies in TGF-β–treated TEL-AML1–positive cell were usually larger than in wild-type populations (Supplemental Figure 9).
Figure 5. Impact of TEL-AML1 expression on B progenitor CFC numbers and their inhibition by TGF-β.
(A) Typical first-round colony morphology in a B progenitor cell colony assay. Colonies (×40) were grown in the presence of IL-7 with additional SCF and Flt-3 ligand, picked after 9 days growth, spread on glass slides, and stained with Giemsa for morphology (×100). (B) First-plate phenotype. Average colony numbers from first plating of BM cells isolated from wild-type and TEL-AML1 transgenic lines. Cells (7 × 105) were plated in methylcellulose for 9 days under B cell growth conditions with additional SCF and Flt-3 ligand. Error bars represent SD from 5 independent experiments. (C) Average resistance to TGF-β. Effect of TGF-β on second-round colonies picked from first-round tight colonies (which yield only tight colonies) and spread colonies (which yield both spread and tight colonies). Average resistance was calculated as the number of colonies in the presence of TGF-β divided by the total number of colonies in its absence in the same experiment. Error bars represent SD from 5 independent experiments.
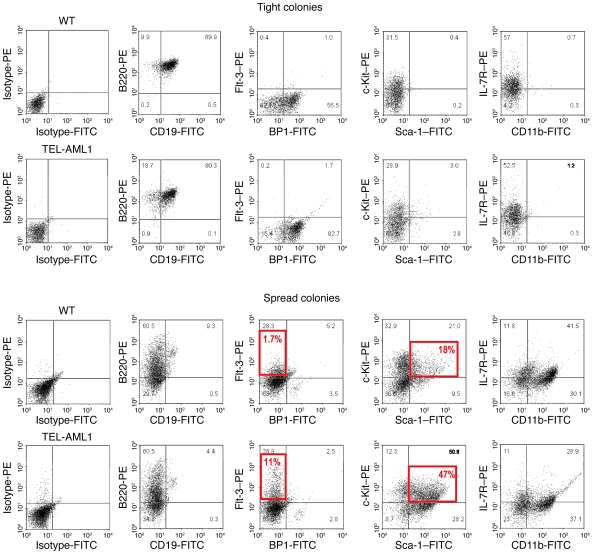
Analysis of cells retrieved from these colonies according to morphology, immunophenotype (Figure 5A and Figure 6), and gene expression (Q-PCR) (Supplemental Figure 10) revealed that the spread colonies were a mixed population of lymphoid and myeloid cells, whereas cells in the tight colonies were all lymphoid. Wild-type and TEL-AML1–positive tight colonies showed a similar pre-B cell phenotype, being B220-, CD19-, and BP1-positive but negative for Flt-3, Sca-1, and CD11b (Figure 6). In contrast, the spread colonies were positive for Flt-3, Sca-1, and c-Kit, indicative of a more immature, progenitor-like phenotype. In line with the morphological variety of cells within the spread colonies, we also observed distinct and separate populations of cells either positive or negative for CD11b (Figure 6). These were flow sorted for future analysis. The CD11b-negative cells derived from TEL-AML1 transgenic mice were exclusively lymphoid in morphology and expressed RAG1, CD79a, and TEL-AML1 (Supplemental Figure 10). The CD11b-positive cells had myeloid morphology, expressed low levels of CD79a but not RAG1, and had low or negligible levels of TEL-AML1 and most closely resemble the pre-pro-B cells of Hardy’s fraction A (32). These data indicate that the spread colonies are derived from a common lympho-myeloid progenitor with (in the assay used) B lineage progeny. We also observed a substantial increase in the number of c-Kit+Sca-1+ cells in spread colonies in the presence of TEL-AML1 expression, which is in keeping with previous findings in BM transplantation models with TEL-AML1 (6, 8) and indicative of a selective impact on both multipotent (lympho-myeloid) and very early B cell progenitor cells.
Figure 6. Colony phenotypes from Eμ TEL-AML1–transgenic lines and wild-type controls.
Analysis of cell immunophenotype by flow cytometry. Tight and spread colonies isolated from wild-type and TEL-AML1–transgenic lines were compared by surface phenotype using monoclonal antibodies against B220, CD19, Flt-3, BP1 (63), c-Kit, Sca-1, IL-7R, and CD11b, as well as isotype controls. The percentage of cells within each quadrant is shown in gray, and the percentage within a specific gate is shown in red.
TEL-AML1 expression provides selective advantage to candidate “preleukemic” stem cells in the presence of TGF-β.
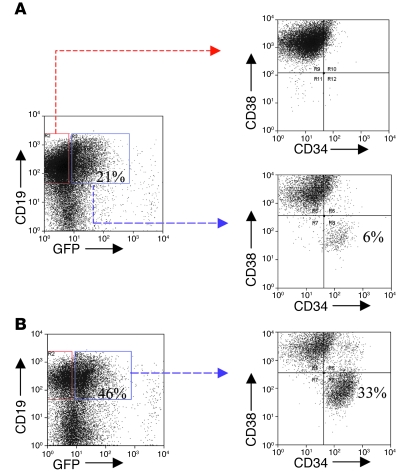
Human cord blood CD34+ cells transduced with TEL-AML1 via a lentiviral vector produce a preleukemic-like phenotype in NOD/SCID mice (13). This is dependent upon the expansion of a putative pre-LSC stem cell population with the immunophenotype CD34+CD38–CD19+ (13). We have assessed whether abrogation of TGF-β sensitivity by TEL-AML1 protein in murine models can be replicated in a system that more approximates clinical preleukemia in patients and, in particular, whether the candidate stem cell can be selected. Candidate pre-LSCs were generated in vitro by culture of TEL-AML1–transduced CD34+CD38–CD19– cord blood cells on MS-5 stroma (13). From an initial average inoculum of 93.3 TEL-AML1–expressing (GFP-positive) cells (SD 15.3, SEM 8.82, range 80–110), 5,146.7 CD34+CD38–CD19+ (candidate pre-LSCs) were generated (SD 147, SEM 85.1, range 4,980–5,260). These cells were then replated on MS-5 stroma in the presence of TGF-β. After an additional 3 weeks, they had proliferated to give rise to 19,667 pre-LSCs (SD 4,400.4, SEM 2,540, range 15,300–24,100). Thus, in the presence of TGF-β, there is an absolute increase in the number of TEL-AML1–expressing pre-LSCs of approximately 4-fold (average 3.82, range 3.1- to 4.6-fold). Interestingly, this net increase in pre-LSCs that occurs in the presence of TGF-β was accompanied by an absolute decrease in cell number of more mature TEL-AML1–expressing and wild-type cells, and this was not seen in the absence of TGF-β. The combination of increased numbers of pre-LSCs and decreased numbers of mature cells resulted in the 12-fold proportionate increase in pre-LSCs presented in Figure 7.
Figure 7. TGF-β “selects” candidate human pre-LSCs.
Cord blood stem cell encoded (CD34+CD38–) populations were transduced with lentiviral TEL-AML1 and plated on MS-5 stroma for 3 weeks to generate pre-LSCs (CD34+CD38–CD19+) (13). Cells were replated on MS-5 with or without TGF-β. (A) Cell populations without TGF-β. (B) Cell populations with TGF-β. The percentage of cells in each particular gate or quadrant is shown from 1 experiment but was replicated and consistent in 2 separate experiments (i.e., 3 in total).
Discussion
We have provided evidence from both murine and human model systems that TEL-AML1 expression has an inhibitory impact on the response to TGF-β. A consequence of this is that although cells expressing TEL-AML1 may actually proliferate more slowly than normal progenitors, they are not further restrained by the normally inhibitory effect of TGF-β and can proliferate continuously, albeit slowly, in its presence, thereby maintaining proliferative parity.
TGF-β is a member of a superfamily of 30 or more critical regulatory ligands that have pleiotypic and profound effects on biological responses. TGF-β signaling contributes to self-renewal and differentiation decisions (33, 34) and to regulation of immunological and inflammatory reactions (34). In cancer, TGF-β has somewhat paradoxical effects. It functions as a potent tumor suppressor via its Smad-dependent inhibitory impact on c-Myc and its activation of p15INK4b/p15 and p21/p27KIP1 (35, 36), but once emergent mutants escape this control, TGF-β can promote progression of disease, metastasis, and epithelial-mesenchymal transition (18, 35). Superimposed on this complexity are networked interactions of TGF-β signaling with other key signaling pathways and cell type variation in the specifics of target genes and phenotypic responses (23). The major role of disrupted TGF-β signaling in cancer is further reinforced by the presence of mutants in components of its signaling pathway, including TGF-β receptors (37) and Smads (18), as well as mutations in transcriptional cofactors (38), Smad phosphorylating proteins (e.g., RAS/ERKs), or TGF-β target genes (39) or the presence of viral genes that inhibit Smad protein expression (40).
There are some prior data indicating dysregulation of TGF-β signaling in leukemia (41, 42). Several myeloid leukemia-associated fusion genes, including AML1-ETO (27), AML1-EVI1 (43), and PML-RARα (41), appear to block the growth-inhibiting properties of TGF-β1, and there may be a loss of TGF-β1 type I receptors in a fraction of chronic lymphocytic leukemias (44).
Our data indicate that loss of sensitivity to TGF-β could be an important component of the function of TEL-AML1, the most frequent fusion gene in pediatric cancer. Regulated expression of TEL-AML1 protein in murine progenitor cells inhibits activation of a target promoter (CD79a, Igα) and blocks the ability of TGF-β to suppress proliferation via the activation of p27KIP1, the latter cell cycle regulator being a selective target for TGF-β signaling in B lineage cells (23).
The precise mechanism by which TEL-AML1 inhibits the TGF-β signaling pathway remains to be established, though our data indicate that this appears to occur downstream of TGF-β receptor signaling of Smad2/3 phosphorylation. We also show that, in common with normal AML1 protein and AML1-ETO and AML1-EVI1 fusion proteins (45), TEL-AML1 binds Smad3 (27). One possibility, therefore, is that TEL-AML1 sequesters Smad away from its target sites in the nucleus. A more likely alternative is that TEL-AML1, in addition to binding Smad3, binds corepressors NcoR and SIN3A (15, 16), and this may then compromise the ability of the Smad complex to transcriptionally activate the key cell cycle negative regulators, including p27KIP1. In accordance with this, our data indicate that TEL-AML1 reduces the competence of TGF-β to activate a synthetic promoter, which itself lacks AML1 binding sites. Supershift assays also suggest that TEL-AML1 and Smad3 are both part of the same complex bound to a TGF-β–responsive promoter sequence (Igα).
These data should be interpreted in the context of the known natural history of childhood ALL with TEL-AML1 fusions. The t(12;21) chromosome translocation and resultant TEL-AML1 chimeric fusion gene arise predominantly prenatally in fetal hemopoiesis (4, 10, 46, 47). The consequence of this genetic event alone is the generation of a persistent but clinically covert preleukemic clone with a low probability (~1%) of conversion to overt ALL following additional genetic changes (4, 5, 10). Murine in vivo models with TEL-AML1 are compatible with this notion, as they initiate a relatively subtle preleukemic-like expansion of early B cell progenitors but not full-blown leukemia (6–8). Epidemiological studies support the contention that a key promotional event that facilitates this transition may be an aberrant immune response to common infections (11). The issues are then to understand both how TEL-AML1 itself may initiate and sustain the preleukemic state and how the presumptive pre-ALL stem cells involved might be further promoted or selected by immune or inflammatory reactions. The current data suggest that interference with the TGF-β pathway might contribute to both processes.
Fusion to 5′ TEL sequences converts AML1 to a transcriptional inhibitor via co-option of corepressor proteins including N-CoR (16) and Sin3A (15). As with AML1-ETO, a primary impact of this appears to be a partial inhibition of differentiation. Animal models with TEL-AML1 replicate this phenotype (6–8). More significantly, a broadly equivalent phenotype can be engineered in NOD/SCID mice with human cord blood cells expressing lentivirally transfected TEL-AML1 (13). The critical AML1 target genes for differentiation inhibition are currently not known, and neither is it known whether this mechanism is sufficient for transformation. One can nevertheless envisage a mechanism by which loss of sensitivity to cell proliferation restraint by TGF-β–induced p27KIP1 might be a route via which TEL-AML1–driven preleukemic cells can expand initially to the level observed at birth (10–3–10–4; ref. 4) and be sustained at a similar level for many years despite their slower proliferation rate (5, 13). Other signal pathways dysregulated by TEL-AML1 might also contribute critically to this phenotype (48). Maintenance of a preleukemic clone of the 10–3–10–4 level by itself would likely be insufficient to explain the evolutionary and clinical emergence of ALL with additional genetic abnormalities on board (12). Some additional selective pressure and clonal advantage may be required.
Studies on identical twins discordant for ALL indicate that candidate preleukemic cells persist at the same relative level (10–3–10–4) observed in cord blood (13). In this same twin context, the candidate pre-LSC population (CD34+CD38–CD19+) represents around 1% of the identifiable (TEL-AML1–positive) total preleukemic population (13). These are clearly very modest levels of candidate pre-LSCs, which raises the question of how multiple secondary genetic changes necessary for transition to overt leukemia (10, 12) are ever acquired, even if TEL-AML1, in the context of TGF-β, maintains proliferative parity. The implication is that some selective expansion of the target pre-LSCs is necessary.
It has been suggested that inflammatory cytokines might provide selective pressure for the outgrowth of pre-ALL clones (11). Indirect support for this idea comes from the observation that B cell precursor ALL cells have a gene expression signature indicative of exposure to interferons (49) and from epidemiological studies implicating dysregulated immune responses to common but “delayed” infections as possible triggers of leukemia (reviewed in ref. 11). Under conditions of a potent immune or inflammatory response, TGF-β is consistently engaged as a negative feedback regulator or dampener of T cell responses (34).
TGF-β is a negative regulator of human early B cell progenitors (19), and we find that it facilitates the selective expansion of modeled pre-LSCs expressing TEL-AML1. The 12-fold relative increase comes at the expense of normal early B progenitors and more differentiated TEL-AML1–expressing B precursors. We speculate that this proportionate increase could place pre-LSCs at a competitive advantage in limited niche space within the marrow after TGF-β exposure, thereby further amplifying the effect of increased pre-LSC numbers. Secondary mutations necessary for overt ALL, including most frequently deletion of the normal TEL allele (50) and additional gene deletions (12), might then be more likely to occur, either stochastically in the expanding clone or via the mutagenic impact of either oxidative stress of the inflammatory milieu (51, 52) or “off-target” lymphoid enzymes RAG and AID (53, 54). The expansion involved with human pre-LSCs is more dramatic than the maintenance of proliferative parity observed in the cell mixing experiments with murine cells. This could reflect a species difference but might also be an indicator that TEL-AML1 exerts its critical effect on TGF-β signaling at the stem cell level. In this respect, our data are reminiscent of the observation that hemopoietic stem cells can be released from quiescence by a combination of p27 antisense and TGF-β signaling (55).
Methods
Cell culture and TEL-AML1 expression.
All BaF3 clones were cultured in RPMI medium supplemented with 10% FCS, 2% MoIL-3–conditioned medium containing IL-3 (56), 10 μM 2-mercaptoethanol, and 0.2 mg/ml hygromycin B (Invitrogen). In addition inducible clones were cultured in the presence of 0.05 mg/ml Zeocin (Invitrogen) (see Supplemental Methods).
The expression of TEL-AML1 was induced by adding to the culture medium 0.0125 nM mifepristone (Invitrogen)/3.8 × 105 cells for 3 days and then in 0.01 nM mifepristone to maintain long-term expression of TEL-AML1. Efficiency of TEL-AML1 induction was verified by immunocytochemistry, flow cytometry, and Western blot analysis using an anti-V5 antibody (Invitrogen). Hereafter, control clones are referred to as “inducible control” and test clones as “inducible TEL-AML1.”
Analysis of TGF-β effects on cell growth profiles.
The BaF3 clones (2.5 × 104) were grown in their standard culture conditions and in the presence or absence of 10 ng/ml rhTGF-β (R&D Systems). In pilot experiments, proliferation of parental BaF3 cells was inhibited by about 90% over the dose range 1–100 ng/ml (Supplemental Figure 4A). We elected to use a concentration of 10 ng/ml in all subsequent experiments. Some experiments were also carried out in the presence of the TGF-β inhibitor SB-43154 (26), a gift of Caroline Hill (Cancer Research UK, London Research Institute, London, United Kingdom). Cell growth was measured at the indicated incubation times by determining the number of viable cells, using trypan blue dye exclusion.
Cell cycle analysis.
BaF3 clones with and without 3-day induction of TEL-AML1 expression were incubated with 20 μM BrdU (Sigma-Aldrich) for 30 minutes in standard culture conditions. Cells were washed and cultured in fresh medium, without BrdU, in the presence or absence of 10 ng/ml rhTGF-β and 0.01 nM mifepristone. After 10 hours, cells were harvested, and 1 × 106 cells were fixed with ice-cold 70% ethanol for 30 minutes at 4°C, washed with PBS, and treated with 0.1% pepsin (Sigma-Aldrich) in 2 M HCl for 20 minutes at room temperature. The acid was neutralized with 0.1 M Na2B4O7×10H2O, pH 8.5. Cells were stained with FITC-conjugated anti-BrdU antibody (BD) for 30 minutes at room temperature. Samples were resuspended in 10 μg/ml propidium iodide (Sigma-Aldrich) and 100 μg/ml RNase (Sigma-Aldrich) in PBS for 20 minutes at 37°C and then analyzed by flow cytometry.
Coculture experiment.
After induction of TEL-AML1 for 3 days, clones were cocultured with negative control clones (at a ratio of about 85% to 15%) in RPMI medium with 10% FCS, 2% MoIL-3 conditioned medium, 10 μM 2-mercaptoethanol, 0.2 mg/ml hygromycin B, and 0.01 nM mifepristone and in the presence or absence of 10 ng/ml rhTGF-β. The percentage of TEL-AML1–positive cells was measured by flow cytometry after staining for intracellular V5 as described in Supplemental Methods.
Transgenic mice.
Lines of TEL-AML1 transgenic mice were generated by insertion of a full-length TEL-AML1 cDNA into the TOK4 expression vector (a gift of Michael Elverstein, Cambridge Laboratory of Molecular Biology, Medical Research Council, Cambridge, United Kingdom) and injection into blastocysts of B6CBAF-1 mice. The construct was driven by the human β-globin promoter and lymphoid lineage specificity achieved via the immunoglobulin (IGH) heavy chain enhancer. All experiments were approved by and conform to the standards of the animal use ethics committee, the Institute of Cancer Research, London, United Kingdom, and Home Office license requirements (PPL 70/5949). Mice were genotyped by PCR using previously published primers (46). Nontransgenic littermates were used as age-matched wild-type controls.
Isolation of human hematopoietic progenitor cells.
Cord blood from anonymized healthy donors was purchased from either the UK National Blood Service or from StemCell Technologies (CB003F) in accordance with local ethical procedures (NHS Oxfordshire Local Research Ethics Committee approval 04/Q1605/22). Anonymized peripheral blood and BM samples were from routine clinical specimens taken from patients at Great Ormond Street Hospital, London. Fully informed written parental consent was obtained in accordance with national guidelines. Total mononuclear cells (MNCs) were isolated by Ficoll gradient centrifugation. CD34+ cell fractionation from cord blood was achieved initially by magnetic bead separation (StemCell Technologies or Miltenyi Biotec). CD34-enriched cells from cord blood or MNCs from peripheral blood and BM were stained with anti-CD19–APC (Dako), -CD34-PE (BD Biosciences — Pharmingen), and -CD38-FITC (Dako).
Preparation of lentivirus, hematopoietic progenitor cell transduction, and identification of pre-LSCs.
The previously described TEL-AML1 myc-tag fusion construct (6) was cloned into the pHR-SINCSGW lentivirus (57), which carries an emerald-GFP reporter. Lentiviruses were pseudotyped with the vesicular stomatitis virus G (VSVG) protein by transient transfection of 293T cells. High-titer viral stocks were prepared by ultracentrifugation (58). Cord blood hematopoietic progenitor cells were infected for 24 hours by lentivirus (either TEL-AML1 or control virus) with an MOI of 100 (or 5 × 107 IU/ml for small numbers of cells) in serum-free medium (StemSpan; StemCell Technologies) supplemented with SCF (100 ng/ml), Flt-3 ligand (100 ng/ml), TPO (20 ng/ml), and IL-6 (20 ng/ml) (all growth factors were from Peprotech) (59, 60). Transduced cells were inoculated onto MS-5 cells. Candidate pre-LSCs were defined as cells expressing TEL-AML1 (i.e., GFP-positive) with the immunophenotype CD34+CD38–CD19– (13).
MS-5–supported culture.
Lentivirus-transduced CD34+ cord blood cell populations were maintained on MS-5 stromal cells. The latter cells were maintained in α-MEM medium complemented with 2 mM l-glutamine, 2 mM sodium pyruvate, 10% FBS (StemCell Technologies). After addition of transduced CD34+ cells, SCF and G-CSF (Peprotech) were added, and weekly half-medium changes were performed for the duration of the culture (61, 62) (further details are given in ref. 13).
Supplementary Material
Acknowledgments
We are grateful for the support of the Leukaemia Research Fund UK (A.M. Ford, D. Hong, D. Knight, T. Enver, M. Greaves), the Kay Kendall Leukaemia Fund UK (P. Cardus, M. Greaves), Ministero Italiano dell’Università e della Ricerca (MiUR), Associazione Italiana per la Ricerca sul Cancro (AIRC), Fondazione Cariplo (C. Palmi), and the Jose Carreras Foundation Fellowship (E.D. Thomas 05) (C. Bueno). We thank Caroline Bennett for help in initiating the BaF3 inducible system, Jackie Kelly-Barrett and Vladimir Grigoriev for generating the transgenic mice, Barbara Deverson for help with the manuscript, and Ian Titley for flow cytometry assistance.
Footnotes
Authorship note: Anthony M. Ford and Chiara Palmi contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: ALL, acute lymphoblastic leukemia; Cdk, cyclin-dependent kinase; pre-LSC, preleukemic stem cell; Q-PCR, real-time quantitative PCR.
Citation for this article: J. Clin. Invest. 119:826–836 (2009). doi:10.1172/JCI36428
References
- 1.Romana S.P., et al. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 2.Golub T.R., et al. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. . Proc. Natl. Acad. Sci. U. S. A. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shurtleff S.A., et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. . Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 4.Mori H., et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaves M.F., Maia A.T., Wiemels J.L., Ford A.M. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 6.Tsuzuki S., Seto M., Greaves M., Enver T. Modelling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. . Proc. Natl. Acad. Sci. U. S. A. 2004;101:8443–8448. doi: 10.1073/pnas.0402063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrow M., Horton S., Kioussis D., Brady H.J.M., Williams O. TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood. 2004;103:3890–3896. doi: 10.1182/blood-2003-10-3695. [DOI] [PubMed] [Google Scholar]
- 8.Fischer M., et al. Defining the oncogenic function of the TEL/AML1 (ETV6/RUNX1) fusion protein in a mouse model. . Oncogene. 2005;24:7579–7591. doi: 10.1038/sj.onc.1208931. [DOI] [PubMed] [Google Scholar]
- 9.Andreasson P., Schwaller J., Anastasiadou E., Aster J., Gilliland D.G. The expression of ETV6/CBF2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. . Cancer Genet. Cytogenet. 2001;130:93–104. doi: 10.1016/S0165-4608(01)00518-0. [DOI] [PubMed] [Google Scholar]
- 10.Greaves M.F., Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat. Rev. Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 11.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat. Rev. Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 12.Mullighan C.G., et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 13.Hong D., et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. . Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 14.Speck N.A., Gilliland D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 15.Hiebert S.W., et al. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol. Cell. Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidez F., et al. Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood. 2000;96:2557–2561. [PubMed] [Google Scholar]
- 17.Vignali D.A.A., Collinson L.W., Workman C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massagué J., Blain S.W., Lo R.S. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 19.Ichii M., et al. Regulation of human B lymphopoiesis by the transforming growth factor-β superfamily in a newly established coculture system using human mesenchymal stem cells as a supportive microenvironment. Exp. Hematol. 2008;36:587–597. doi: 10.1016/j.exphem.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., O’Malley B.W., Jr., Tsai S.Y., O’Malley B.W. A regulatory system for use in gene transfer. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palacios R., Steinmetz M. IL3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/S0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 22.Mahmud N., et al. Possible involvement of bcl-2 in regulation of cell-cycle progression of haemopoietic cells by transforming growth factor-1. Br. J. Haematol. 1999;105:470–477. doi: 10.1111/j.1365-2141.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 23.Kamesaki H., Nishizawa K., Michaud G.Y., Cossman J., Kiyono T. TGF-β1 induces the cyclin-dependent kinase inhibitor p27Kip1 mRNA and protein in murine B cells. . J. Immunol. 1998;160:770–777. [PubMed] [Google Scholar]
- 24.Lin Y.C., Stavnezer J. Regulation of transcription of the germ-line Ig alpha constant region gene by an ATF element and by novel transforming growth factor-beta 1 responsive elements. J. Immunol. 1992;149:2914–2925. [PubMed] [Google Scholar]
- 25.Dennler S., et al. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inman G.J., et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 27.Jakubowiak A., et al. Inhibition of the transforming growth factor β1 signaling pathway by the AML1/ETO leukemia-associated fusion protein. J. Biol. Chem. 2000;275:40282–40287. doi: 10.1074/jbc.C000485200. [DOI] [PubMed] [Google Scholar]
- 28.Mitani K. Molecular mechanisms of leukemogenesis by AML1/EVI-1. Oncogene. 2004;23:4263–4269. doi: 10.1038/sj.onc.1207777. [DOI] [PubMed] [Google Scholar]
- 29.Pardali E., et al. Smad and AML proteins synergistically confer transforming growth factor beta1 responsiveness to human germ-line IgA genes. J. Biol. Chem. 2000;275:3552–3560. doi: 10.1074/jbc.275.5.3552. [DOI] [PubMed] [Google Scholar]
- 30.Ford A.M., Watt S.M., Furley A.J.W., Molgaard H.V., Greaves M.F. Cell lineage specificity of chromatin configuration around the immunoglobulin heavy chain enhancer. EMBO J. 1988;7:2393–2399. doi: 10.1002/j.1460-2075.1988.tb03084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernardin F., et al. TEL-AML1, expressed from t(12;21) in human acute lymphocytic leukemia, induces acute leukemia in mice. Cancer Res. 2002;62:3904–3908. [PubMed] [Google Scholar]
- 32.Rumfelt L.L., Zhou Y., Rowley B.M., Shinton S.A., Hardy R.A. Lineage specification and plasticity in CD19- early B cell precursors. J. Exp. Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra L., Derynck R., Mishra B. Transforming growth factor-β signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt-Weber C.B., Blaser K. Regulation and role of transforming growth factor-β in immune tolerance induction and inflammation. Curr. Opin. Immunol. 2004;16:709–716. doi: 10.1016/j.coi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Siegel P.M., Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 36.Bierie B., Moses H.L. TGFβ: the molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 37.Wang D., et al. Analysis of specific gene mutations in the transforming growth factor-β signal transduction pathway in human ovarian cancer. Cancer Res. 2000;60:4507–4512. [PubMed] [Google Scholar]
- 38.Gomis R.R., Alarcón C., Nadal C., Van Poznak C., Massagué J. C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Ito Y., Miyazono K. RUNX transcription factors as key targets of TGF-β superfamily signaling. Curr. Opin. Genet. Dev. 2003;13:43–47. doi: 10.1016/S0959-437X(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 40.Flavell J.R., et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111:292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- 41.Lin H.-K., Bergmann S., Pandolfi P.P. Deregulated TGF-β signaling in leukemogenesis. Oncogene. 2005;24:5693–5700. doi: 10.1038/sj.onc.1208923. [DOI] [PubMed] [Google Scholar]
- 42.Dong M., Blobe G.C. Role of transforming growth factor-β in hematologic malignancies. Blood. 2006;107:4589–4596. doi: 10.1182/blood-2005-10-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sood R., Talwar-Trikha A., Chakrabarti S.R., Nucifora G. MDS1/EVI1 enhances TGF-β1 signaling and strengthens its growth-inhibitor effect, but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t(3;21), abrogates growth-inhibition in response to TGF-β1. Leukemia. 1999;13:348–357. doi: 10.1038/sj/leu/2401360. [DOI] [PubMed] [Google Scholar]
- 44.DeCoteau J.F., et al. Loss of functional cell surface transforming growth factor β (TGF-β) type 1 receptor correlates with insensitivity to TGF-β in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5877–5881. doi: 10.1073/pnas.94.11.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurokawa M., et al. The t(3;21) fusion product, AML1/Evi-1, interacts with Smad3 and blocks transforming growth factor-β-mediated growth inhibition of myeloid cells. Blood. 1998;92:4003–4012. [PubMed] [Google Scholar]
- 46.Ford A.M., et al. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. . Proc. Natl. Acad. Sci. U. S. A. 1998;95:4584–4588. doi: 10.1073/pnas.95.8.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiemels J.L., et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/S0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 48.Diakos C., et al. RNAi-mediated silencing of TEL/AML1 reveals a heat-shock protein- and survivin-dependent mechanism for survival. Blood. 2007;109:2607–2610. doi: 10.1182/blood-2006-04-019612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Einav U., et al. Gene expression analysis reveals a strong signature of an interferon-induced pathway in childhood lymphoblastic leukemia as well as in breast and ovarian cancer. Oncogene. 2005;24:6367–6375. doi: 10.1038/sj.onc.1208797. [DOI] [PubMed] [Google Scholar]
- 50.Cavé H., et al. ETV6 is the target of chromosome 12p deletions in t(12;21) childhood acute lymphoblastic leukemia. Leukemia. 1997;11:1459–1464. doi: 10.1038/sj.leu.2400798. [DOI] [PubMed] [Google Scholar]
- 51.Balkwill F. Cancer and the chemokine network. Nat. Rev. Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 52.Meira L.B., et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Invest. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullighan C.G., et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. . Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 54.Pasqualucci L., et al. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 55.Dao M.A., Taylor N., Nolta J.A. Reduction in levels of the cyclin-dependent kinase inhibitor p27(kip-1) coupled with transforming growth factor beta neutralization induces cell-cycle entry and increases retroviral transduction of primitive human hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13006–13011. doi: 10.1073/pnas.95.22.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karasuyama H., Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using a modified cDNA expression vectors. Eur. J. Immunol. 1988;18:97–104. doi: 10.1002/eji.1830180115. [DOI] [PubMed] [Google Scholar]
- 57.Demaison C., et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of immunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 58.Guenechea G., et al. Transduction of human CD34+ CD38- bone marrow and cord blood-derived SCID-repopulating cells with third-generation lentiviral vectors. Mol. Ther. 2000;1:566–573. doi: 10.1006/mthe.2000.0077. [DOI] [PubMed] [Google Scholar]
- 59.Piacibello W., et al. Engraftment in nonobese diabetic severe combined immunodeficient mice of human CD34(+) cord blood cells after ex vivo expansion: evidence for the amplification and self-renewal of repopulating stem cells. Blood. 1999;93:3736–3749. [PubMed] [Google Scholar]
- 60.Ailles L., et al. Molecular evidence of lentiviral vector-mediated gene transfer into human self-renewing, multi-potent, long-term NOD/SCID repopulating hematopoietic cells. Mol. Ther. 2002;6:615–626. doi: 10.1016/S1525-0016(02)90720-3. [DOI] [PubMed] [Google Scholar]
- 61.Nishihara M., et al. A combination of stem cell factor and granulocyte colony-stimulating factor enhances the growth of human progenitor B cells supported by murine stromal cell line MS-5. Eur. J. Immunol. 1998;28:855–864. doi: 10.1002/(SICI)1521-4141(199803)28:03<855::AID-IMMU855>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 62.Ohkawara J.I., et al. Culture system for extensive production of CD19+IgM+ cells by human cord blood CD34+ progenitors. Leukemia. 1998;12:764–771. doi: 10.1038/sj.leu.2401004. [DOI] [PubMed] [Google Scholar]
- 63.Cooper M.D., Mulvaney D., Coutinho A., Cazenave P.-A. A novel cell surface molecule on early B-lineage cells. Nature. 1986;321:616–618. doi: 10.1038/321616a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.