Abstract
Background
Short-read high-throughput DNA sequencing technologies provide new tools to answer biological questions. However, high cost and low throughput limit their widespread use, particularly in organisms with smaller genomes such as S. cerevisiae. Although ChIP-Seq in mammalian cell lines is replacing array-based ChIP-chip as the standard for transcription factor binding studies, ChIP-Seq in yeast is still underutilized compared to ChIP-chip. We developed a multiplex barcoding system that allows simultaneous sequencing and analysis of multiple samples using Illumina's platform. We applied this method to analyze the chromosomal distributions of three yeast DNA binding proteins (Ste12, Cse4 and RNA PolII) and a reference sample (input DNA) in a single experiment and demonstrate its utility for rapid and accurate results at reduced costs.
Results
We developed a barcoding ChIP-Seq method for the concurrent analysis of transcription factor binding sites in yeast. Our multiplex strategy generated high quality data that was indistinguishable from data obtained with non-barcoded libraries. None of the barcoded adapters induced differences relative to a non-barcoded adapter when applied to the same DNA sample. We used this method to map the binding sites for Cse4, Ste12 and Pol II throughout the yeast genome and we found 148 binding targets for Cse4, 823 targets for Ste12 and 2508 targets for PolII. Cse4 was strongly bound to all yeast centromeres as expected and the remaining non-centromeric targets correspond to highly expressed genes in rich media. The presence of Cse4 non-centromeric binding sites was not reported previously.
Conclusion
We designed a multiplex short-read DNA sequencing method to perform efficient ChIP-Seq in yeast and other small genome model organisms. This method produces accurate results with higher throughput and reduced cost. Given constant improvements in high-throughput sequencing technologies, increasing multiplexing will be possible to further decrease costs per sample and to accelerate the completion of large consortium projects such as modENCODE.
Background
Novel high-throughput DNA sequencing technologies have allowed the generation of millions of short reads and have empowered a wide variety of studies such as genome-wide analysis of transcriptomes (RNA-Seq) [1-4], transcription factor binding sites (ChIP-Seq) [5,6] and whole-genome sequencing and analysis [7,8]. However, these studies have often been limited by a high cost per sample and low throughput. A typical sequencing run on an Illumina Genome Analyzer II currently costs about $500 in reagents per flowcell lane and requires ~4 days to complete both the sequencing and Illumina analysis pipeline phases. Moreover, the number of mapped reads (up to 10 M per lane) is often significantly higher than required for the experiment, especially for organisms of small genome sizes such as yeasts, worms and flies.
Multiplex DNA sequencing has been pursued since the beginning of Sanger sequencing [9] and has been applied to Roche's 454 platform [10]. Here we describe a multiplexing strategy for Illumina sequencing to process multiple DNA samples simultaneously. The strategy was applied to analyze the targets of three yeast DNA binding proteins (Cse4, Ste12 and RNA polymerase II) using chromatin immunoprecipitation (ChIP) and was shown to yield accurate and high quality results. We also included a reference sample for ChIP-Seq termed input DNA.
ChIP followed by high-throughput sequencing (ChIP-Seq) has been developed to map the protein-DNA interactions at a genome-wide level [5,6]. It allows characterization of transcription factor binding and other DNA-binding proteins during development [5,11], under different environmental conditions [6,12] or in different cell types or tissues. ChIP-Seq has also been used to study the epigenome by mapping the distribution of histone modifications and chromatin-modifying complexes [12-14]. Combination of multiple ChIP-Seq experiments can help to determine transcriptional networks [15].
Cse4 is a centromeric variant of histone H3 [16] and its human homolog is the centromeric protein A (CENP-A) [17]. Yeast centromeres span 126 base pairs and are divided in three centromeric DNA elements (CDEI, CDEII and CDEIII); Cse4 binds CDEII [18]. Cse4 participates in the formation of a specialized hexameric nucleosome with histone H4 and Scm3 that diverges from the standard H2A-H2B-H3-H4 octamer [19-22]. The kinetochore assembles at the centromere and Cse4 is required for normal kinetochore assembly and function [23-26]. Cse4 mutants display strong chromosome missegregation due to incorrect kinetochore structure [25,27,28].
Ste12 is a transcription factor that regulates two sets of genes: those involved in invasive growth (pseudohyphal growth) and those implicated in the mating response (pheromone stimulation) [29-31]. Pseudohyphal growth is a polarized invasion of media by S. cerevisiae upon nitrogen starvation. It integrates signals from a MAP kinase cascade and the cAMP-dependant pathway [30,32-35]. During pseudohyphal growth, Ste12 associates as a dimer with Tec1 on transcription factor binding sites (TFBS) upstream of invasive genes [33,36-38]. During the mating response, Tec1 is phosphorylated by Fus3 upon pheromone stimulation, leading to its degradation and the binding of Ste12 on the pheromone response elements [39-41]. Thus, the dual role of Ste12 depends mainly on different phosphorylation events [42-46].
RNA polymerase II (PolII) transcribes most protein-coding genes and is conserved among metazoans. It is recruited to a particular transcription start site depending on the chromatin structure and the presence of preinitiation complexes and transcription factors. In budding yeast, the PolII holoenzyme consists of 12 subunits (Rpb1-12) [47]. Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12 are shared with the two other major RNA polymerases and Rpb5 contacts DNA to promote transcriptional activation [48,49]. PolII directly interacts with the mediator complex, histones, transcription factors, elongation factors and many other proteins [50]. Although yeast RNA polymerase III genome-wide distribution has been studied extensively using microarrays [51-53], only one similar study has been performed to characterize PolII distribution across the yeast genome [54].
Using multiplex ChIP-Seq, we mapped the targets of Cse4, Ste12 and PolII across the yeast genome. We found a large number of binding sites for each factor. Cse4 localized predominantly to centromeres and we also observed other Cse4 binding sites with lower signal intensity elsewhere in the genome. Ste12 and PolII binding sites were found in close proximity to genes.
Results and discussion
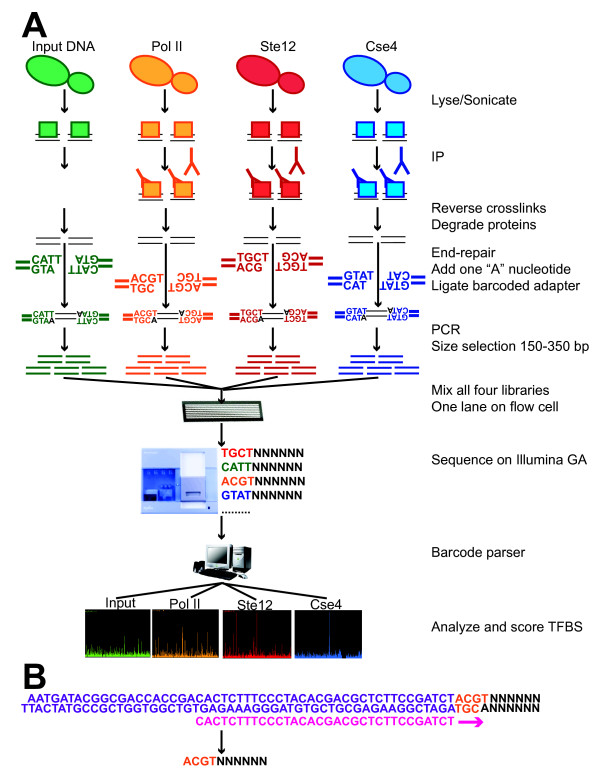
The barcoded ChIP-Seq workflow is illustrated in Figure 1. We first generated standard Illumina genomic DNA adapter sequences augmented with one of four barcodes (ACGT, CATT, GTAT and TGCT; Table 1). The first three bases uniquely tag or "barcode" a given sample preparation and are separated by a Hamming distance [55] of three which will prevent one barcode being miscalled as another barcode when allowing for one- or two-base sequencing errors. The final 'T' at the fourth base anneals with the 'A' overhang from the end-repaired DNA sample for ligation of ChIP or other DNA fragments. Illumina PCR primers for genomic DNA are used without any changes after adapter ligation to generate the sequencing libraries. Four libraries are pooled together in equimolar ratios and sequenced in a single lane. After sequencing, four distinct sequencing profiles are obtained by parsing the barcodes into distinct bins for subsequent analyses.
Figure 1.
Scheme for yeast barcoded ChIP-Seq. (a) Barcoded ChIP-Seq workflow. Ovals depict yeast cells and squares depict proteins. An aliquot of sheared cell lysate is not immunoprecipitated but is otherwise processed normally (green). This DNA, termed input DNA, is a reference sample for ChIP-Seq. Illumina DNA libraries are generated from both ChIP and input DNA samples. In multiplex ChIP-Seq, a barcoded adapter is ligated to an individual DNA sample. The barcode has 3 unique bases followed by a 'T' to anneal with the end-repaired DNA. Four libraries are then pooled together and applied to a single flowcell lane. After sequencing on the Genome Analyzer, reads are separated according to the first four bases and aligned to the yeast genome. Reads are stacked to generate a signal profile and scored against a pool of input DNA to determine significant transcription factor binding sites. (b) The barcode (orange) is located between Illumina adapter sequences (purple) and ChIP or input DNA inserts (black). The sequencing primer (pink) anneals to the adapter sequences and short reads start with the four bases of the barcode (orange) followed by DNA inserts (black). For the sequencing primer and Illumina adapter, oligonucleotide sequences were given by the manufacturer © 2006 Illumina, Inc. All rights reserved.
Table 1.
Oligonucleotide sequences for generation of the four barcoded adapters
| Oligo name | Barcode | Modification | Sequence (5'→3') |
| MPLEXA1F | GTAT | None | ACACTCTTTCCCTACACGACGCTCTTCCGATCTGTAT |
| MPLEXA1R | GTAT | 5' phosphate | TACAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| MPLEXA6F | CATT | None | ACACTCTTTCCCTACACGACGCTCTTCCGATCTCATT |
| MPLEXA6R | CATT | 5' phosphate | ATGAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| MPLEXA8F | ACGT | None | ACACTCTTTCCCTACACGACGCTCTTCCGATCTACGT |
| MPLEXA8R | ACGT | 5' phosphate | CGTAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
| MPLEXA9F | TGCT | None | ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCT |
| MPLEXA9R | TGCT | 5' phosphate | GCAAGATCGGAAGAGCTCGTATGCCGTCTTCTGCTTG |
Similar barcode behavior with the same DNA sample
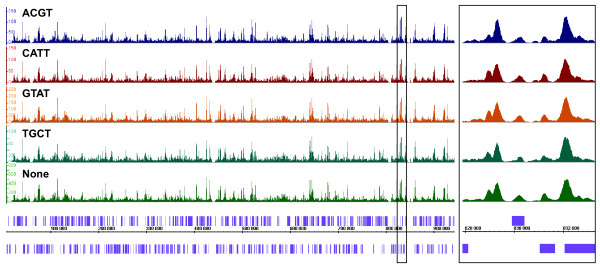
To first test whether barcodes generate similar results when added to the same population of DNA fragments, we divided an input DNA sample into five aliquots. The input sample consists of Saccharomyces cerevisiae DNA prepared from crosslinked and sonicated chromatin without immunoprecipation and it represents breaks at open chromatin regions (R.K. Auerbach, G.M. Euskirchen, unpublished). Four of the samples were differentially barcoded and the libraries were mixed together in the same flowcell lane. As a control, standard non-barcoded Illumina genomic DNA adapters were added to the fifth aliquot and this library was sequenced in another lane. After sorting reads by barcodes, the barcode sequences were stripped and 26 base pairs were aligned back to the yeast genome. Similar numbers of reads were obtained for each barcoded library. As shown in Figure 2, signal tracks are very similar for the five aliquots. The normalized average tag counts in 500 bp windows are also highly comparable among aliquots (Table 2, R2 = 0.93–0.97). The results for each barcoded library were scored against the control without a barcode to find significant differences; only 36,992 out of 12,156,677 unique bps (0.304%) from the S. cerevisiae genome differ significantly for all four barcoded libraries (Figure 2; See Methods for analysis). In addition, comparison of peaks among the different libraries revealed high degrees of similarity. These characteristics highly resemble those of a non-barcoded input library analyzed in two different sequencing reactions. Thus, overall biases are not observed in efficiencies of various barcoded libraries and barcoding does not appear to induce differences relative to non-barcoded libraries.
Figure 2.
Comparison of input DNA signal tracks among all four barcoded adapters relative to standard Illumina adapters. An input sample was split in five aliquots. Four were barcoded differentially (top four lanes) and one had non-barcoded, Illumina adapters (fifth lane, labeled 'None'). Barcoded inputs were scored against non-barcoded input. IGB signal tracks of yeast chromosome 16 are shown for each sample, with ORF locations on the x-axis. ORFs are depicted in purple. On the y-axis, a normalized scale represents the number of read counts at a particular location. Each scale is normalized according to the number of mapped reads (Table 10). A box in the left panel depicts the enlarged section shown in the right panel for positions between 828,000 and 833,000 to demonstrate the overlap among all signal tracks.
Table 2.
Comparisons of the average tag count in 500 bp windows across the yeast genome among different samples
| Experiment A | Experiment B | ||||
| Experiment # | Experiment ID | Experiment # | Experiment ID | R2 | p-value |
| 10 | Input_NB | 14 | Input_NB | 0.9455 | < 2.2e-16 |
| 14 | Input_NB | 15 | Input_ACGT | 0.9556 | < 2.2e-16 |
| 14 | Input_NB | 16 | Input_CATT | 0.9473 | < 2.2e-16 |
| 14 | Input_NB | 17 | Input_GTAT | 0.9685 | < 2.2e-16 |
| 14 | Input_NB | 18 | Input_TGCT | 0.9325 | < 2.2e-16 |
| 15 | Input_ACGT | 16 | Input_CATT | 0.9422 | < 2.2e-16 |
| 17 | Input_GTAT | 18 | Input_TGCT | 0.9304 | < 2.2e-16 |
| 1 | PolII_Rep1_ACGT | 8 | PolII_Rep2_TGCT | 0.8904 | < 2.2e-16 |
| 1 | PolII_Rep1_ACGT | 9 | PolII_Rep3_ACGT | 0.9104 | < 2.2e-16 |
| 1 | PolII_Rep1_ACGT | 13 | PolII_Rep4_NB | 0.9040 | < 2.2e-16 |
| 8 | PolII_Rep2_TGCT | 9 | PolII_Rep3_ACGT | 0.9288 | < 2.2e-16 |
| 8 | PolII_Rep2_TGCT | 13 | PolII_Rep4_NB | 0.9464 | < 2.2e-16 |
| 9 | PolII_Rep3_ACGT | 13 | PolII_Rep4_NB | 0.9302 | < 2.2e-16 |
| 4 | Ste12_Rep1_TGCT | 7 | Ste12_Rep2_GTAT | 0.9655 | < 2.2e-16 |
| 4 | Ste12_Rep1_TGCT | 12 | Ste12_Rep3_NB | 0.9058 | < 2.2e-16 |
| 7 | Ste12_Rep2_GTAT | 12 | Ste12_Rep3_NB | 0.8963 | < 2.2e-16 |
Correlation coefficients on the average tag count in 500 bp bins between two experiments among barcoded and non-barcoded input DNA samples from the section "Similar barcode behavior with the same DNA sample" (Comparison 1–7), among barcoded and non-barcoded PolII replicates (Comparison 8–13) and among barcoded and non-barcoded Ste12 replicates (Comparison 14–16). p-values are inferior to 2.2e-16, the lowest p-value that R can calculate. See Methods for details of the analysis.
Experimental design for multiplex yeast ChIP-Seq
We next analyzed the distributions of three diverse DNA-binding proteins across the yeast S. cerevisiae genome using three to four biological replicates per factor. This study allowed us to 1) further examine the reproducibility of a specified barcode using a different biological replicate 2) determine whether there was crossover signal between barcodes and 3) use the technique to map the binding distributions of three DNA-binding proteins. The proteins selected were the centromeric histone H3 variant Cse4 [16], RNA PolII (each grown in vegetative growth) and the transcription factor Ste12 [31] (incubated in pseudohyphal growth conditions). Cse4 is expected to have a very distinct binding distribution from that of Ste12 which binds near promoters. We also included input DNA as a reference for ChIP-Seq as a fourth sample. Initially we compared three biological replicates for each factor using barcoded sequences for two of the replicates and non-barcoded sequences for the third one. Each biological replicate was made into its own DNA library. The barcoded libraries were pooled and sequenced with one pool per lane (Table 3) whereas the non-barcoded libraries were sequenced in individual lanes. Two lanes were run using the barcoded adapters: Pol II-ACGT, Input-CATT, Cse4-GTAT and Ste12-TGCT (Pool #1, Table 3) and Input-ACGT, Cse4-CATT, Ste12-GTAT and PolII-TGCT (Pool #2, Table 3). To compare reproducibility of alternative barcodes relative to reproducibility between biological replicates, we sequenced another replicate of Pol II ChIP DNA in a separate pool using the same barcode as one of the other replicates (Pol II-ACGT) (Pool #3, Table 3). After sorting reads by barcodes, the barcodes were stripped and 26 base pairs were aligned to the yeast genome. We scored ChIP samples against 8 million input reads derived equally from barcoded and non-barcoded input DNA. Rank-order comparisons between the top targets for two different biological replicates were performed to assess similarity. We obtained 148 targets for Cse4, 823 targets for Ste12 and 2508 targets for PolII (FDR = 0.05; Additional Files 1, 2, 3).
Table 3.
DNA libraries pooled for multiplex sequencing
| Pool | ACGT | CATT | GTAT | TGCT |
| 1 | PolII_Rep1 | Input_CATT | Cse4_Rep1 | Ste12_Rep1 |
| 2 | Input_ACGT | Cse4_Rep2 | Ste12_Rep2 | PolII_Rep2 |
| 3 | PolII_Rep3 | Input_CATT | Cse4_Rep1 | Ste12_Rep1 |
Comparison of PolII biological replicates
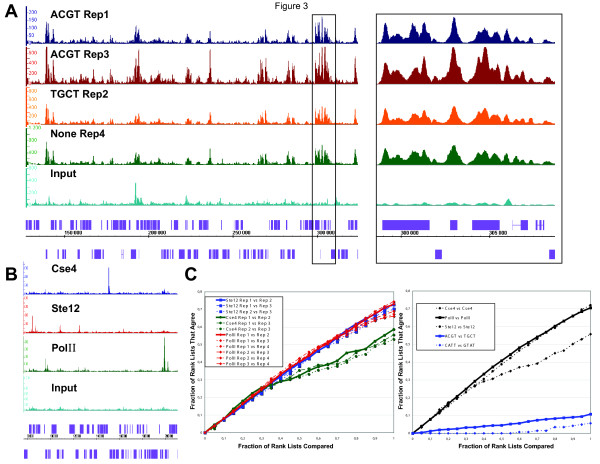
By comparing two biological replicates of PolII ChIP DNA with the same barcode (ACGT) we found a close correlation between the target lists (Figure 3a and 3c). A similar close correlation was observed by comparing the targets of the individual biological replicates using different barcodes among each of the different factors (Figure 3c). Furthermore, comparison of normalized average tag counts in 500 bp windows between biological replicates for PolII and Ste12 revealed strong correlations (Table 2, R2 = 0.89–0.97). These values are similar to the correlation coefficients between biological replicates found in a recent yeast RNA-Seq study (R2 = 0.87–0.90) [1]. Thus, consistent with barcoded input DNA, the different barcodes give similar results when analyzing similar libraries. Our results strongly suggest that the barcode has negligible impact on binding site detection.
Figure 3.
Barcoded adapters perform similarly to standard Illumina adapters and do not crossover to other samples in the same lane. (a) RNA PolII binding profiles from different biological replicates with the same barcode (PolII_Rep1, dark blue; PolII_Rep3, red), with different barcodes (PolII_Rep2, orange) or without barcode (PolII_Rep4, green) strongly overlap. See also Table 3. Input DNA serves as a reference (light blue). IGB signal tracks of chromosome 5 between 130,000 and 320,000 are shown for each library. A box in the left panel depicts the enlarged section shown in the right panel between positions 298,000 and 309,000 to illustrate the overlap among all PolII signal tracks. (b) Binding profiles from four different libraries pooled and sequenced in the same flowcell lane show very little resemblance. Shown here are the binding profiles for Cse4_Rep2 (dark blue), Ste12_Rep2 (red), PolII_Rep2 (green) and Input_ACGT (light blue). IGB signal tracks of chromosome 12 between 80,000 and 210,000 are shown for each sample. For (a) and (b), axis and scale normalizations are similar to Figure 2. (c) Left: Rank-rank comparison of target lists between all pairwise barcoded replicates for Cse4, PolII and Ste12. The horizontal axis shows the fraction of the two lists being compared and the vertical axis shows the fraction of those targets that agree between a given pair of target lists. All comparisons show strong agreement, although the rank lists for Cse4 differ more than PolII or Ste12 for the second half of their length. Right: Rank-rank comparison between barcoded replicates from the same factors (averaged over all pairwise comparisons) compared to rank-rank comparisons for barcoded replicates between different factors: PolII_Rep1 (ACGT) vs. Ste12_Rep1 (TGCT) and Cse4_Rep2 (CATT) vs. Ste12_Rep2 (GTAT).
Comparison of ChIP-Seq data to previously published ChIP-chip data
We also compared the binding site distributions for Ste12, Cse4 and PolII to previously published data [16,36,37,54,56]. Our Ste12 ChIP-Seq targets overlap with 67% (232/346) of the targets found in Borneman et al. [37] while our PolII ChIP-Seq targets overlap with 72% (667/929) of the PolII ChIP-chip targets from the Steinmetz et al. data [54]. These overlaps between ChIP-Seq and ChIP-chip do not differ significantly from those determined in other studies [6], typically in the 64–71% range. However, we observe 2- to 4-fold more targets using ChIP-Seq as compared to ChIP-chip, consistent with the observation that ChIP-Seq is more sensitive than ChIP-chip [6].
Non-overlapping binding profiles among samples sequenced simultaneously
By comparing binding profiles obtained from a single lane containing four different barcoded libraries, we observed that many targets belong uniquely to a single signal track and their distributions are consistent with the biological roles of the factors studied (Figure 3b and 3c). This is particularly the case for Ste12 and Cse4 which have highly dissimilar binding patterns and almost no target overlap (Figure 3c). Note that some peaks are present in both ChIP and input samples and are removed after scoring. Thus, analysis of different ChIP samples with distinct barcodes indicates that the signal from one barcoded library does not crossover to those of other barcoded libraries (Figure 3b). We define signal crossover as the erroneous assignment of a barcoded read from a particular ChIP sample to the wrong ChIP sample during multiplex ChIP-Seq, generating a signal profile for one barcoded sample with reads from other ChIPs sequenced simultaneously.
Ste12, PolII and Cse4 binding distributions
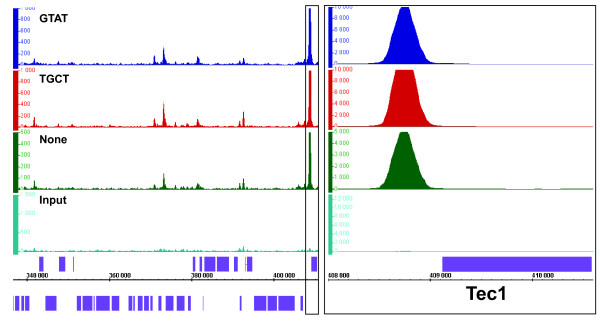
Analysis of the binding targets of Ste12, Pol II and Cse2 reveals expected and novel results. Ste12p binds upstream of genes implicated in pseudohyphal growth such as Tec1, Mga1, Phd1 and Flo8 (Figure 4 and [31,36]). As expected, Ste12 targets are enriched for GO categories including pseudohyphal growth, cell wall organization, and stress response (Table 4 and Additional File 5), which is consistent with the role of Ste12p in regulating the pseudohyphal pathway. The PolII distribution in mid-log phase intersects ~2500 genes, which is fewer than that reported by recent RNA-Seq studies [1]. This may be due to differences in the analysis of ChIP-Seq and RNA-Seq data. Enriched regions in ChIP-Seq data are determined using a threshold for a minimal read count difference at a particular region between the ChIP sample and input DNA while RNA-Seq data is analyzed using a global threshold for a minimal number of mapped reads. Overall, we recapitulated all the binding profiles displayed in the genome-wide PolII ChIP-chip study by Steinmetz et al. (Figure 5) [54]. As expected, PolII targets are overrepresented in GO categories such as translation, transcription, RNA processing and primary metabolism, which are crucial for exponential growth in rich media (Table 4 and Additional File 5). Cse4p binds strongly to all yeast 16 centromeres as shown previously (Figure 6 and[16]). Our data indicate that Cse4 tightly occupies a narrow region around the centromere. In addition, we also detected significant binding relative to background at 132 novel non-centromeric sites; Cse4p is less abundant at these sites relative to centromeric regions (Additional File 1). These non-centromeric sites are enriched for metabolic genes, in particular those involved in glucose metabolism (e.g. glycolysis, hexose and glucose catabolism), which are highly expressed under our conditions (Table 4 and Additional File 5). In addition, nearly all non-centromeric Cse4 target genes are present among the 100 highest ranked PolII targets. To explain this novel finding, we hypothesize that Cse4 transiently integrates at regions of high histone turnover which could now be detected using ChIP-Seq. Cse4p might temporarily integrate at these non-centromeric sites before being outcompeted by histone H3 since it has been demonstrated by ChIP-qPCR that a Cse4 mutant protein can localize to non-centromeric locations when the stoichiometry between histone H3 and Cse4p is perturbed [27].
Figure 4.
Ste12 distribution during pseudohyphal growth is similar across three different biological replicates. Two barcoded replicates (Ste12_Rep2, dark blue; Ste12_Rep1, red) and a non-barcoded replicate (Ste12_Rep3, green) were compared to input DNA (light blue). Ste12 ChIP samples were scored against a pool of input DNA. IGB signal tracks of chromosome 2 between 340,000 and 410,000 are shown for each sample. Axis and scale normalizations are similar to Figure 2. A box in the left panel containing the TEC1 gene and its surrounding intergenic region was enlarged in panel B and rescaled to emphasize the strong signal at the TEC1 promoter. The same normalization as in Figure 2 was applied. Ste12p and Tec1p act as a dimer during pseudohyphal growth [31].
Table 4.
GO process analysis of gene targets for Cse4, Ste12 and PolII. For a complete GO analysis, please refer to Additional File 5.
| GO term | Cluster frequency | Background frequency | p-value |
| GO process categories for Cse4 | |||
| glycolysis | 8 out of 132 genes, 6.1% | 22 out of 7158 background genes, 0.3% | 8.32e-07 |
| glucose catabolic process | 8 out of 132 genes, 6.1% | 32 out of 7158 background genes, 0.4% | 2.34e-05 |
| alcohol metabolic process | 16 out of 132 genes, 12.1% | 179 out of 7158 background genes, 2.5% | 4.49e-05 |
| hexose catabolic process | 8 out of 132 genes, 6.1% | 37 out of 7158 background genes, 0.5% | 7.96e-05 |
| cellular biosynthetic process | 57 out of 132 genes, 43.2% | 1689 out of 7158 background genes, 23.6% | 0.00013 |
| monosaccharide catabolic process | 8 out of 132 genes, 6.1% | 42 out of 7158 background genes, 0.6% | 0.00023 |
| alcohol catabolic process | 8 out of 132 genes, 6.1% | 45 out of 7158 background genes, 0.6% | 0.00039 |
| pyruvate metabolic process | 7 out of 132 genes, 5.3% | 39 out of 7158 background genes, 0.5% | 0.00174 |
| hexose metabolic process | 9 out of 132 genes, 6.8% | 78 out of 7158 background genes, 1.1% | 0.00349 |
| biosynthetic process | 58 out of 132 genes, 43.9% | 1929 out of 7158 background genes, 26.9% | 0.00496 |
| hexose biosynthetic process | 6 out of 132 genes, 4.5% | 31 out of 7158 background genes, 0.4% | 0.00529 |
| glucose metabolic process | 8 out of 132 genes, 6.1% | 64 out of 7158 background genes, 0.9% | 0.00601 |
| GO process categories for Ste12 | |||
| biological regulation | 193 out of 823 genes, 23.5% | 1132 out of 7158 background genes, 15.8% | 7.46e-07 |
| regulation of cellular process | 158 out of 823 genes, 19.2% | 889 out of 7158 background genes, 12.4% | 2.02e-06 |
| regulation of biological process | 160 out of 823 genes, 19.4% | 904 out of 7158 background genes, 12.6% | 2.13e-06 |
| monosaccharide transport | 15 out of 823 genes, 1.8% | 24 out of 7158 background genes, 0.3% | 3.48e-06 |
| hexose transport | 15 out of 823 genes, 1.8% | 24 out of 7158 background genes, 0.3% | 3.48e-06 |
| multi-organism process | 40 out of 823 genes, 4.9% | 136 out of 7158 background genes, 1.9% | 9.27e-06 |
| response to pheromone | 32 out of 823 genes, 3.9% | 96 out of 7158 background genes, 1.3% | 1.03e-05 |
| response to chemical stimulus | 86 out of 823 genes, 10.4% | 412 out of 7158 background genes, 5.8% | 1.18e-05 |
| response to stimulus | 144 out of 823 genes, 17.5% | 818 out of 7158 background genes, 11.4% | 2.41e-05 |
| cellular carbohydrate metabolic process | 47 out of 823 genes, 5.7% | 187 out of 7158 background genes, 2.6% | 0.00011 |
| growth | 40 out of 823 genes, 4.9% | 148 out of 7158 background genes, 2.1% | 0.00012 |
| regulation of transcription from RNA Pol II | 56 out of 823 genes, 6.8% | 243 out of 7158 background genes, 3.4% | 0.00017 |
| regulation of cell size | 34 out of 823 genes, 4.1% | 117 out of 7158 background genes, 1.6% | 0.00017 |
| sexual reproduction | 34 out of 823 genes, 4.1% | 120 out of 7158 background genes, 1.7% | 0.00033 |
| conjugation | 34 out of 823 genes, 4.1% | 120 out of 7158 background genes, 1.7% | 0.00033 |
| conjugation with cellular fusion | 34 out of 823 genes, 4.1% | 120 out of 7158 background genes, 1.7% | 0.00033 |
| response to abiotic stimulus | 32 out of 823 genes, 3.9% | 111 out of 7158 background genes, 1.6% | 0.00047 |
| cellular structure morphogenesis | 40 out of 823 genes, 4.9% | 156 out of 7158 background genes, 2.2% | 0.00057 |
| anatomical structure development | 40 out of 823 genes, 4.9% | 156 out of 7158 background genes, 2.2% | 0.00057 |
| cell morphogenesis | 40 out of 823 genes, 4.9% | 156 out of 7158 background genes, 2.2% | 0.00057 |
| anatomical structure morphogenesis | 40 out of 823 genes, 4.9% | 156 out of 7158 background genes, 2.2% | 0.00057 |
| signal transduction | 51 out of 823 genes, 6.2% | 222 out of 7158 background genes, 3.1% | 0.00067 |
| carbohydrate metabolic process | 49 out of 823 genes, 6.0% | 210 out of 7158 background genes, 2.9% | 0.00068 |
| external encapsulating structure organization | 48 out of 823 genes, 5.8% | 206 out of 7158 background genes, 2.9% | 0.00093 |
| cell wall organization and biogenesis | 48 out of 823 genes, 5.8% | 206 out of 7158 background genes, 2.9% | 0.00093 |
| carboxylic acid metabolic process | 67 out of 823 genes, 8.1% | 332 out of 7158 background genes, 4.6% | 0.00195 |
| organic acid metabolic process | 67 out of 823 genes, 8.1% | 332 out of 7158 background genes, 4.6% | 0.00195 |
| carbohydrate transport | 15 out of 823 genes, 1.8% | 35 out of 7158 background genes, 0.5% | 0.00254 |
| cell growth | 26 out of 823 genes, 3.2% | 87 out of 7158 background genes, 1.2% | 0.00268 |
| reproduction | 65 out of 823 genes, 7.9% | 323 out of 7158 background genes, 4.5% | 0.00309 |
| hexose metabolic process | 24 out of 823 genes, 2.9% | 78 out of 7158 background genes, 1.1% | 0.00367 |
| glucose metabolic process | 21 out of 823 genes, 2.6% | 64 out of 7158 background genes, 0.9% | 0.00478 |
| pseudohyphal growth | 21 out of 823 genes, 2.6% | 65 out of 7158 background genes, 0.9% | 0.00631 |
| cell communication | 56 out of 823 genes, 6.8% | 271 out of 7158 background genes, 3.8% | 0.00665 |
| nitrogen compound metabolic process | 51 out of 823 genes, 6.2% | 241 out of 7158 background genes, 3.4% | 0.00860 |
| establishment and maintenance of cell polarity | 32 out of 823 genes, 3.9% | 126 out of 7158 background genes, 1.8% | 0.00953 |
| Top 40 GO process categories for PolII | |||
| cellular process | 1864 out of 2508 genes, 74.3% | 4710 out of 7158 background genes, 65.8% | 8.82e-27 |
| ribonucleoprotein complex biogenesis | 289 out of 2508 genes, 11.5% | 494 out of 7158 background genes, 6.9% | 3.36e-25 |
| ribosome biogenesis | 246 out of 2508 genes, 9.8% | 407 out of 7158 background genes, 5.7% | 7.01e-24 |
| cellular component organization and biogenesis | 785 out of 2508 genes, 31.3% | 1725 out of 7158 background genes, 24.1% | 4.02e-22 |
| organelle organization and biogenesis | 621 out of 2508 genes, 24.8% | 1331 out of 7158 background genes, 18.6% | 2.59e-19 |
| primary metabolic process | 1276 out of 2508 genes, 50.9% | 3187 out of 7158 background genes, 44.5% | 1.59e-12 |
| maturation of SSU-rRNA from tricistronic rRNA | 49 out of 2508 genes, 2.0% | 60 out of 7158 background genes, 0.8% | 1.86e-10 |
| RNA metabolic process | 486 out of 2508 genes, 19.4% | 1082 out of 7158 background genes, 15.1% | 2.71e-10 |
| nucleotide and nucleic acid metabolic process | 649 out of 2508 genes, 25.9% | 1512 out of 7158 background genes, 21.1% | 6.60e-10 |
| maturation of SSU-rRNA | 49 out of 2508 genes, 2.0% | 62 out of 7158 background genes, 0.9% | 1.97e-09 |
| rRNA metabolic process | 143 out of 2508 genes, 5.7% | 257 out of 7158 background genes, 3.6% | 7.39e-09 |
| ribosomal large subunit biogenesis | 50 out of 2508 genes, 2.0% | 65 out of 7158 background genes, 0.9% | 7.46e-09 |
| ribonucleoprotein complex assembly | 96 out of 2508 genes, 3.8% | 156 out of 7158 background genes, 2.2% | 1.23e-08 |
| macromolecule metabolic process | 1096 out of 2508 genes, 43.7% | 2752 out of 7158 background genes, 38.4% | 1.71e-08 |
| biopolymer metabolic process | 1033 out of 2508 genes, 41.2% | 2580 out of 7158 background genes, 36.0% | 2.55e-08 |
| ribosomal subunit assembly | 44 out of 2508 genes, 1.8% | 56 out of 7158 background genes, 0.8% | 4.08e-08 |
| cellular localization | 303 out of 2508 genes, 12.1% | 644 out of 7158 background genes, 9.0% | 4.38e-08 |
| ribosome assembly | 49 out of 2508 genes, 2.0% | 65 out of 7158 background genes, 0.9% | 4.46e-08 |
| cellular metabolic process | 1340 out of 2508 genes, 53.4% | 3448 out of 7158 background genes, 48.2% | 4.55e-08 |
| metabolic process | 1356 out of 2508 genes, 54.1% | 3496 out of 7158 background genes, 48.8% | 6.00e-08 |
| rRNA processing | 137 out of 2508 genes, 5.5% | 250 out of 7158 background genes, 3.5% | 9.13e-08 |
| ncRNA processing | 172 out of 2508 genes, 6.9% | 335 out of 7158 background genes, 4.7% | 3.92e-07 |
| cellular macromolecular complex organization | 184 out of 2508 genes, 7.3% | 364 out of 7158 background genes, 5.1% | 4.79e-07 |
| establishment of localization in cell | 278 out of 2508 genes, 11.1% | 594 out of 7158 background genes, 8.3% | 6.51e-07 |
| RNA processing | 234 out of 2508 genes, 9.3% | 487 out of 7158 background genes, 6.8% | 9.22e-07 |
| gene expression | 704 out of 2508 genes, 28.1% | 1708 out of 7158 background genes, 23.9% | 9.27e-07 |
| intracellular transport | 260 out of 2508 genes, 10.4% | 551 out of 7158 background genes, 7.7% | 9.36e-07 |
| regulation of translation | 38 out of 2508 genes, 1.5% | 49 out of 7158 background genes, 0.7% | 1.88e-06 |
| maturation of 5.8S rRNA | 37 out of 2508 genes, 1.5% | 48 out of 7158 background genes, 0.7% | 4.22e-06 |
| maturation of 5.8S rRNA from tricistronic rRNA | 37 out of 2508 genes, 1.5% | 48 out of 7158 background genes, 0.7% | 4.22e-06 |
| posttranscriptional regulation of gene expression | 44 out of 2508 genes, 1.8% | 61 out of 7158 background genes, 0.9% | 4.99e-06 |
| macromolecular complex subunit organization | 197 out of 2508 genes, 7.9% | 404 out of 7158 background genes, 5.6% | 5.30e-06 |
| nuclear export | 64 out of 2508 genes, 2.6% | 102 out of 7158 background genes, 1.4% | 1.28e-05 |
| localization | 448 out of 2508 genes, 17.9% | 1046 out of 7158 background genes, 14.6% | 1.35e-05 |
| nuclear transport | 76 out of 2508 genes, 3.0% | 128 out of 7158 background genes, 1.8% | 1.86e-05 |
| nucleocytoplasmic transport | 76 out of 2508 genes, 3.0% | 128 out of 7158 background genes, 1.8% | 1.86e-05 |
| cleavages during rRNA processing | 29 out of 2508 genes, 1.2% | 36 out of 7158 background genes, 0.5% | 3.89e-05 |
| RNA 5'-end processing | 26 out of 2508 genes, 1.0% | 31 out of 7158 background genes, 0.4% | 4.30e-05 |
| transcription from RNA polymerase I promoter | 30 out of 2508 genes, 1.2% | 38 out of 7158 background genes, 0.5% | 5.25e-05 |
| regulation of cellular biosynthetic process | 46 out of 2508 genes, 1.8% | 68 out of 7158 background genes, 0.9% | 5.66e-05 |
GO process analysis (p < 0.01) for Cse4 combined targets, for Ste12 combined targets and for PolII combined targets.
For a complete GO analysis, please refer to Additional File 5.
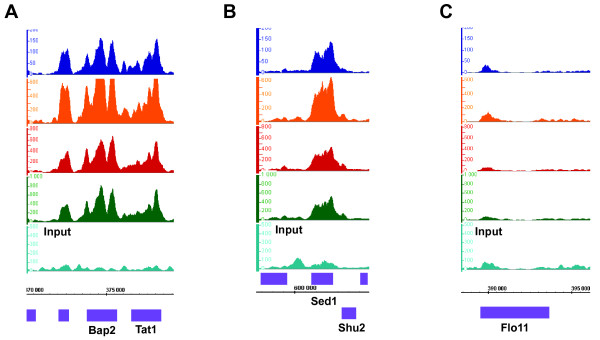
Figure 5.
PolII signal profiles recapitulate findings from Steinmetz et al. PolII ChIP-Seq signal profiles resemble very closely to those published in Figure 3 of Steinmetz et al [54]. We obtained consistent binding at the Bap2-Tat1 loci (a) and at the Sed1-Shu2 loci (b). As expected, we did not observe binding at the Flo11 locus (c). For PolII ChIP-Seq experiments, two biological replicates were barcoded with ACGT (PolII_Rep1, dark blue; PolII_Rep2, orange), one was barcoded with TGCT (PolII_Rep3, red) and a fourth replicate had non-barcoded adapters (PolII_Rep4, green). Input DNA serves as a reference (light blue). Axis and scale normalizations are similar to Figure 2. ORFs above the coordinates axis are on the Watson strand while ORFs below this axis are on the Crick strand.
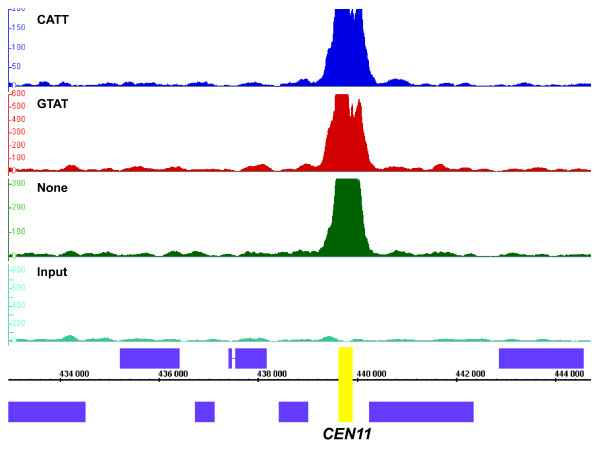
Figure 6.
Cse4p is found robustly at centromeres. All biological replicates were strongly and tightly bound to centromeres, as it is depicted here in the case of CEN11. Two barcoded replicates (Cse4_Rep2, dark blue; Cse4_Rep1, red) and a non-barcoded replicate (Cse4_Rep3, green) were compared to input DNA (light blue). Cse4 ChIP samples were scored against a pool of input DNA. IGB signal tracks of the CEN11 on chromosome 11 are shown for each sample. CEN11 is highlighted in a yellow box. Axis and scale normalizations are similar to Figure 2.
Artifacts absent in barcoded libraries but present in non-barcoded libraries
Finally, we did observe one set of differences between barcoded and non-barcoded libraries. A curious rare artifact was found in non-barcoded libraries and excluded in barcoded libraries. ~31 abnormally-shaped peaks were present in non-barcoded ChIP libraries in which the same sequences was obtained many times resulting in block-shaped peaks (Additional File 4). This artifact was not observed in barcoded libraries.
Although the source of this artifact is not clear, it might be due to specific priming from the non-barcoded adaptor on yeast DNA. Regardless, these artifacts are readily eliminated by removing non-unique reads. We did not find additional areas of dissimilarity between barcoded and non-barcoded libraries.
Conclusion
Our multiplex strategy for ChIP-Seq allows reliable and accurate analysis of multiple samples simultaneously. We processed four times as many samples in the same amount of time and reduced our cost per sample by 65% (Table 5). Barcoded ChIP-Seq will greatly facilitate large-scale studies in organisms with large genomes for which smaller numbers of reads are required and will be particularly valuable for organisms with small genomes such as bacteria, viruses, other yeasts, algae such as Chlamydomonas reinhardtii and plants such as A. thaliana. The relatively small genomes of two model organisms, the worm C. elegans and the fruit fly D. melanogaster, are under intensive characterization by the modENCODE consortium [57]. modENCODE (Model organism ENCyclopedia Of DNA Elements) aims to identify all functional elements, including transcription factor binding sites and transcribed regions, in the genomes of C. elegans and D. melanogaster. Thus the application of barcoded ChIP-Seq to globally map transcription factor binding sites in C. elegans and D. melanogaster will be valuable for cost-effective completion of this project. We estimate for yeast that we would need 260,000, 90,000 and 18,000 mapped reads to saturate transcription factor binding sites, given fold-enrichments of 5×, 10× and 50× respectively (Table 6). With an enrichment of 10-fold, 800,000 mapped reads would be needed for C. elegans, 1,000,000 reads for D. melanogaster and 1,500,000 reads for A. thaliana (Table 7). These numbers can be easily achieved with barcoded ChIP-Seq. Moreover, considering the high degree of ChIP-Seq target overlap between different biological replicates (Figure 3c), we estimate that two biological replicates are sufficient to achieve statistical significance for a particular ChIP-Seq experiment if the biological replicates are very similar. Otherwise, three biological replicates should be used if there is a higher level of divergence among different biological replicates.
Table 5.
Cost per sample and number of mapped reads with increasing multiplexing
| Number of barcoded adapters | Fraction of non-barcoded cost per sample* | Number of mapped reads** |
| 1 | 100% | 10.00 M |
| 4 | 34.61% | 2.50 M |
| 8 | 23.71% | 1.25 M |
| 12 | 20.08% | 0.83 M |
| 16 | 18.26% | 0.63 M |
| 40 | 15.00% | 0.25 M |
*Includes Illumina DNA library preparation (fixed cost), sequencing run
*Excludes ChIP reagents, Illumina GAII sequencer, HR and data storage
*For a non-barcoded sample, the cost per sample is $498.23 USD as of October 2008.
**Assumes optimal mapping values for a lane run on a GA II (10 million mapped reads)
Table 6.
ChIP-Seq simulations of depth of sequencing for S. cerevisiae
| Fold Enrichment | 2× | 5× | 10× | 50× |
| Number of Reads | 2,500,000 | 260,000 | 90,000 | 18,000 |
Number of reads required in order to identify greater than 95% of 600 simulated binding sites distributed evenly over the 12 Mb S. cerevisiae genome with average fold enrichments of 2×, 5×, 10× and 50×. See Methods for details of the simulation.
Table 7.
ChIP-Seq simulations of depth of sequencing for three model organisms
| Fold Enrichment of Simulated Sites | |||
| Genome Size (Mb) | 5× | 10× | |
| C. elegans | 100 | 2,400,000 | 800,000 |
| D. melanogaster | 123 | 3,500,000 | 1,000,000 |
| A. thaliana | 157 | 6,800,000 | 1,500,000 |
Number of reads required in order to identify greater than 95% of 1,000 simulated binding sites distributed over the genomes of C. elegans, D. melanogaster and A. thaliana for average fold enrichments of 5× and 10×. See Methods for details of the simulation.
Our general multiplexing approach is expected to be useful for a variety of other applications including bacterial genome re-sequencing and RNA-Seq; it will also be valuable for other short read DNA sequencing platforms. Using the current technology, genome re-sequencing in yeasts, with a 25× coverage for SNP detection, is not amenable to multiplexing as it would require about 9.50 M mapped reads of length 32 (36 bp minus 4-bp barcode). This is equivalent to one or two Illumina flowcell lanes. RNA-Seq experiments could also be multiplexed in the future although considerable sequencing depth is required to detect at a significant level low-abundance transcripts or rare splice variants. In general, to detect binding sites or transcripts that are present in low abundance by barcoded ChIP-Seq or RNA-Seq, it is important to determine by calculations or computer simulations the minimal number of mapped reads per sample to establish the appropriate level of multiplexing. With the new Illumina Genome Analyzer II, there is an increase in the number of reads that can be obtained (currently ~15 million per lane) and it will be possible to further multiplex ChIP-Seq by creating many other different barcoded adapters, ligating them to individual samples and pooling all of these barcoded libraries in the same flow cell lane. Sequencing reagent costs per sample would further decrease from 65% (4 barcodes) to 76% (8 barcodes), 80% (12 barcodes), 82% (16 barcodes) and 85% (40 barcodes), in comparison to non-barcoded samples (Table 5).
To maintain base balance at any given sequencing cycle, indexing adapters should be designed in increments of four. We prefer to keep the barcodes distinct from one another to avoid crossover due to sequencing errors; for a 3-base barcode this leads to 16 different combinations. To increase multiplexing beyond this figure one can either use barcodes with one base difference and correct for sequencing errors or increase the size of the barcode. Massively barcoded ChIP-Seq, bearing constant improvements in ultra-throughput sequencing technologies, will become widely accessible and is expected to supplant array-based technologies for gene regulation and expression studies in organisms with both small and larger genomes.
Methods
Data accession and supplementary methods
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [58] and are accessible through GEO Series accession number GSE13322 [59]. Sgr files and other data can be accessed at this URL address: http://archive.gersteinlab.org/proj/Lefrancois_et_al_2008/
Cell growth
Yeast strains are described in Table 8. Yeast strains CMY288-1B, CMY8058-3-4 and CMY8082-20-3 were grown in YPAD rich media to exponential mid-log phase (OD600 = 1.0, 500 mL culture). For yeast strains YJM339 and CMY291, cells were grown overnight to mid-log phase to OD600 = 0.6 and then pseudohyphal growth was conducted in nitrogen-depleted SLAD for 4 h as described [36].
Table 8.
Yeast strains used in this study
| Strain ID | Genotype | Parent strain | Source |
| CMY 288-1B | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | BY 4741 | Christopher Yellman, unpublished |
| YJM 339 | MATa/α HO/HO | Clinical isolate | McCusker et al., 1994 |
| CMY 291 | MATa/α HO/HO STE12-13MYC-kanMX6/STE12-13MYC-kanMX6 | YJM 339 | Christopher Yellman, unpublished |
| CMY 8058-3-4 | MATa ade2-1 bar1::loxP can1H cyh2H gal1H his3Δ1 leu2-3,112 trp1-289 ura3-52 | A364a | Christopher Yellman, unpublished |
| CMY 8082-20-3 | MATa ade2-1 bar1::loxP can1H cyh2H gal1H his3Δ1 leu2-3,112 trp1-289 ura3-52 CSE4-3HA-URA3 | A364a | Christopher Yellman, unpublished; Meluh et al., 1998 |
Chromatin immunoprecipation (ChIP)
Chromatin immunoprecipitations were performed as described [60]. All ChIP experiments were completed as biological triplicates for Cse4 and Ste12 and as biological quadruplicates for PolII. For the immunoprecipitations of Ste12 and Cse4, Ste12-13X Myc (160 uL of a 50% anti-Myc EZiew affinity gel; Sigma) and Cse4-3HA (160 uL of a 50% anti-HA EZiew affinity gel; Sigma) were pre-washed three times in lysis/IP buffer containing protease inhibitors and PMSF and added to the lysates from their respective epitope-tagged strains and untagged controls. For the immunoprecipitation of native RNA polymerase II from strain CMY288-1B, 20 μL of mouse ascites containing RNA polymerase II 8WG16 mouse monoclonal antibody (Covance, Cat. #MMS-126R) was added to one set of triplicates. The other set was left without antibody as a control.
Immunoprecipitations were carried out with inverting at 4°C for 14–16 h. For the PolII samples, 250 uL of a 50% Protein G agarose slurry (washed twice in lysis/IP buffer) was added to each of the PolII samples as well as to the no antibody controls, and incubated for 1 h at 4°C with gentle inverting.
Input DNA preparation
To isolate input DNA, 250 uL of clarified CMY288-1B lysate was reserved before immunoprecipitation and combined with 250 μL of 1× TE [pH 8.0]/1% SDS. Crosslinks were reversed by an overnight incubation at 65°C and Proteinase K treatment was carried out as described above. Instead of direct DNA precipitation as for ChIP samples, input DNA was extracted three times using phenol:chloroform:isoamyl alcohol (25:24:1) (Fluka) and once with chloroform alone. To precipitate DNA, 50 μL of 5 M LiCl and 1 mL of 100% ethanol were mixed with the upper phase isolated from the chloroform extraction and precipitation occurred for 1 h at -20°C. Samples were centrifuged for 20 min at 14,000 RPM at 4 °C and were air-dried for 10 min. DNA was resuspended in 30 μL 1× TE [pH 8.0] and input DNA was incubated at 37°C for 30 min with 2 μL of DNase-free RNase A. Finally, input DNA was further purified using a Qiagen MinElute column.
Real-time quantitative polymerase chain reaction
Real time quantitative PCR (qPCR) with SYBR green dye was performed using a Roche LightCycler 480 qPCR machine to confirm enrichment of control regions. A two-fold enrichment between experimental samples (epitope-tagged strains for Ste12 and Cse4, primary antibody for PolII) and reference samples (untagged strains for Ste12 and Cse4, protein G beads only for PolII) for known binding sites was set as the threshold for samples to continue forward towards Illumina library preparation. For each DNA-binding protein studied, four primer pairs were designed and these included at least two primer pairs amplifying known binding sites and at least one primer pair amplifying a random region as a negative control. Primers were generated using Primer3 http://primer3.sourceforge.net/ with the following criteria: fragment size between 200 and 250 bp, primer length 20 and melting temperature between 59°C and 61°C. Other settings were kept as default. Primer sequences are given in Table 9. A standard curve with a dilution series of genomic DNA was generated for each primer pair to determinate primer pair efficiency [61]. Each 10 μL reaction contained 2 μL of nuclease-free water (Gibco), 5 μL of LightCycler 480 SYBR Green I Master mix (Roche, Cat. #04 707 516 001), 0.5 μL of each primer (10 μM stocks) and 2 μL of either water (negative control), diluted genomic DNA (standard curve) or ChIP DNA (1/21 dilution). For ChIP DNA, reactions were done in duplicates on the 384-well plate. Phenol-chloroform extracted genomic DNA was diluted to 8-1, 8-2, 8-3, 8-4, 8-5, 8-6 and 8-7 to generate a dilution series for determination of the standard curve. Each qPCR reaction was run using the same program: pre-incubation (95°C for 5 min), 45 cycles of amplification (95°C for 20 s, 54°C for 30 s, 72°C for s), melting curves using a heat ramp and cool down. Crossing point values (Cp values) were obtained using the second derivative maximal analysis tool included in the Roche LightCycler480 software. To determine enrichment, the 2-ΔΔCp method was used by comparing enrichment values for positive primer pairs to a negative primer pair between experimental and reference ChIP experiments. The negative primer pair should not give differences between experimental samples and control samples. Average enrichment for all biological replicates was determined for each primer pair and standard deviation to the mean was indicated as error bars. Mean enrichments over negative primer pair (+/- SD) were plotted in Excel (Figure 7).
Table 9.
Primer sequences for qPCR analysis of Cse4, PolII and Ste12 ChIPs
| qPCR experiment | Pair name | Primer name | Sequence |
| Cse4 | Cse4P1 | Cse4P1for | GATCAGCGCCAAACAATATGGAAAATCC |
| Cse4 | Cse4P1 | Cse4P1rev | AACTTCCACCAGTAAACGTTTCATATATCC |
| Cse4 | Cse4P2 | Cse4P2for | CGTATTACAATGGCCGAAGGC |
| Cse4 | Cse4P2 | Cse4P2rev | GCGACAACAAGAGGGAAATGA |
| Cse4 | Cse4P3 | Cse4P3for | CGTCCAAACATGAAAGTGCTCC |
| Cse4 | Cse4P3 | Cse4P3rev | CAGCGATTGACTTTCTCCCATT |
| Cse4 | Cse4P4 | Cse4P4for | GAAGCGTTATGGAACCTGTCGAA |
| Cse4 | Cse4P4 | Cse4P4rev | GTCGGTCGTCCAATATCATCGTAAA |
| PolII | Pol2P1 | Pol2P1for | ACCGGTACAAGGACAAGACG |
| PolII | Pol2P1 | Pol2P1rev | GTTCGTTCTCACGCACTTCA |
| PolII | Pol2P2 | Pol2P2for | AAGACGCTCGAAACCAAGTG |
| PolII | Pol2P2 | Pol2P2rev | GCTCACGTTTTGCAATGATG |
| PolII | Pol2P3 | Pol2P3for | GACCGTTGCAAGGATTGATAA |
| PolII | Pol2P3 | Pol2P3rev | TCAACCGAAGGAAGGAGAAA |
| PolII | Pol2P4 | Pol2P4for | ATTGCCTGGTTCTTGTCCTG |
| PolII | Pol2P4 | Pol2P4rev | CGTTGGCATATCACACCTTG |
| Ste12 | Ste12P1 | Pseudo35 | CCCGTAGTCCGGTTTAATCA |
| Ste12 | Ste12P1 | Pseudo36 | AACTGTGCATGAGCCAAGAG |
| Ste12 | Ste12P2 | Pseudo37 | AAAAGGAGATAGGGCCCAGA |
| Ste12 | Ste12P2 | Pseudo38 | CCAGAACAGCCAGCTAGACC |
| Ste12 | Ste12P3 | Pseudo39 | TCGGGCTTCTAAGGCAAATA |
| Ste12 | Ste12P3 | Pseudo40 | TCCTTTAAATGATGTTGCGATG |
| Ste12 | Ste12P4 | Pseudo41 | TGTAGCCCAACGGATTCTTC |
| Ste12 | Ste12P4 | Pseudo42 | AGAAGCTTTGCCAGGTGAAA |
Figure 7.
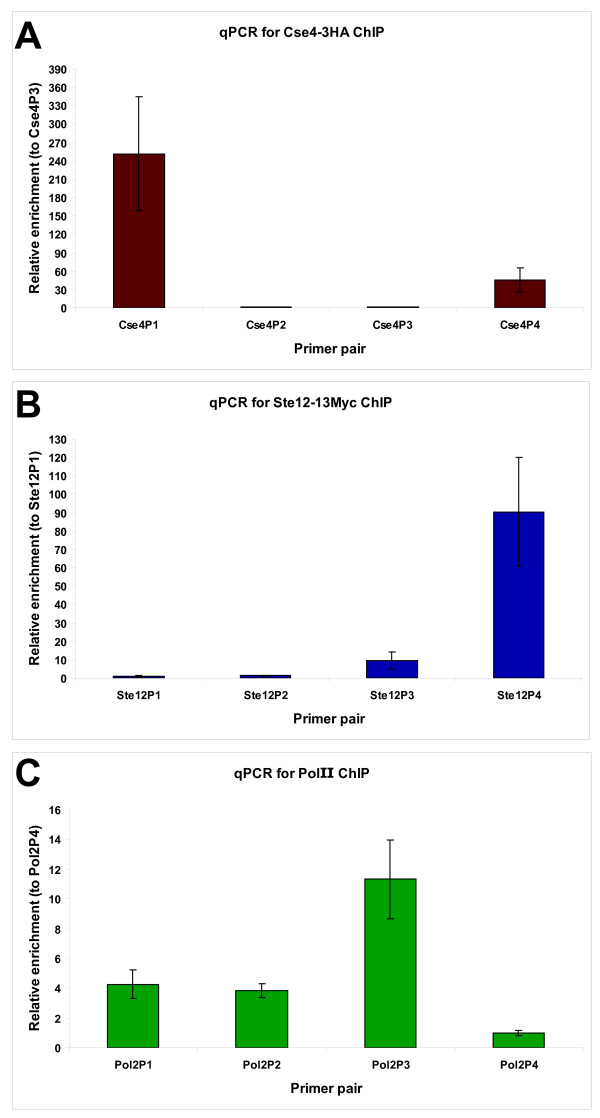
qPCR analysis for ChIP samples Cse4-3HA (a), Ste12-13Myc (b) and RNA polymerase PolII (c). For all qPCR analysis, normalization using the 2-ΔΔCp method was used to compare results from a given primer pair to a negative primer pair (respectively Cse4P2, Ste12P1 and Pol2P4) (Table 9). Error bars represent standard deviation across three biological replicates for relative enrichment to the negative primer pair. (a) Cse4p is enriched preferentially at the centromeres (Cse4P1 for CEN3 and Cse4P4 for CEN6) but not at two random non-centromeric locations (Cse4P2 and Cse4P3). (b) Ste12p binds known target sites in pseudohyphal growth. Ste12P1, Ste12P2, Ste12P3 and Ste12P4 represent respectively sites with no enrichment, low enrichment, medium enrichment and high enrichment as determined by ChIP-chip studies (Christopher M. Yellman, unpublished data). While the low enrichment site was not found to be significantly enriched for this ChIP sample, the medium and high enrichment sites were strongly present in our samples used for qPCR. (c) PolII primer pairs were selected using Steinmetz microarray data [54,64] to have three positive pairs (Pol2P1, Pol2P2 and Pol2P3) and one negative pair (Pol2P4). Positive targets were all significantly enriched over the negative control.
Adapter design and annealing
Each barcode has a 3 base index followed by a 'T' (position 4) to promote pairing with the 'A'-overhang that is added to the samples as part of the Illumina genomic DNA library preparation. Barcodes were created in such a way that no barcode had the same base at positions 1, 2 and 3 for two reasons. First, we wanted to have a balanced base composition. Second, since ELAND alignment software allows 2 mismatches to map any read, we wanted to make sure that sequencing errors would not influence mapping. These four barcodes were used for this study: GTAT, CATT, ACGT and TGCT.
Oligos were synthesized by MWG at a 0.05 μmol scale with HPLC purification. Oligo sequences are given in Table 1. Each oligo was resuspended in annealing buffer (10 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA) to 200 μM. The forward and reverse oligos for each pair were mixed in equal volumes to 100 μM and denatured for 5 min at 95°C in a wet heat block. The heat block was then removed to room temperature and allowed to cool slowly over 45 min to promote annealing. For input DNA, Illumina genomic DNA adapters were diluted 1:20 using RNase-free DNase-free water (Gibco) and our adapters were diluted 1:30. For ChIP DNA libraries, the DNA concentrations of the barcoded-adapters were measured by a Nanodrop spectrophotometer and adjusted to the same working concentration as Illumina's adapters. The following dilutions were therefore determined: 1:500 for barcode GTAT, 1:450 for barcode CATT, 1:750 for barcode ACGT and 1:330 for barcode TGCT.
Illumina DNA library generation
We followed the manufacturer's protocol for creating genomic DNA libraries with a few modifications based on experience gained from ChIP-Sequencing [6]. Here we present our modifications for generating barcoded libraries. ChIP DNA and input DNA were first band-isolated on a 2% agarose to obtain fragments between 150 and 350 base pairs and DNA was extracted using the QIAquick gel extraction kit (Qiagen) and eluted in 34 μL. For input DNA, because DNA amounts are higher than DNA recovered by ChIP, input DNA after gel extraction was diluted 1:5. After end-repair and addition of a single adenosine ("A") nucleotide, adapters were ligated to samples for 15 min at room temperature in the following fashion: the samples eluted from the MinElute column in 10 μL were ligated to 1 μL of adapters using 1.3 μL of LigaFast T4 DNA Ligase (3 Units/μL; Promega) and 12.3 μL Rapid Ligation Buffer (Promega). For input DNA libraries, the Illumina genomic DNA adapters were diluted 1:20 and all barcoded adapters were diluted 1:30. For ChIP DNA libraries, the Illumina genomic DNA adapters were diluted 1:40 and barcoded adapter dilutions are described in the previous section. After 15 min, samples were purified with the MinElute PCR purification kit (Qiagen).
We found that size-selection and purification of the ligation products by agarose gel before amplification of the library by PCR resulted in better libraries and decreased the incidence and intensity of an adapter-adapter band at ~120 bp. To eliminate those adapters that lack a fragment insert, samples were run on a 2% agarose E-Gel (Invitrogen) for 20 min, together with Track-It 50 bp DNA ladder (Invitrogen). DNA fragments ranging from 150 base pairs to 500 base pairs were extracted and recovered in 28 μL EB with a QIAquick gel extraction kit (Qiagen). To amplify the library, PCR was performed using Illumina genomic DNA primer "1.1" and Illumina genomic DNA primer "2.1" as described [6,62] with 15 cycles (Input DNA) or 17 cycles (ChIP DNA) of amplification. A final size selection was performed using a 2% agarose E-Gel to obtain a library with a median length of ~230 bp which is within the recommended size range for cluster generation on Illumina's flowcell. The library was recovered in 20 μL EB using MinElute Gel Extraction kit (Qiagen). Finally, DNA concentrations and purities (A260/280 nm ratios) were measured on a Nanodrop spectrophotometer and are given in Table 10.
Table 10.
Experimental design and values for ChIP replicates and input samples
| Experiment # | Experiment ID | Barcode | Sample | DNA Concentration (ng/φL) | A260/280 | Total reads | Mapped reads | % Mapping | Threshold |
| 1 | PolII_Rep1 | ACGT | PolII | 16.6 | 1.83 | 1110177 | 623328 | 56.15 | 14 |
| 2 | Input_CATT | CATT | Input | 51.6 | 1.85 | 2526689 | 1719855 | 68.07 | Not being scored |
| 3 | Cse4_Rep1 | GTAT | Cse4 | 12.0 | 1.99 | 2606497 | 1080537 | 41.46 | 81 |
| 4 | Ste12_Rep1 | TGCT | Ste12 | 11.6 | 1.85 | 2020432 | 1072428 | 53.08 | 42 |
| 5 | Input_ACGT | ACGT | Input | 66.9 | 1.80 | 4678579 | 2857615 | 61.08 | Not being scored |
| 6 | Cse4_Rep2 | CATT | Cse4 | 8.6 | 1.85 | 679315 | 357384 | 52.61 | 25 |
| 7 | Ste12_Rep2 | GTAT | Ste12 | 9.4 | 1.86 | 2316023 | 1067739 | 46.10 | 44 |
| 8 | PolII_Rep2 | TGCT | PolII | 15.5 | 2.00 | 3991833 | 2561147 | 64.16 | 12 |
| 9 | PolII_Rep3 | ACGT | PolII | 9.7 | 1.97 | 2661304 | 1725793 | 64.85 | 42 |
| 10 | Input_NB | None | Input | 60.4 | 1.83 | 2432221 | 1582226 | 65.05 | Not being scored |
| 11A* | Cse4_Rep3 | None | Cse4 | 10.7 | 1.69 | 1168583 | 239580 | 20.50 | 60 |
| 11B* | Cse4_Rep3 | None | Cse4 | 10.7 | 1.69 | 3245287 | 583876 | 17.99 | |
| 12A* | Ste12_Rep3 | None | Ste12 | 9.1 | 1.64 | 2106917 | 473965 | 22.50 | 39 |
| 12B* | Ste12_Rep3 | None | Ste12 | 9.1 | 1.64 | 2668067 | 551040 | 20.65 | |
| 13 | PolII_Rep4 | None | PolII | 18.8 | 1.94 | 4649077 | 3151068 | 67.78 | 40 |
| 14 | Input_NB | None | Input | 60.4 | 1.83 | 4373256 | 2455181 | 56.14 | Not being scored |
| 15 | Input_ACGT | ACGT | Input | 66.9 | 1.80 | 873037 | 529038 | 60.60 | 5 |
| 16 | Input_CATT | CATT | Input | 51.6 | 1.85 | 799430 | 479546 | 59.99 | 5 |
| 17 | Input_GTAT | GTAT | Input | 34.6 | 1.69 | 1380820 | 850262 | 61.58 | 6 |
| 18 | Input_TGCT | TGCT | Input | 68.1 | 1.78 | 611123 | 362458 | 59.31 | 4 |
NB (No Barcode) indicates that standard non-barcoded Illumina adapters were used. DNA concentrations of Illumina DNA libraries and their A260/280 ratios are given in columns 5 and 6.
Mapping values after sequencing (total sequencing reads, total mapped reads and percentage of mapped reads) are shown in columns 7, 8 and 9.
Threshold values for PeakSeq scoring algorithm are displayed in column 10.
* Sample ran twice on sequencer and reads pooled together from both lanes for scoring as a single experiment
Mapping values and scoring
Raw data from the Illumina Genome Analyzer I and II were analyzed with Illumina's Firecrest, Bustard and GERALD modules for image analysis, basecalling and run metrics respectively, and a PhiX174 control lane was used for matrix and phasing estimations, as per the manufacturer's instructions. At this stage, a Perl script was used to partition the reads and remove the barcodes, i.e. the first four bases of each read. The next 26 bases of each read were aligned against the reference genome S288c Saccharomyces cerevisiae, using Illumina's ELAND program in standalone mode. For each barcode from each flowcell lane of barcoded libraries, the numbers of total and mapped reads were determined. Reads lacking a fully-intact barcode were discarded in a fifth bin called unclassified (not shown) and were not used in individual barcode mapping analysis, although they are calculated in the global lane mapping analysis. Mapping values and thresholds for scoring are given in Table 10. Uniquely-mapped reads with ChIP factors and barcoding schemes in common were pooled to produce 19 different data sets (Table 10). A full lane of non-barcoded input DNA comprising 2,455,181 mapped reads was used as a control when scoring barcoded input DNA. For scoring ChIP DNA relative to input DNA, a reference set consisting of two full lanes of input DNA and two barcoded input DNA data sets were combined, adding up to 13,198,172 total reads. Signal files were created using a 200 bp sliding window and scoring was performed with the PeakSeq program [63] using a mappability fraction of 1.0. Significant "ChIP hits" were determined relative to the corresponding input DNA by PeakSeq and further filtered by requiring a hit length of at least 100 bp, a p-value of < 0.05, a ratio of at least 2.0 between ChIP DNA and input DNA read counts and a difference of at least 10 between the ChIP DNA and input DNA read counts.
In the section "Similar barcode behavior with the same DNA sample", each barcoded input DNA (Table 10, experiments #15–18) was scored against the non-barcoded input DNA (Table 10, experiment #14) using PeakSeq. Similar criteria were applied to filter barcoded input DNA hits. In these significant "input hits" from all four barcoded input DNA samples, the number of overlapping nucleotides was determined. Finally, the total number of nucleotide positions from all significant "input hits" was calculated and compared to the S. cerevisiae genome size.
Signal tracks and browser
Signal tracks were produced from uniquely-mapped reads and the S. cerevisiae genome using a 200 base pair sliding window. The Integrated Genome Browser (IGB, Affymetrix) was used to view images of signal tracks and to overlay them against the October 2003 version of the Saccharomyces cerevisiae genome. For ORFs and other annotations, the Saccharomyces cerevisiae genome was imported from SGD, the Saccharomyces Genome Database http://www.yeastgenome.org. After barcode parsing, each barcoded sample was compared over input DNA signal. To build signal tracks for comparing samples with different number of sequencing reads, the y-axis was normalized for each sample according to the total number of mapped reads.
Comparisons of the average tag counts in 500 bp bins between two experiments
We divided the yeast genome into 500 bp bins. For each experiment (Table 10), we calculated the average tag count for each bin by summing the tag counts at each nucleotide position within a bin and dividing the total by 500. We then normalized each experiment by dividing the average tag count for each bin by the total tag count for the experiment. Normalized average tag count for each 500 bp bin were compared between two experiments among barcoded and non-barcoded samples for a particular factor. Linear regression was performed using R and the R2 (R-squared) and p-values recorded.
Target list annotation
For RNA PolII, target lists were overlapped with SGD ORFs and then annotated using SQL operation chains. Targets that did not overlap with any ORF were manually annotated to the closest gene by comparing PolII bed files in IGB. If we could not discriminate between two genes, then both were selected.
For Cse4, target lists were manually curated. CENs and ORFs were identified manually by looking at Cse4 signal tracks and Cse4 bed files in IGB and associating them to overlapping features. If one target overlapped two ORFs, both ORFs were included in subsequent analyses. If one target was located between two ORFs, the closest ORF was selected if the target's signal was noticeably higher towards this ORF and if the target was positioned closer to this ORF by at least 200 bp. Otherwise, both ORFs were considered.
For Ste12, target lists were manually curated. Targets located within 1.5 kb upstream of an ORF's transcription start site were assigned uniquely to that ORF. In the case of two ORFs with divergent promoters, both ORFs were considered if they were located less than 1 kb away from the target. For all other targets, the closest ORF was selected except when distances between a target and its two closest ORFs were less than 200 bp. In this case, both ORFs were included. Targets whose signals were marginally above background and that were distal to annotated regions of the genome were discarded.
Comparisons between ChIP-Seq and published ChIP-chip data
Comparisons between PolII ChIP-Seq signal tracks and PolII ChIP-chip signal profiles around genes were made using data from Steinmetz et al. [54] and were performed visually in IGB. Comparisons between Ste12 ChIP-Seq targets and Ste12 ChIP-chip targets were done manually using IGB. If a ChIP-chip target was located less than 150 bp away from a ChIP-Seq target in at least one of the biological replicates, they were considered as overlapping for analysis purposes. ChIP-chip targets located more than 2 kb from a gene were excluded from comparisons. For Ste12, the Ste12 ChIP-chip target list from Borneman et al. [37] was used. For PolII ChIP-chip data from Steinmetz et al. [54], readily available on SGD and GEO (Series GSE6293, Sample GSM144667) [64], similar analyses and criteria as in Borneman et al. [37] were used to determine PolII ChIP-chip targets for consistency. 929 PolII ChIP-chip targets were visually inspected in IGB to ensure that they matched the PolII ChIP-chip signal profile [54]. Comparison with PolII ChIP-Seq data was done manually using IGB with similar criteria as those described for the Ste12 comparison between ChIP-Seq and ChIP-chip targets.
Rank-rank correlations
In order to demonstrate that the rank-order lists of target binding sites for different samples are correlated, we performed the following comparisons. Lists of target binding sites are ranked first by p-value from most significant to least significant and in second by the difference between the ChIP DNA and input DNA read counts from the highest to the lowest. For a pairwise comparison of two target lists, we can vary the fraction of each list that is compared. The number of targets that agree (i.e. overlap by at least one bp) between the two lists can be plotted as a function of the fraction of the two lists compared. Ideally for targets that are identical the rank-rank plot will be linear having a value of 100% agreement when the entire rank lists are compared. Deviation for this linear correlation shows where the rank lists being compared begin to differ.
Gene Ontology (GO) analysis
GO analyses were performed on SGD http://db.yeastgenome.org/cgi-bin/GO/goTermFinder.pl. We chose the GO Process Ontology with a p-value cutoff at p < 0.01. Other settings were set at default. For all three factors, ORFs were required to be present in at least one biological replicate. For Cse4 and PolII, all ORFs were used for the analyses. For Ste12, only those ORFs that were downstream of a target were analyzed.
Simulation of depth of sequencing
To investigate the sequencing depth necessary to saturate the number of detectable binding sites, we performed the following computational simulation. We simulated a genome of size SG, with Nsites binding sites of average fold enrichment f, using Nreads mapped sequence reads. In order to perform the simulation efficiently, we scaled the genome size to 1 Mb, and correspondingly scaled the number of binding sites and mapped reads. We randomly generated sequence reads using a uniform distribution over the 1 Mb interval for both a simulated input DNA sample as well as for a ChIP DNA sample with regions with enriched numbers of sequence reads corresponding to binding sites. These simulated datasets were scored in the same manner as ChIP DNA data was scored against input using a false discovery rate of 5%. The sensitivity (i.e. the fraction of simulated binding sites identified) as well as the positive predictive value (i.e. the accuracy of the target sites identified) of the scored results was computed using the simulated data. The simulation was repeated 50 times in order to accurately estimate the sensitivity and positive predictive values. By varying the number of simulated mapped sequence-reads Nreads, we can compute the minimum number of reads required in order to achieve a sensitivity of 95% (i.e. to be able to identify 95% of the simulated binding sites).
Authors' contributions
The laboratories of MS and MG contributed to this publication. PL, GME, RKA, MS and CMY drafted the manuscript. GME, PL and MS designed the multiplexing scheme. PL performed benchwork experiments, target list analyses and comparisons. GME designed the barcoded adapters, performed Illumina sequencing and contributed to troubleshooting and analysis. RKA adapted PeakSeq for the barcoded yeast ChIP-Seq context, created a pipeline for barcoded ChIP-Seq, scored the data and contributed to troubleshooting. JR created the scoring algorithm (PeakSeq), the sequencing depth algorithm and performed the rank-rank correlations. TG performed sequencing depth simulations. CMY generated strains and contributed to data analysis. All authors read and approved the final manuscript.
Supplementary Material
Cse4 target lists. This data correspond to the annotated filtered target lists and the raw 'hits' returned from PeakSeq for three biological replicates of Cse4 ChIP DNA processed by ChIP-Seq.
Ste12 target lists. This data correspond to the annotated filtered target lists and the raw 'hits' returned from PeakSeq for three biological replicates of Ste12 ChIP DNA processed by ChIP-Seq.
PolII target lists. This data correspond to the annotated filtered target lists and the raw 'hits' returned from PeakSeq for four biological replicates of PolII ChIP DNA processed by ChIP-Seq.
Complete GO analysis for Cse4, Ste12 and PolII. This data represent the complete GO process analysis for Cse4 combined targets, Ste12 combined targets and PolII combined targets, with a p-value cutoff < 0.01.
Sequencing artifacts present in non-barcoded samples and absent in barcoded samples. These IGB signal tracks show abnormally-shaped peaks present in non-barcoded biological replicates at a particular genomic location that are absent in barcoded biological replicates.
Acknowledgments
Acknowledgements
We thank N. Carriero and R. Bjornson from the Yale Center for High Performance Computation in Biology and Biomedicine for their dedication in providing Illumina pipeline support, M. Wilson for valuable help with GEO data submission and T. Gianoulis for thoughtful suggestions.
Contributor Information
Philippe Lefrançois, Email: philippe.lefrancois@yale.edu.
Ghia M Euskirchen, Email: ghia.euskirchen@yale.edu.
Raymond K Auerbach, Email: raymond.auerbach@yale.edu.
Joel Rozowsky, Email: joel.rozowsky@yale.edu.
Theodore Gibson, Email: theodore.gibson@yale.edu.
Christopher M Yellman, Email: christopher.yellman@yale.edu.
Mark Gerstein, Email: mark.gerstein@yale.edu.
Michael Snyder, Email: michael.snyder@yale.edu.
References
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett C, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley R, Tonti-Filippini J, Gregory B, Berry C, Millar A, Ecker J. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Johnson D, Mortazavi A, Myers R, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith O, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi A, Gibbs R, Wang J, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K, Parkhill J, Mazzoni C, Roumagnac P, Weill F, Goodhead I, Rance R, Baker S, Maskell D, Wain J, Dolecek C, Achtman M, Dougan G. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G, Kieffer-Higgins S. Multiplex DNA sequencing. Science. 1988;240:185–188. doi: 10.1126/science.3353714. [DOI] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Myles S, Prüfer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wederell E, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, Ingham M, Hirst M, Robertson G, Marra M, Jones S, Hoodless P. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes A, Wederell E, Cullum R, Euskirchen G, Krzywinski M, Birol I, Snyder M, Hoodless P, Hirst M, Marra M, Jones S. Genome wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008 doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko A, Peng S, Larschan E, Gorchakov A, Lee O, Kharchenko P, McGrath S, Wang C, Mardis E, Park P, Kuroda M. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega V, Wong E, Orlov Y, Zhang W, Jiang J, Loh Y, Yeo H, Yeo Z, Narang V, Govindarajan K, Leong B, Shahab A, Ruan Y, Bourque G, Sung W, Clarke N, Wei C, Ng H. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Meluh P, Yang P, Glowczewski L, Koshland D, Smith M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/S0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K, Fitzgerald-Hayes M. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics. 2000;156:973–981. doi: 10.1093/genetics/156.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson S, Washburn M, Gerton J. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith M, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker R. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mellone B, Karpen G. A specialized nucleosome has a "point" to make. Cell. 2007;129:1047–1049. doi: 10.1016/j.cell.2007.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Castillo A, Tatsutani S, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly: the kinetochore connection. Curr Biol. 2002;12:R256–258. doi: 10.1016/S0960-9822(02)00786-8. [DOI] [PubMed] [Google Scholar]
- Smith M, Yang P, Santisteban M, Boone P, Goldstein A, Megee P. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Cheeseman I, Anderson S, Yates Jr, Drubin D, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W, Crisp M, DeLuca S, Rando O, Basrai M. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Camahort R, Seidel C, Gerton J, Biggins S. The overexpression of a Saccharomyces cerevisiae centromeric histone H3 variant mutant protein leads to a defect in kinetochore biorientation. Genetics. 2007;175:513–525. doi: 10.1534/genetics.106.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- Madhani H, Fink G. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Roberts R, Fink G. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Wesche S, Taheri N, Braus G, Mösch H. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot Cell. 2002;1:673–686. doi: 10.1128/EC.1.5.673-686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S, Summers E, Lo H, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A, Leigh-Bell J, Yu H, Bertone P, Gerstein M, Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A, Gianoulis T, Zhang Z, Yu H, Rozowsky J, Seringhaus M, Wang L, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Oehlen L, Cross F. The mating factor response pathway regulates transcription of TEC1, a gene involved in pseudohyphal differentiation of Saccharomyces cerevisiae. FEBS Lett. 1998;429:83–88. doi: 10.1016/S0014-5793(98)00568-7. [DOI] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Chou S, Zhao S, Song Y, Liu H, Nie Q. Fus3-triggered Tec1 degradation modulates mating transcriptional output during the pheromone response. Mol Syst Biol. 2008;4:212. doi: 10.1038/msb.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick J, Aparicio O, Jennings E, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert T, Wilson C, Bell S, Young R. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Hung W, Olson K, Breitkreutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur J Biochem. 1997;245:241–251. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- Kusari A, Molina D, Sabbagh WJ, Lau C, Bardwell L. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J Cell Biol. 2004;164:267–277. doi: 10.1083/jcb.200310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- Song D, Dolan J, Yuan Y, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dohlman H. Pheromone-regulated sumoylation of transcription factors that mediate the invasive to mating developmental switch in yeast. J Biol Chem. 2006;281:1964–1969. doi: 10.1074/jbc.M508985200. [DOI] [PubMed] [Google Scholar]
- Young R. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T, Woychik N. RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc Natl Acad Sci USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff D, Abruzzi K, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci USA. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Gendrel C, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Stewart A, Huff J, Cairns B. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E, Warren C, Kuehner J, Panbehi B, Ansari A, Brow D. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Hamming RW. Error Detecting and Error Correcting Codes. Bell System technical journal. 1950;26:147–160. [Google Scholar]
- Harbison C, Gordon D, Lee T, Rinaldi N, Macisaac K, Danford T, Hannett N, Tagne J, Reynolds D, Yoo J, Jennings E, Zeitlinger J, Pokholok D, Kellis M, Rolfe P, Takusagawa K, Lander E, Gifford D, Fraenkel E, Young R. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Project: Model Organism ENCyclopedia Of DNA Elements (modENCODE) http://www.modencode.org/
- Edgar R, Domrachev M, Lash A. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Gene Expression Omnibus, Series GSE13322 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13322
- Aparicio O, Geisberg J, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004;Chapter 17 doi: 10.1002/0471143030.cb1707s23. Unit 17.17. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G, Rozowsky J, Wei C, Lee W, Zhang Z, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein M, Ruan Y, Snyder M. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach R, Zhang Z, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein M. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Gene Expression Omnibus, Series GSE6293 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6293
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cse4 target lists. This data correspond to the annotated filtered target lists and the raw 'hits' returned from PeakSeq for three biological replicates of Cse4 ChIP DNA processed by ChIP-Seq.
Ste12 target lists. This data correspond to the annotated filtered target lists and the raw 'hits' returned from PeakSeq for three biological replicates of Ste12 ChIP DNA processed by ChIP-Seq.
PolII target lists. This data correspond to the annotated filtered target lists and the raw 'hits' returned from PeakSeq for four biological replicates of PolII ChIP DNA processed by ChIP-Seq.
Complete GO analysis for Cse4, Ste12 and PolII. This data represent the complete GO process analysis for Cse4 combined targets, Ste12 combined targets and PolII combined targets, with a p-value cutoff < 0.01.
Sequencing artifacts present in non-barcoded samples and absent in barcoded samples. These IGB signal tracks show abnormally-shaped peaks present in non-barcoded biological replicates at a particular genomic location that are absent in barcoded biological replicates.