Abstract
Kinase Gcn2 is activated by amino acid starvation and downregulates translation initiation by phosphorylating the α subunit of translation initiation factor 2 (eIF2α). The Gcn2 kinase domain (KD) is inert and must be activated by tRNA binding to the adjacent regulatory domain. Previous work indicated that Saccharomyces cerevisiae Gcn2 latency results from inflexibility of the hinge connecting the N and C lobes and a partially obstructed ATP-binding site in the KD. Here, we provide strong evidence that a network of hydrophobic interactions centered on Leu-856 also promotes latency by constraining helix αC rotation in the KD in a manner relieved during amino acid starvation by tRNA binding and autophosphorylation of Thr-882 in the activation loop. Thus, we show that mutationally disrupting the hydrophobic network in various ways constitutively activates eIF2α phosphorylation in vivo and bypasses the requirement for a key tRNA binding motif (m2) and Thr-882 in Gcn2. In particular, replacing Leu-856 with any nonhydrophobic residue activates Gcn2, while substitutions with various hydrophobic residues maintain kinase latency. We further provide strong evidence that parallel, back-to-back dimerization of the KD is a step on the Gcn2 activation pathway promoted by tRNA binding and autophosphorylation. Remarkably, mutations that disrupt the L856 hydrophobic network or enhance hinge flexibility eliminate the need for the conserved salt bridge at the parallel dimer interface, implying that KD dimerization facilitates the reorientation of αC and remodeling of the active site for enhanced ATP binding and catalysis. We propose that hinge remodeling, parallel dimerization, and reorientation of αC are mutually reinforcing conformational transitions stimulated by tRNA binding and secured by the ensuing autophosphorylation of T882 for stable kinase activation.
Protein synthesis is downregulated under various adverse conditions by phosphorylation of serine 51 in the α subunit of translation initiation factor 2 (eIF2α). The eIF2 forms a ternary complex (TC) with GTP and methionyl-tRNAMet, which delivers methionyl-tRNAMet to the small (40S) ribosomal subunit. Phosphorylated eIF2 inhibits its guanine nucleotide exchange factor, eIF2B, reducing formation of TC and inhibiting subsequent steps on the initiation pathway (28). The eIF2α-Ser-51 kinase Gcn2 is activated by amino acid starvation in the budding yeast Saccharomyces cerevisiae and mammals (16, 18) and was first identified as a translational inducer of GCN4, a transcriptional activator of amino acid biosynthetic enzymes in yeast. Four short open reading frames (uORFs) in the GCN4 mRNA leader prevent scanning ribosomes from reaching the GCN4 start codon under nonstarvation conditions when TC is abundant. The reduction in TC levels evoked by eIF2α phosphorylation in starved cells allows ribosomes that have translated the first uORF to scan past the subsequent uORFs and reinitiate further downstream at GCN4. Thus, Gcn2 induces GCN4 and its amino acid biosynthetic target genes to replenish amino acid pools while reducing general protein synthesis and the rate of amino acid consumption. This dual response to amino acid starvation by eIF2α phosphorylation also operates in mammalian cells, involving transcription factors ATF4 (16, 34) and ATF5 (42). Translational control by mammalian Gcn2 is important for lipid homeostasis under starvation conditions (14), in behavioral aversion to amino acid-deficient diets (15), and in learning and memory (5). Mammals also contain three other eIF2α-Ser-51 kinases, which share extensive sequence similarities in their kinase domains (KDs) but are activated by different stresses through their distinct regulatory regions: PKR (virus infection), PERK (endoplasmic reticulum stress), and HRI (hemin starvation) (8).
Because eIF2α kinases act by inhibiting translation, their functions must be tightly regulated to permit high-level kinase activity only under adverse conditions. We have shown previously that the Gcn2 KD is intrinsically inert and depends on stimulatory interactions with adjacent domains in the protein (29). Binding of uncharged tRNA to a region related to histidyl-tRNA synthetase (HisRS), located C-terminally to the KD, is required to activate Gcn2 in amino acid-starved cells (35). Mutations in conserved residues of the HisRS domain (the m2 motif) destroy tRNA binding in vitro and impair Gcn2 function in vivo (11, 37). An N-terminal segment in the HisRS domain (HisRS-N) interacts with a portion of the KD containing the hinge that connects the N- and C-terminal lobes, and this interaction is thought to participate in kinase activation by tRNA (27).
As with other kinases, autophosphorylation of the activation loop is additionally required to activate Gcn2. Thr-882 and Thr-887 are the sites of autophosphorylation by Gcn2 in vitro, and mutating them to Ala impairs (T882A) or abolishes (T887A) Gcn2 function (32). There is also evidence that dimerization of the KD is important for Gcn2 activation. The KD, HisRS region, and extreme C terminus of Gcn2 (C-term) are all capable of self-interaction as isolated domains. However, only the C-term is essential for dimerization by full-length Gcn2 (28), and C-term dimerization is critical for Gcn2 activation (24). Since Gcn2 dimerizes efficiently without the HisRS domain, and thus, binding of uncharged tRNA is not required, Gcn2 probably dimerizes constitutively in vivo through the self-interaction of the C-term (28).
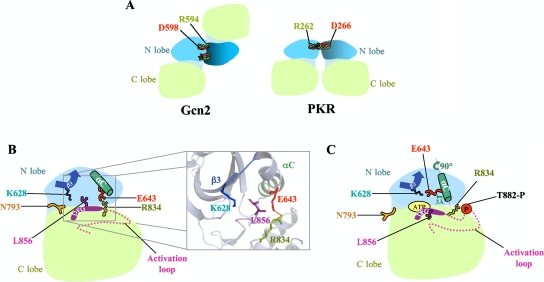
The GCN2 KD crystallized as a symmetric homodimer, with the monomers arranged in an antiparallel orientation (25) (Fig. 1A, left). However, genetic evidence suggests that the active form of the GCN2 KD dimerizes in a different orientation. In the case of PKR, the crystal structure of the active KD revealed a symmetric homodimer involving back-to-back, parallel interaction of the N lobes (6) (Fig. 1A, right). Substitutions at R262 and D266 that disrupt a salt bridge at the PKR dimer interface impair dimerization and kinase activity, and both functions are recovered by a double substitution that reconstitutes a salt bridge of opposite polarity (10). The salt bridge residues are conserved in GCN2 but are too remote to interact between protomers in the antiparallel dimer of the inactive KD (Fig. 1A). However, analogous genetic experiments showed that GCN2 activity requires either the wild-type (WT) salt bridge (R594-D598) or a reconstituted one of opposite polarity (D594-R598) (11). This suggested that the active GCN2 KD adopts the parallel dimer configuration displayed in the PKR KD structure. It is possible that the GCN2 KD isomerizes from the antiparallel to parallel dimer configuration within the full-length GCN2 dimer (stabilized by C-term interactions) when tRNA binding to the HisRS domain elicits kinase activation.
FIG. 1.
(A) Schematic representation of distinct dimer conformations in the KD crystal structures of inactive Gcn2 and active PKR. The Gcn2 KD dimer (left) has protomers arranged in antiparallel orientation, whereas the PKR KD dimer (right) displays a back-to-back, parallel mode of dimerization. The KD N and C lobes are colored light blue and green, respectively. The conserved R262-D266 salt bridge is depicted in the PKR dimer, whereas the corresponding residues in Gcn2 cannot interact in the antiparallel dimer. (B) Schematic representation of the inactive Gcn2 KD. L856 obstructs reorientation of αC and blocks formation of the K628-E643 salt bridge; improper rotation of αC is stabilized by the inappropriate E643-R843 salt bridge. The N793 “molecular flap” partially occludes the ATP-binding pocket. The inset shows the relevant residues in the ribbon diagram of the Gcn2 KD crystal structure, with stick representations of side chains, displayed using PyMOL software (http://www.pymol.org) and Protein Data Bank file 1ZYC. (C) Schematic depiction of the predicted active conformation of the Gcn2 KD. Phosphorylated T882 neutralizes the positive charge of R834 and disrupts the inhibitory R834-E643 salt bridge, allowing axial rotation and translation of αC (as shown) and formation of the critical K628-E643 salt bridge.
Given the relatively strong sequence similarities among the KDs of different eIF2α kinases (10), the PKR crystal structure may provide a good model for the active conformations of the KDs in other eIF2α kinases. However, detailed information about the inactive conformation of the KD exists only for Gcn2. We previously isolated two substitutions in the KD (R794G and F842L) that constitutively activate Gcn2 in the absence of tRNA binding to the HisRS domain (29). The crystal structure of the inactive form of the WT Gcn2 KD revealed that residues affected by these GCN2c mutations participate in a network of interactions that rigidify the hinge to partially close the active site cleft and occlude the ATP-binding pocket. The crystal structure of the activated R794G mutant, by contrast, revealed a greater separation between the N and C lobes, plus an opening of the “molecular flap” contributed by Asn-793, to yield a more accessible ATP-binding site. This led us to propose that interaction of the KD with the tRNA-bound HisRS domain provokes a remodeling of the hinge, mimicked by R794G, that stimulates ATP binding and increases the flexibility of the N and C lobes (25). Our isolation of the GCN2c mutation M788V (mutations that affect the phenotype of a strain are in italics; substitutions that affect protein function are roman) (30) in the “gatekeeper” residue of the ATP-binding pocket (1) is also consistent with the idea that ATP binding is impeded in the latent Gcn2 KD. Kinase activation produced by combining the hinge mutations R794G and F842L (dubbed GCN2Hyper) leads to hyperphosphorylation of T882, while mutation of both autophosphorylation sites nearly inactivates the GCN2Hyper protein. Accordingly, we proposed that the remodeling of the hinge to facilitate ATP binding and catalysis is the first stage in activation by uncharged tRNA, which is followed by (and facilitates) autophosphorylation of the activation loop (29).
The structure of the inactive Gcn2 KD revealed a second feature that likely contributes to kinase latency, involving incorrect orientation of the αC helix in the N lobe of the KD (25). This prevents the invariant Glu in αC (Glu-643) from forming the salt bridge with the invariant Lys in the β3 strand (Lys-628) that is crucial for positioning the triphosphate moiety of ATP in the active site. Instead, Glu-643 makes a salt bridge with Arg-834 in the HRD motif of the catalytic loop, immediately preceding the catalytic aspartate (Asp-835) (Fig. 1B). A nonproductive configuration of αC occurs in other kinases of the “RD” subclass and is thought to be corrected by autophosphorylation of the activation loop. It was proposed that charge neutralization of the autophosphorylated residues by a cluster of basic residues, including Arg of the HRD motif, drives a structural rearrangement to the active configuration (20). In the case of Gcn2, interaction of Arg-834 with phosphorylated Thr-882 (or Thr-887) could release Glu-643 from the inhibitory salt bridge and allow it to form the productive salt bridge with Lys-628 instead (Fig. 1C). This rearrangement requires an ∼90° axial rotation and ∼3-Å translation of αC that, interestingly, appears to be obstructed by the invariant Leu residue immediately following the DFG motif in the beginning of the activation loop (L856 in Gcn2) (25) (Fig. 1B). If this last prediction of the crystal structure is correct, then L856 and any residues that stabilize its inhibitory disposition in the inactive state should be critical for maintaining Gcn2 latency in nonstarved cells.
In this report, we demonstrate that nearly all amino acid substitutions of L856 except those involving bulky hydrophobic residues constitutively activate Gcn2, and we provide in vivo evidence that the hydrophobic side chain of L856 must interact with hydrophobic residues in αC, β3, and β5 to oppose the rotation of αC to the active orientation in nonstarved cells. Remarkably, disrupting this hydrophobic network eliminates the requirements for tRNA binding and Thr-882 for high-level kinase activity, implying that tRNA binding and T882 autophosphorylation promote reorientation of αC to the active conformation. We also present in vivo evidence that achieving the “PKR mode” of parallel, back-to-back dimerization of the KD is a step on the activation pathway and helps to overcome impediments to ATP binding or catalysis imposed by the L856 hydrophobic network and hinge rigidity. We propose a model wherein tRNA binding elicits mutually reinforcing conformational changes in the KD, including hinge remodeling, parallel dimerization, and reorientation of αC, to stimulate ATP binding and autophosphorylation, which then “locks in” this productive conformation to achieve stable kinase activation. Our results have interesting implications for the latency mechanisms of other important protein kinases, including PKR, the EFG receptor, and CDK2.
MATERIALS AND METHODS
The phenotypes of plasmid-borne GCN2 alleles were tested in strains H1149 (MATα ura3-52 inol leu2-3 leu2-112 gcn2-Δ63-3284::LEU2 [HIS4::lacZ integrated at ura3-52]) (36) and H1817 (MATa ura3-52 leu2-3 leu2-112 gcn2Δ sui2Δ trp1-Δ63 [GCN4-lacZ TRP1 integrated at trp1-Δ63] p1098[SUI2-S51A LEU2]) (9). Strain HQY346 is a GAL2 derivative of GP3299 (MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ gcd2Δ::hisG pAV1033[GCD2-K627T, TRP1]) (26). The plasmids and mutagenic primers employed in this work are listed in Tables 1 and 2. All GCN2 mutations were constructed by a PCR fusion technique (39) using p722 or its derivative (p722-NruI) containing an NruI site introduced by the “silent” substitution of TCA for TCG at codon 593 in GCN2 and primers listed in Table 2. For constructing the majority of the mutations, two PCRs were carried out using p722 or p722-NruI as a template and (i) primer 189 and the appropriate mutagenic primer and (ii) primer 199 and the primer complementary to the mutagenic primer used in the first reaction. A third PCR was conducted using the products of the first two reactions and primers 189 and 199. The final PCR fragment was purified and digested with different pairs of restriction enzymes, Blp1 and BspEI, NruI and Asp718, or BspEI and Asp718, depending on the location of the mutation and whether p722 or p722-NruI was employed. The digested PCR fragment was ligated with p722 (or p722-NruI) and digested with the same pair of enzymes and, in some cases, with XhoI to eliminate the recovery of clones containing nonrecombinant, undigested p722 (or p722-NruI). (XhoI digestion was not possible when cloning BlpI/BspEI fragments, and in those cases, the digested vector was dephosphorylated with calf intestine phosphatase to reduce the background.) A similar approach was used to introduce the mutations contained in plasmids pHQ1746 and pHQ1747, except that primer 290 replaced primer 199, and the final PCR product was digested with BlpI and XhoI and used to replace the corresponding fragment in p722. DNA sequence analysis of the region encompassing the entire subcloned PCR fragment was conducted to identify recombinant plasmids containing the desired mutations and no unintended mutations. Sensitivities of yeast transformants to 3-aminotriazole (3-AT) or 5-fluorotryptophan (5-FT) were determined as previously described (30) and as indicated in the figure legends.
TABLE 1.
Plasmids employed in this work
| Plasmid | Description | Source or reference |
|---|---|---|
| p722 | GCN2 CEN6 URA3 | 36 |
| p722-NruI | p722 derivative with NruI site at codon 593 in GCN2 | This study |
| p1253 | GCN2-E803V in p722 backbone | 30 |
| p299 | gcn2-m2 in p722 backbone | 37 |
| pC2746 | gcn2-R594D in p722 backbone | 11 |
| pC2747 | gcn2-D598R in p722 backbone | 11 |
| pC2748 | GCN2-R594D,D598R in p722 backbone | 11 |
| pB9 | gcn2-T882A CEN4 URA3 | 32 |
| pHQ1140 | GCN2-R794G,F842L,m2 in p722 derivative pHQ644 | 29 |
| pAG1 | GCN2-L856A in p722 backbone | This study |
| pAG2 | GCN2-L856G in p722 backbone | This study |
| pAG3 | GCN2-L856V in p722 backbone | This study |
| pAG4 | GCN2-L856I in p722 backbone | This study |
| pAG5 | GCN2-L856M in p722 backbone | This study |
| pAG6 | gcn2-L856P in p722 backbone | This study |
| pAG7 | GCN2-L856F in p722 backbone | This study |
| pAG8 | GCN2-L856Y in p722 backbone | This study |
| pAG9 | GCN2-L856W in p722 backbone | This study |
| pAG10 | GCN2-L856D in p722 backbone | This study |
| pAG11 | GCN2-L856E in p722 backbone | This study |
| pAG12 | GCN2-L856N in p722 backbone | This study |
| pAG13 | GCN2-L856Q in p722 backbone | This study |
| pAG14 | GCN2-L856H in p722 backbone | This study |
| pAG15 | GCN2-L856K in p722 backbone | This study |
| pAG16 | GCN2-L856R in p722 backbone | This study |
| pAG17 | GCN2-L856S in p722 backbone | This study |
| pAG18 | GCN2-L856T in p722 backbone | This study |
| pAG19 | GCN2-L856C in p722 backbone | This study |
| pAG120 | GCN2-I630A in p722-NruI backbone | This study |
| pAG21 | GCN2-V644A in p722-NruI backbone | This study |
| pAG22 | GCN2-I640A in p722-NruI backbone | This study |
| pHQ1746 | GCN2-I640L in p722 backbone | This study |
| pHQ1747 | GCN2-I640V in p722 backbone | This study |
| pAG23 | GCN2-I786A in p722-NruI backbone | This study |
| pAG24 | GCN2-L647A in p722-NruI backbone | This study |
| pAG25 | GCN2-L647I in p722-NruI backbone | This study |
| pAG26 | GCN2-L647V in p722-NruI backbone | This study |
| pAG27 | GCN2-I630A,I640A in p722-NruI backbone | This study |
| pAG28 | GCN2-I640A,V644A in p722-NruI backbone | This study |
| pAG29 | GCN2-I640A,L647A in p722-NruI backbone | This study |
| pAG30 | GCN2-I640A,L647A,m2 in p722-NruI backbone | This study |
| pAG31 | GCN2-I640A,L647A,R594D in p722-NruI backbone | This study |
| pAG32 | GCN2-L856A,T882A in p722 backbone | This study |
| pAG33 | GCN2-L856A,m2 in p722 backbone | This study |
| pAG34 | GCN2-L650F in p722 backbone | This study |
| pAG35 | GCN2-Y825H in p722 backbone | This study |
| pAG36 | GCN2-K850R in p722 backbone | This study |
| pAG37 | GCN2-L650F,K850R in p722 backbone | This study |
| pAG38 | GCN2-L650F,K850R,R594D,D598R in p722 backbone | This study |
| pAG39 | GCN2-L650F,K850R,T882A in p722 backbone | This study |
| pAG40 | GCN2-L650F,K850R,m2 in p722 backbone | This study |
| pAG41 | GCN2-R794G,F842L,R594D in p722 backbone | This study |
| pAG42 | GCN2-L856A,R594D in p722 backbone | This study |
| pAG43 | GCN2-N618A in p722 backbone | This study |
| pAG44 | GCN2-L856A,N618A in p722 backbone | This study |
| pAG45 | GCN2-R794G,F842L,N618A in p722 backbone | This study |
| pAG48 | GCN2-Y625A in p722 backbone | This study |
| pAG73 | GCN2-N651A in p722 backbone | This study |
| pAG74 | GCN2-Y658A in p722 backbone | This study |
| pAG75 | GCN2-S597A in p722 backbone | This study |
| pAG76 | GCN2-L646A in p722 backbone | This study |
| pAG77 | GCN2-S649A in p722 backbone | This study |
| pAG78 | GCN2-L663A in p722 backbone | This study |
| pAG79 | GCN2-R768A in p722 backbone | This study |
| pAG80 | GCN2-S597A,R768A in p722 backbone | This study |
TABLE 2.
Selected oligonucleotides used for mutagenesis
| Name of oligonucleotide | Sequencea | Substitution |
|---|---|---|
| 189 | GGATCCGCCATGGACAGCTATTTCAACACAAGTAATGG | None |
| 199 | CCGGTC GACCTATCGGTAAACGTGCTGCATCCTATA | None |
| 290 | CCGGTCGACTACTTAACAGCAGTCATCGGTTT | None |
| 856A | CGGTGATTTTGGGGCAGCTAAGAACGTCCATAGATCTCTGG | L856A |
| 856Ar | CCAGAGATCTATGGACGTTCTTAGCTGCCCCAAAATCACCG | L856A |
| 856G | CGAGAAATGTTAAAATCGGTGATTTTGGGGGAGCTAAGAACG | L856G |
| 856V | CGAGAAATGTTAAAATCGGTGATTTTGGGGTAGCTAAGAACG | L856V |
| 856I | CGAGAAATGTTAAAATCGGTGATTTTGGGATAGCTAAGAACG | L856I |
| 856 M | CGAGAAATGTTAAAATCGGTGATTTTGGGATGGCTAAGAACG | L856M |
| 856P | CGAGAAATGTTAAAATCGGTGATTTTGGGCCAGCTAAGAACG | L856P |
| 856F | CGAGAAATGTTAAAATCGGTGATTTTGGGTTTGCTAAGAACG | L856F |
| 856Y | CGAGAAATGTTAAAATCGGTGATTTTGGGTATGCTAAGAACG | L856Y |
| 856W | CGAGAAATGTTAAAATCGGTGATTTTGGGTGGAGCTAAGAACG | L856W |
| 856D | CGAGAAATGTTAAAATCGGTGATTTTGGGGATGCTAAGAACG | L856D |
| 856E | CGAGAAATGTTAAAATCGGTGATTTTGGGGAAGCTAAGAACG | L856E |
| 856N | CGAGAAATGTTAAAATCGGTGATTTTGGGAATGCTAAGAACG | L856N |
| 856Q | CGAGAAATGTTAAAATCGGTGATTTTGGGCAAGCTAAGAACG | L856Q |
| 856H | CGAGAAATGTTAAAATCGGTGATTTTGGGCATGCTAAGAACG | L856H |
| 856K | CGAGAAATGTTAAAATCGGTGATTTTGGGAAAGCTAAGAACG | L856K |
| 856R | CGAGAAATGTTAAAATCGGTGATTTTGGGCGAGCTAAGAACG | L856R |
| 856S | CGAGAAATGTTAAAATCGGTGATTTTGGGTCAGCTAAGAACG | L856S |
| 856T | CGAGAAATGTTAAAATCGGTGATTTTGGGACTGCTAAGAACG | L856T |
| 856C | CGAGAAATGTTAAAATCGGTGATTTTGGGTGTGCTAAGAACG | L856C |
| 856r | CCCAAAATCACCGATTTTAACATTTCTCG | Reverse primer for mutations at position 856 |
| 630A | ATAGCAGATACTATGCGATCAAGAAGGCTAGACATACAG | I630A |
| 630Ar | CTGTATGTCTAGCCTTCTTGATCGCATAGTATCTGCTAT | I630A |
| 640A | CAGAAGAAAAGTTATCTACTGCATTGAGTG | I640A |
| 640Ar | CACTCAATGCAGTAGATAACTTTTCTTCTG | I640A |
| I640L-f | GACATACAGAAGAAAAGTTATCTACTTTATTGAGTGAAGTAATGC | I640L |
| I640L-r | GCATTACTTCACTCAATAAAGTAGATAACTTTTCTTCTGTATGTC | I640L |
| I640V-f | GACATACAGAAGAAAAGTTATCTACTGTATTGAGTGAAGTAATGC | I640V |
| I640V-r | GCATTACTTCACTCAATACAGTAGATAACTTTTCTTCTGTATGTC | I640V |
| 644A | CTATATTGAGTGAAGCAATGCTGTTAGCAAGC | V644A |
| 644Ar | GCTTGCTAACAGCATTGCTTCACTCAATATAG | V644A |
| 647A | GAGTGAAGTAATGCTGGCAGCAAGCTTAAATCATC | L647A |
| 647Ar | GATGATTTAAGCTTGCTGCCAGCATTACTTCACTC | L647A |
| 647I | GAGTGAAGTAATGCTGATAGCAAGCTTAAATCATC | L647I |
| 647ir | GATGATTTAAGCTTGCTATCAGCATTACTTCACTC | L647I |
| 647V | GAGTGAAGTAATGCTGGTAGCAAGCTTAAATCATC | L647V |
| 647vr | GATGATTTAAGCTTGCTACCAGCATTACTTCACTC | L647V |
| 640A647A | AAGTTATCTACTGCATTGAGTGAAGTAATGCTGGCAGCAAGCTTAAATCATC | I640A L647A |
| 640a647ar | GATGATTTAAGCTTGCTGCCAGCATTACTTCACTCAATGCAGTAGATAACTT | I640A L647A |
| 786A | CGCTTTTTGCTCAAATGGAGTACTGTG | L786A |
| 786Ar | CACAGTACTCCATTTGAGCAAAAAGCG | L786A |
| 640A644A | AAGTTATCTACTGCATTGAGTGAAGCAATGCTGTTAGCAAGCTTAAATCATC | I640A V644A |
| 640a644ar | GATGATTTAAGCTTGCTAACAGCATTGCTTCACTCAATGCAGTAGATAACTT | I640A V644A |
| 650F | CTGTTAGCAAGCTTCAATCATCAATATGTTGTGC | L650F |
| 650Fr | GCACAACATATTGATGATTGAAGCTTGCTAACAG | L650F |
| 825H | GGAAGCACTGAGTCATATACATTCCCAGGGTATCATTCATAGG | Y825H |
| 825Hr | CCTATGAATGATACCCTGGGAATGTATATGACTCAGTGCTTCC | Y825H |
| 850R | CGAGAAATGTTAGAATCGGTGATTTTGGGTTAGC | K850R |
| 850Rr | GCTAACCCAAAATCACCGATTCTAACATTTCTCG | K850R |
| 618A | GTTGTCAAGGCACGTGCAGCTCTCGATAGCAGATAC | N618A |
| 618Ar | GTATCTGCTATCGAGAGCTGCACGTGCCTTGACAAC | N618A |
| 625A | GCAGATACGCTGCGATCAAGAAGATTAGAC | Y625A |
| 625Ar | GTCTAATCTTCTTGATCGCAGCGTATCTGC | Y625A |
| 794G | GGAGTACTGTGAAAATGGAACGCTATATGATTTGATCC | R794G |
| 794Gr | GGTTCAAATCATATAGCGTTCCATTTTCACAGTACTCC | R794G |
| 842L | GGGATCTGAAGCCAATGAATATTTTAATAGATGAATCG | F842L |
| 842Lr | CGATTCATCTATTAAAATATTCATTGGCTTCAGATCCC | F842L |
| 594D | CCTGCAACGCGATCAGATTATGCTTCTGACTTTGAAGAG | R594D |
| 594Dr | CTCTTCAAAGTCAGAAGCATAATCTGATCGCGTTGCAGG | R594D |
| 598R | CCTGCAACGCGATCACGATATGCTTCTCGCTTTGAAGAG | D598R |
| 598Rr | CTCTTCAAAGCGAGAAGCATATCGTGATCGCGTTGCAGG | D598R |
| 594D598R | CCTGCAACGCGATCAGATTATGCTTCTCGCTTTGAAGAG | R594D D598R |
| 594D598Rr | TCTTCAAAGCGAGAAGCATAATCTGATCGCGTTGCAGG | R594D D598R |
| 651A | GCTGTTAGCAAGCTTAGCTCATCAATATG | N651A |
| 651Ar | CATATTGATGAGCTAAGCTTGCTAACAGC | N651A |
| 658A | CATCAATATGTTGTGCGTGCCTATGCTGCATGGTTAG | Y658A |
| 658Ar | CTAACCATGCAGCATAGGCACGCACAACATATTGATG | Y658A |
| S597 | CGATCACGATATGCTGCTGACTTTGAAGAGATTGC | S597A |
| S597r | GCAATCTCTTCAAAGTCAGCAGCATATCGTGATCG | S597A |
| L646 | GAGTGAAGTAATGGCTTTAGCAAGCTTAAATCATC | L646A |
| L646r | GATGATTTAAGCTTGCTAAAGCCATTACTTCACTC | L646A |
| S649 | GAGTGAAGTAATGCTGTTAGCAGCCTTAAATCATC | S649A |
| S649r | GATGATTTAAGGCTGCTAACAGCATTACTTCACTC | S649A |
| L663 | CGTTACTATGCTGCATGGGCAGAAGAAGACAGTATGG | L663A |
| L663r | CCATACTGTCTTCTTCTGCCCATGCAGCATAGTAACG | L663A |
| R768 | CCAGAACGTTCCAAGAGCCAGGAATTTTGTAAAACC | R768A |
| R768r | GGTTTTACAAAATTCCTGGCTCTTGGAACGTTCTGG | R768A |
Mutated positions are in boldface.
To measure eIF2α phosphorylation, total protein extracts were prepared by TCA extraction (31), resolved by SDS-PAGE, and subjected to Western analysis as described previously (29). Western signals were quantified by video image densitometry using Scion Image software (imported from NIH Image for the Macintosh by Scion Corporation and available on the Internet at http://www.scioncorp.com).
RESULTS
A bulky hydrophobic residue at position 856 is required for the inactive state of Gcn2 in vivo.
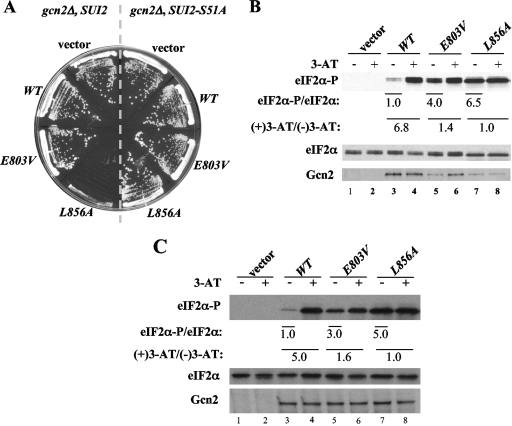
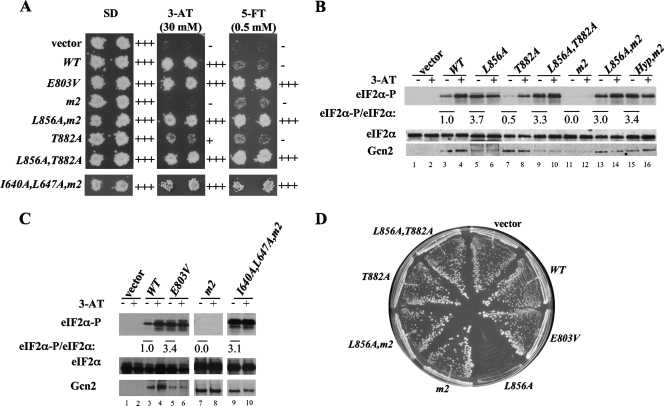
The crystal structure of the inactive Gcn2 KD shows that helix αC is rotated by ∼90° and displaced by 3 Å and that Leu-856 appears to block reorientation of αC to the position required to form the critical E643-K628 salt bridge between αC and β3 (25) (Fig. 1B). To examine whether Leu-856 contributes to Gcn2 latency in vivo, we asked whether replacing it with Ala leads to constitutive eIF2α phosphorylation by Gcn2 in yeast cells. Supporting this possibility, the introduction of the GCN2-L856A allele on a low-copy-number plasmid into gcn2Δ cells conferred a strong slow-growth (Slg−) phenotype on nutrient-replete medium in a strain expressing WT eIF2α (SUI2) but caused no growth defect in cells containing nonphosphorylatable eIF2α with Ala replacing Ser at position 51 (SUI2-S51A) (Fig. 2A). This finding suggests that L856A activates Gcn2 constitutively to produce high-level eIF2α phosphorylated on S51 (eIF2α-P) that inhibits general translation initiation and cell growth. Indeed, Western analysis with phospho-specific antibodies revealed a strong (∼6.5-fold) increase in eIF2α-P relative to that of total eIF2α under nonstarvation conditions (no 3-AT) in L856A versus WT cells (Fig. 2B, lane 3 versus lane 7). Whereas inhibiting histidine biosynthesis with 3-AT elicits an ∼6.8-fold increase in eIF2α-P in the WT, treating L856A cells with 3-AT does not further elevate eIF2α-P (Fig. 2B), indicating that the L856A product is constitutively activated. By contrast, the GCN2c-E803V allele described previously (30) is not fully activated, as 3-AT elicits a significant increase in eIF2α-P (Fig. 2B), and no growth defect is observed in nonstarved cells in this mutant (Fig. 2A).
FIG. 2.
Alanine substitution of L856 constitutively activates Gcn2 in vivo. (A) L856A impairs cell growth dependent on eIF2α phosphorylation. Transformants of gcn2Δ SUI2 strain H1149 or gcn2Δ SUI2-S51A strain H1817 containing empty vector or the indicated plasmid-borne GCN2 alleles were streaked on synthetic complete medium lacking uracil and incubated for 3 days at 30°C. (B) L856A confers constitutive eIF2α phosphorylation in vivo. Duplicate cultures of strains from panel A were grown in synthetic complete medium lacking uracil and histidine to saturation, diluted into fresh medium at an optical density at 600 nm of ∼0.2, and grown for 6 h at 30°C. 3-AT was added at 10 mM to one culture for 1 h before harvesting. Whole cell extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western analysis using the indicated specific antibodies. The Western signals were quantified, and the ratios of eIF2α-P to total eIF2α were calculated. The ratios for the nonstarved cultures were normalized to those measured for the WT strain and indicated as eIF2α-P/eIF2α values below the appropriate lanes for nonstarved cultures; the ratios for the starved cultures were normalized to the nonstarved cultures for each strain and listed as (+)3-AT/(−)3-AT values under the lanes for each strain. (C) Hyperphosphorylation of eIF2α by L856A does not reduce Gcn2 expression in GCD2-K627T cells. Transformants of gcn2Δ GCD2-K627T strain HQY346 carrying the indicated plasmid-borne GCN2 alleles or empty vector were analyzed as described in panel B.
Note that the expression of Gcn2 is reduced by L856A (Fig. 2B, cf. Gcn2 in lanes 3 and 7), which likely reflects the steady-state reduction in general protein synthesis provoked by eIF2α hyperphosphorylation, as observed previously in yeast cells expressing PKR (7). This effect is abolished in GCD2-K627T cells, where inhibition of eIF2B by eIF2(αP) is blunted (26) (Fig. 2C). It is also noteworthy that the level of eIF2α-P declines after several hours in WT but not in L856A cells (data not shown). Together, these observations can likely explain the strong Slg− phenotype of the L856A mutant, with a constitutive level of eIF2α-P similar to that observed in WT cells after 1 h of 3-AT treatment.
Leu-856 operates within a network of hydrophobic residues to maintain Gcn2 latency.
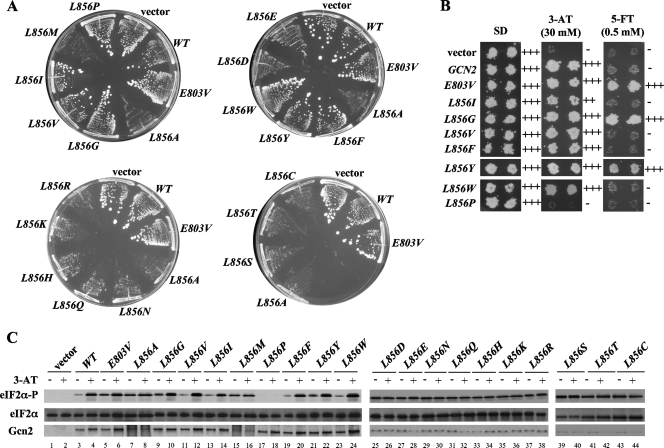
As described in detail below, L856 is embedded in a network of hydrophobic residues that might facilitate its ability to block the reorientation of αC to maintain Gcn2 latency. If so, then a large side chain per se would be insufficient, and only bulky hydrophobic side chains would suffice at position 856 to preserve Gcn2 latency. To test this prediction, we changed L856 to all other 19 amino acids and tested them for activation of Gcn2 in vivo. Remarkably, replacing L856 with 10 out of 11 nonhydrophobic residues (R, K, H, Q, N, D, E, T, S, and C) led to hyperactivation of Gcn2, producing Slg− phenotypes in SUI2 but not SUI2-S51A cells, and constitutive, high-level eIF2α-P in SUI2 cells (Fig. 3A and C and data not shown; summary in Table 3). Note that this list includes a number of residues with bulky side chains, including R, K, D, and E, which despite their large size cannot substitute for Leu at 856 in maintaining Gcn2 latency. Although the glycine substitution did not confer a Slg− phenotype, it did provoke high-level eIF2α-P under nonstarvation conditions (Fig. 3C, lane 9, and Table 3) and confer resistance to the tryptophan analog 5-FT (Fig. 3B). 5-FT resistance (5-FTR) results from the derepression of GCN4 translation and attendant transcriptional induction of tryptophan biosynthetic enzymes under nonstarvation conditions and, hence, is a sensitive indicator of derepressed GCN4 translation (30). Accordingly, GCN2c mutations that do not strongly impair growth, such as the E803V allele (30), confer robust growth on 5-FT medium, whereas GCN2+ cells are 5-FT sensitive (5-FTS) (Fig. 3B). We conclude that L856G also activates Gcn2 under nonstarvation conditions, just not to the extent observed for other nonhydrophobic residues.
FIG. 3.
All but bulky hydrophobic substitutions at L856 constitutively activate Gcn2 in vivo. (A) Transformants of H1149 bearing empty vector or the indicated GCN2 alleles were streaked on synthetic complete medium lacking uracil and incubated for 3 days at 30°C. (B) Duplicate transformants from panel A were replica plated to SD, SD plus 30 mM 3-AT (3-AT), or SD plus 0.5 mM 5-FT (5-FT) medium and incubated for 3 days at 30°C. (C) The indicated strains from panel A were subjected to Western analysis of eIF2α phosphorylation as described in Fig. 2B.
TABLE 3.
Phenotypic analysis of the mutations altering L856a
| GCN2 allele | Growth on SCb | Growth on 3-ATb | Growth on 5-FTc | Relative eIF2α-P/eIF2α ratio value under nonstarvation conditions |
|---|---|---|---|---|
| Vector | ++++ | − | − | None |
| WT | ++++ | +++ | − | 1.0 |
| E803V | ++++ | +++ | +++ | 4.7 |
| L856A | + | ND | ND | 7.0 |
| L856G | ++++ | +++ | +++ | 5.0 |
| L856R | + | ND | ND | 7.0 |
| L856K | + | ND | ND | 10 |
| L856H | + | ND | ND | 8.5 |
| L856D | + | ND | ND | 8.6 |
| L856E | + | ND | ND | 6.7 |
| L856Q | + | ND | ND | 7.4 |
| L856N | + | ND | ND | 6.7 |
| L856T | + | ND | ND | 6.5 |
| L856S | + | ND | ND | 8.0 |
| L856C | + | ND | ND | 6.5 |
| L856P | ++++ | − | − | None |
| L856I | ++++ | ++ | − | 1.0 |
| L856V | ++++ | +++ | − | 1.2 |
| L856M | + | ND | ND | 6.5 |
| L856F | ++++ | +++ | − | 1.3 |
| L856Y | ++++ | +++ | ++ | 4.5 |
| L856W | ++++ | +++ | − | 1.3 |
Transformants of gcn2Δ strain H1149 bearing empty vector or the indicated GCN2 alleles were streaked on synthetic complete medium lacking uracil and scored for average colony size after incubating for 3 days at 30°C. Transformants were replica plated to SD plus 30 mM 3-AT (3-AT) or SD plus 0.5 mM 5-FT (5-FT) and scored for growth after 3 to 4 days at 30°C. The relative eIF2α-P/eIF2α ratios were determined by Western blot analysis of whole cell extracts from cells growing in synthetic complete medium lacking uracil and histidine. The Western blot signals were quantified with a scanner and Scion Image software and normalized to the ratio measured for the WT strain. No eIF2α-P was detectable in the vector and L856P transformants. 3-ATS and 5-FTS could not be measured meaningfully in those mutants with strong Slg− phenotypes; hence, such results are listed as not determined (ND).
++++, colony size or growth of replica-plated patches of cells indistinguishable from that of WT GCN2 cells; −, colony size or patch growth identical to that of gcn2Δ cells; +, ++, and +++, three classes of colony size or patch growth intermediate between that given by gcn2Δ and GCN2 cells.
+++, growth of replica-plated patches indistinguishable from that of GCN2-E803V cells; −, patch growth identical to that of GCN2 cells; + and ++, two classes of patch growth intermediate between that given by GCN2-E803V and GCN2 cells.
Importantly, we found that replacing Leu with the bulky hydrophobic residues Val, Ile, Trp, and Phe preserves essentially WT Gcn2 regulation. Thus, these substitutions do not produce a Slg− phenotype or elevate eIF2α-P in nonstarved cells (Fig. 3A and C and Table 3), nor do they confer 5-FT resistance (Fig. 3B; Table 3). Hence, these four amino acids mimic Leu at 856 and maintain Gcn2 latency under nonstarvation conditions. While L856Y does not confer Slg− (Fig. 3A and Table 3), it does produce 5-FTR (Fig. 3B) and elevates eIF2α-P in nonstarved cells (Fig. 3C, lane 21, and Table 3), indicating a measure of Gcn2 activation comparable to that given by E803V and L856G. By contrast, L856M strongly activates Gcn2 function, conferring Slg− (Fig. 3A) in addition to constitutive eIF2α-P production (Fig. 3C, lane 15; Table 3). We speculate that the polar hydroxyl group of Tyr and the strong conformational flexibility of Met (13, 23) might explain their inability to support the hydrophobic network involving residue 856.
Finally, the proline substitution of L856 differs from all others in that it completely inactivates Gcn2, eliminating eIF2α-P under starvation and nonstarvation conditions alike (Fig. 3C, lanes 17 and 18) and the ability to complement the 3-AT sensitivity (3-ATS) phenotype of the gcn2Δ strain (Fig. 3B and Table 3). Although Pro has a hydrophobic side chain, its inflexibility might prevent the reorientation of its side chain needed to remove the obstruction to αC rotation during kinase activation (cf. L856 in Fig. 1B and C).
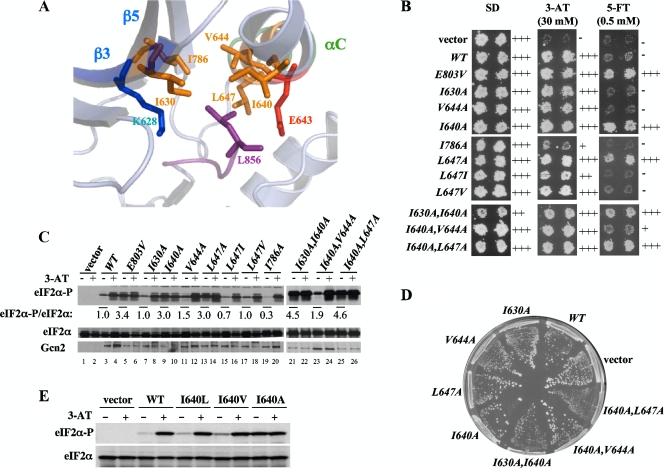
In the inactive Gcn2 KD structure, L856 is surrounded by hydrophobic residues located in β3 (I630), between αB and αC (I640), in αC (V644 and L647), and in β5 (I786) (25) (Fig. 4A). We hypothesized that some of these residues would buttress L856 and facilitate its role in blocking αC rotation. Supporting this idea, we found that the I640A and L647A substitutions in αC conferred 5-FTR and constitutive eIF2α-P levels (Fig. 4B and C, lanes 9 and 13) and that combining these mutations additionally produced a Slg− phenotype in SUI2 (but not in SUI2-S51A) cells (Fig. 4D and data not shown). Although I630A alone had no discernible effect, it conferred Slg− when combined with I640A (Fig. 4B and D). I786A did not activate Gcn2 under nonstarvation conditions; in fact, it increased 3-ATS, indicating defective activation in starved cells. As I786 is only two residues from the gatekeeper residue, this mutation might deform the ATP-binding pocket in the active state, which could obscure its predicted effect in facilitating αC rotation to the active conformation. Indeed, an Asn substitution in the corresponding residue in PKR (I362) also eliminated kinase activity (4). A similar explanation might account for our finding that V644A does not activate Gcn2 (Fig. 4B and D), since this substitution could affect the orientation of the immediately adjacent catalytic residue E643 and its ability to form the salt bridge with K628. Nevertheless, it is striking that Ala substitutions at three of five hydrophobic residues in proximity to L856 confer constitutive activation of Gcn2 function.
FIG. 4.
Leu-856 functions in a network of hydrophobic residues to inhibit Gcn2 activity. (A) Residues making hydrophobic interactions with L856 are displayed on the ribbon diagram of the relevant portion of the Gcn2 KD crystal structure, with stick representations of the side chains (constructed from Protein Data Bank file 1ZYC). (B) Duplicate transformants of H1149 bearing empty vector or the indicated GCN2 alleles were replica plated to SD, SD plus 30 mM 3-AT (3-AT), or SD plus 0.5 mM 5-FT (5-FT) and incubated for 3 days at 30°C. (C) The indicated strains from panel B were subjected to Western analysis of eIF2α phosphorylation as described in Fig. 2B. (D) The indicated strains from panel B were streaked on synthetic complete medium lacking uracil and incubated for 3 days at 30°C. (E) Transformants of H1149 bearing empty vector or the indicated GCN2 alleles were subjected to Western analysis of eIF2α phosphorylation as described in Fig. 2B.
To provide additional evidence that the Leu side chain of L647 makes a hydrophobic interaction with L856 to stabilize the inhibitory conformation, we examined the effects of replacing L647 with other bulky hydrophobic residues. In contrast to the Ala substitution at 647, which activates Gcn2 and confers 5-FTR, both Ile and Val replacements were 5-FTS and did not elevate eIF2α-P in nonstarved cells (Fig. 4B and C, lanes 15 and 17), indicating that they functionally replace Leu in maintaining Gcn2 latency. Similarly, we found that replacing I640 with Leu or Val maintains nearly WT or partial latency, respectively, whereas the Ala substitution strongly activates Gcn2 (Fig. 4E). Thus, residues I640 and L647 both resemble L856 in requiring a bulky hydrophobic side chain to block Gcn2 activation under nonstarvation conditions. Together, our results strongly support the model that the bulky side chain of L856 impedes the rotation of αC to the active position under nonstarvation conditions and that its inhibitory conformation is buttressed by hydrophobic interactions with (at a minimum) I630, I640, and L647.
Disrupting the L856 hydrophobic network bypasses T882 autophosphorylation and tRNA binding for kinase activation.
Besides the obstruction posed by L856, a second feature predicted to stabilize the incorrect rotation of αC is the inappropriate salt bridge between E643 in αC and residue R834 in the catalytic loop of the Gcn2 KD (Fig. 1B). It has been proposed that autophosphorylation of activation loop residue T882 or T887 would displace E643 from R834, destabilizing the inactive orientation of αC and allowing E643 to form the crucial salt bridge with K628 in β3 (25) (Fig. 1C). A prediction for this model is that eliminating the impediment to αC rotation presented by L856 should reduce the requirement for activation loop phosphorylation. To test this prediction, we asked whether the deleterious effect of replacing Thr-882 with nonphosphorylatable Ala can be suppressed by the activating mutation L856A. Indeed, L856A completely suppresses the moderate 3-ATS phenotype of T882A and also confers 5-FTR (signifying constitutive activation) in the L856A T882A double mutant (Fig. 5A). In keeping with its 3-ATS phenotype, the T882A single mutant exhibits reduced levels of eIF2α-P in starved cells (Fig. 5B, cf. lanes 4 and 8). In the L856A T882A double mutant, L856A restores high-level eIF2α-P under starvation conditions and also confers strong eIF2α phosphorylation under nonstarvation conditions (Fig. 5B, lanes 9 and 10). Although L856A completely suppresses the 3-ATS and reduced eIF2α-P phenotypes conferred by T882A, it does not completely bypass the stimulatory effect of T882 phosphorylation, because T882A suppresses the Slg− phenotype conferred by L856A in the double mutant (Fig. 5D). Hence, it appears that T882 is still required for maximum kinase activation, presumably to disrupt the nonproductive salt bridge E643-R834, even when the impediment to αC rotation is diminished by L856A.
FIG. 5.
Disrupting the L856 hydrophobic network reduces the requirement for tRNA binding and autophosphorylation at T882 in Gcn2 activation. (A) H1149 transformants containing the indicated GCN2 alleles or empty vector were replica plated to SD, SD plus 30 mM 3-AT (3-AT), or SD plus 0.5 mM 5-FT (5-FT) and incubated for 3 days at 30°C. (B and C) The indicated strains from panel A were subjected to Western analysis of eIF2α phosphorylation as described in Fig. 2B. (D) Strains from panel A were streaked on synthetic complete medium lacking uracil and incubated for 3 days at 30°C.
Autophosphorylation by Gcn2 is stimulated by tRNA binding to the HisRS domain (43). Accordingly, we asked next whether facilitating αC rotation by L856A would also suppress the activation defect produced by the m2 substitutions (Y1119L R1120L) in conserved motif 2 of the HisRS domain, which impair tRNA binding to Gcn2 (12, 37). Indeed, L856A suppresses the strong 3-ATS phenotype provoked by m2 and confers 5-FTR (Fig. 5A) and elevated eIF2α-P under starvation and nonstarvation conditions (Fig. 5B, lanes 13 and 14 versus lanes 11 and 12) in the L856A m2 double mutant. Importantly, the double mutation I640A L647A, which disrupts the L856 hydrophobic network in a different way, also restores eIF2α phosphorylation and confers 5-FTR in the m2 background (Fig. 5A and C, lanes 7 to 10). The fact that the requirements for motif 2 and T882 in Gcn2 activation are diminished by disrupting the L856 hydrophobic network is consistent with the idea that tRNA binding and autophosphorylation are required to overcome the hydrophobic network and achieve reorientation of αC to the active conformation.
As in the case of the L856A T882A double mutant discussed above, the m2 mutations suppress the Slg− phenotypes of L856A and I640A L647A (Fig. 5D and data not shown). Hence, although L856A and I640A L647A permit kinase activation without robust tRNA binding, a higher level of function can be achieved by these activated mutants if their tRNA binding is intact. This is not surprising considering our previous finding that the GCN2Hyper mutations, which enhance hinge flexibility in the KD, also restore function to the m2 product (Fig. 5B, lanes 15 and 16), and the lethality of these mutations is, in turn, suppressed by m2 (29). We propose that the L856A and Hyper mutations contribute to the active conformation of the KD by facilitating αC rotation and hinge remodeling, respectively, and that both conformational transitions are promoted by T882 phosphorylation and tRNA binding.
Promoting the parallel mode of dimerization constitutively activates Gcn2.
Activation of PKR requires parallel, back-to-back dimerization of the KD (Fig. 1A, right), as the conserved salt bridge or one with opposite polarity at the dimer interface is necessary for PKR function (10). Additional evidence that KD dimerization is crucial for PKR came from the isolation of activating substitutions (L315F, Y404H, and K429R) that restored kinase function in the absence of the extrinsic dimerization domain (the double-stranded RNA binding motifs). These activating substitutions are close to the parallel dimer interface, and two of them (Y404H and K429R) were shown to promote KD dimerization (10). We investigated whether making the equivalent substitutions in the conserved residues of Gcn2 (i.e., L650F, Y825H, and K850R) would activate its function in nonstarved cells, as this would provide evidence that parallel dimerization of the KD is a regulated step in Gcn2 activation that is enhanced during amino acid starvation.
Remarkably, L650F and K850R both conferred 5-FTR phenotypes (Fig. 6A) and significantly increased eIF2α phosphorylation, and combining the mutations produced an even larger increase in eIF2α-P levels under nonstarvation conditions (Fig. 6B, lanes 7, 11, and 13). Furthermore, these mutations increase the kinase activity of the reciprocal salt bridge R594D D598R mutant. Thus, whereas D598R clearly rescues the ability of the R594D mutant to phosphorylate eIF2α-P and grow on 3-AT, the resulting R594D D598R double mutant performs below the WT level in both respects (Fig. 6A and B). Importantly, the activating double mutation L650F K850R restored high-level 3-AT resistance and eIF2α-P in the R594D D598R L650F K850R quadruple mutant (Fig. 6A and B, lanes 15 to 18). These findings strongly suggest that the L650F K850R double mutation activates Gcn2 by enhancing parallel KD dimerization and that strengthening the parallel dimer interface bypasses the need for amino acid starvation for activation of Gcn2.
FIG. 6.
Enhancing the parallel dimer interface reduces the requirement for T882 autophosphorylation and tRNA binding in Gcn2 activation. (A) H1149 transformants containing the indicated GCN2 alleles or empty vector were replica plated to SD, SD plus 30 mM 3-AT (3-AT), or SD plus 0.5 mM 5-FT (5-FT) and incubated for 3 days at 30°C. (B and C) Strains indicated in panel A were subjected to Western analysis of eIF2α phosphorylation as described in Fig. 2B.
We reasoned that if tRNA binding and autophosphorylation promote Gcn2 function in part by enhancing parallel KD dimerization, then the L650F K850R mutation should diminish the requirements for the m2 motif and T882 for high-level Gcn2 activation. Confirming this prediction, the L650F K850R mutation restored nearly WT function to the otherwise deficient T882A product (Fig. 6A and B, lanes 20 and 22) and partially rescued the kinase activity of the m2 product (Fig. 6A and B, lanes 24 and 26). These results fit with the idea that parallel KD dimerization is a step on the Gcn2 activation pathway that is stimulated by tRNA binding and T882 autophosphorylation.
In addition to the R594-D598 salt bridge, we identified two other amino acids in Gcn2 corresponding to residues involved in PKR dimerization that are important for Gcn2 function. Y625 of Gcn2 corresponds to Y293 in PKR, one of three residues that comprise the critical H bond triad D289-Y293-Y323 at the PKR dimer interface (6, 10). N618 of Gcn2 corresponds to H286 in PKR, which makes backbone and side chain contacts with C326 in the other protomer of the PKR dimer (6). Consistent with the idea that parallel KD dimerization is important for Gcn2 activation, the GCN2 mutations N618A and Y625A strongly impair kinase function, conferring a 3-ATS phenotype and eliminating detectable eIF2α-P (Fig. 6A and C).
Mutating residues at the antiparallel dimer interface do not constitutively activate Gcn2.
We concluded above that parallel KD dimerization is a step on the GCN2 activation pathway that is stimulated by tRNA binding and T882 autophosphorylation. This implies that the two KDs in the inactive form of a full-length Gcn2 dimer would exist in a monomeric state or be dimerized in a different conformation. The inactive form of the Gcn2 KD formed stable dimers in solution and repeatedly crystallized as a symmetric homodimer in the antiparallel configuration, burying an extensive (∼2,600-Å) solvent-accessible surface at the dimer interface (25). This led us to consider that the inactive form of full-length Gcn2 might contain the KDs dimerized in antiparallel configuration and that activation would require isomerization to the parallel mode of KD dimerization. If so, then mutations that destabilize the antiparallel dimer interface should confer constitutive activation of Gcn2. To test this possibility, we examined the effects of Ala substitutions in seven different residues involved in side chain interactions at the antiparallel dimer interface including N651, Y658, S597, L646, S649, L663, and R768 (25). We also examined the S597A R768A double mutant because substituting these two residues should eliminate four different interactions at the antiparallel dimer interface (25). Importantly, none of these substitutions conferred 5-FTR or altered growth on 3-AT medium (data not shown). These results do not support the possibility that antiparallel dimerization promotes Gcn2 latency. Perhaps antiparallel KD dimerization would be disfavored in full-length Gcn2 dimers, owing to parallel dimerization of the adjacent HisRS domains, for which there is considerable evidence (27). In this event, activation would likely involve the transition from monomeric KDs to the parallel KD dimer, similar to that proposed previously for PKR activation (10).
Disrupting the L856 hydrophobic network or reducing hinge rigidity reduces the requirement for a parallel KD dimer interface.
We hypothesized that forming a parallel dimer interface would facilitate reorientation of αC or remodeling of the hinge to achieve the proper alignment of active site residues and facilitate ATP binding. Parallel KD dimerization could directly influence αC reorientation because αC residues participate in the KD dimer interface of PKR (6). If our hypothesis is correct, then the L856A, I640A, L647A, and Hyper activating mutations should suppress mutations that weaken the dimer interface. Remarkably, L856A suppressed the 3-ATS and failure to induce eIF2α-P on 3-AT treatment conferred by the destruction of the salt bridge by R594D and by the N618A mutation at the predicted parallel dimer interface (Fig. 7A and B, cf. lanes 10 and 12 and lanes 16 and 18). Similarly, disrupting the hydrophobic network with the I640A L647A mutations fully suppressed the R594D mutation (data not shown). The Hyper mutations restored even higher levels of kinase activation in the presence of R594D and N618A, conferring 5-FTR and constitutively elevated eIF2α-P (Fig. 7A and B, lanes 13 and 19 versus lane 3). Thus, the requirement for parallel KD dimerization is greatly reduced by substitutions that facilitate reorientation of αC (L856A, I640A, and L647A) or increase flexibility of the hinge (Hyper). This finding supports the idea that parallel dimerization promotes these activating conformational changes in the Gcn2 KD. Like the findings above, the R594D and N618A mutations at the predicted dimer interface suppressed the Slg− phenotype of L856A and the lethality of the Hyper mutations, indicating that parallel dimerization is still required to realize the full potential of these activating mutations (Fig. 7C).
FIG. 7.
The L856A and Hyper activating mutations reduce the requirement for the parallel dimer interface in Gcn2 activation. (A) H1149 transformants containing the indicated GCN2 alleles or empty vector were replica plated to SD, SD plus 30 mM 3-AT (3-AT), or SD plus 0.5 mM 5-FT (5-FT) and incubated for 3 days at 30°C. (B) The indicated strains from panel A were subjected to Western analysis of eIF2α phosphorylation as described in Fig. 2B. (C) Strains from panel A were streaked on synthetic complete medium lacking uracil and incubated for 3 days at 30°C.
DISCUSSION
In this report, we provide strong evidence that the latency of Gcn2 in nonstarved cells depends on a network of hydrophobic residues comprised of L856 in the activation loop and hydrophobic residues in strand β3, strand β5, or helix αC. The crystal structure of inactive Gcn2 reveals a ∼90° axial rotation of helix αC, which blocks formation of the E643-K628 salt bridge crucial for positioning the triphosphate moiety of ATP. L856 appears to impede the rotation of αC to the position observed in active KD structures that is permissive for the E643-K628 salt bridge (25). The bulky hydrophobic side chain of L856 packs against the hydrophobic side chains of I640, V644, and L647 in αC, I630 in β3, and I786 in β5 (Fig. 4A), leading us to predict that the ability of L856 to block reorientation of αC depends on its insertion into a hydrophobic pocket comprised of these other residues. Supporting this prediction, our exhaustive substitution analysis of L856 revealed that replacing this residue with Ala, Gly, or any amino acid with a polar or charged side chain leads to hyperactivation of Gcn2 in vivo, bypassing the amino acid starvation signal and producing a high constitutive level of eIF2α phosphorylation that inhibits cell growth in nutrient-replete medium. Remarkably, the replacement of L856 with Ile, Val, Phe, and Trp produced little or no increase in Gcn2 function, indicating that only these four bulky hydrophobic residues can substitute for Leu-856 to maintain WT latency. Substitutions of residues comprising the hydrophobic pocket also activate Gcn2, as the double substitutions L647A I640A and I630A I640A elicited eIF2α hyperphosphorylation and Slg− phenotypes approaching those of L856A. These results provide compelling in vivo evidence that the L856 hydrophobic network is crucial for Gcn2 latency in nonstarved cells.
It could be argued that the activating substitutions we identified in residues comprising the hydrophobic pocket stimulate Gcn2 function by a mechanism unrelated to their proposed role in buttressing the inhibitory orientation of L856. We consider this unlikely for several reasons. First, it should be recalled that we targeted these residues for mutagenesis precisely because they make direct interactions with L856 in the crystal structure of the inactive Gcn2 KD (25). Second, we found that substitutions at L647 conform to the rule we established for activating substitutions at L856, in that replacing L647 with Ala activates Gcn2, whereas its replacement with another bulky hydrophobic residue, Ile or Val, maintains Gcn2 latency. Similarly, we found that replacing I640 with Leu or Val maintains nearly WT or partial latency, respectively, whereas the Ala substitution strongly activates Gcn2. Thus, residues I640 and L647 resemble L856 in requiring a bulky hydrophobic side chain to block Gcn2 activation under nonstarvation conditions. Third, we found that Ala substitutions at L647 and I640 individually conferred 5-FTR and no Slg− phenotype, but they produced a synthetic Slg− phenotype when combined in the same mutant. I640A and I630A also produced a Slg− phenotype only in the double mutant. Such synthetic interactions frequently indicate that both single mutations partially impair the same pathway or function (17). Finally, we showed that the I640A L647A double mutation resembles L856A by strongly suppressing the activation defect conferred by the m2 mutation. Such suppression is not a general feature of activating mutations in GCN2, as most GCN2c mutations remain dependent on the m2 motif for efficient tRNA binding for strong kinase activation (29). Hence, the weight of the evidence supports the idea that the inhibitory conformation of L856 is buttressed by its hydrophobic interactions with (at a minimum) I630, I640, and L647.
The crystal structure of inactive Gcn2 predicts that the inactive orientation of αC is also stabilized by the incorrect salt bridge formed between E643 (in αC) and R834 in the catalytic loop (25) (Fig. 1B). As proposed previously (20), autophosphorylation of T882 in the activation loop should destabilize the incorrect rotation of αC by neutralizing the positive charge of R834 and disrupting the inhibitory E643-R834 salt bridge (Fig. 1B). Supporting this hypothesis, the L856A mutation suppresses the activation defect conferred by the autophosphorylation site mutation T882A. Thus, it appears that decreasing the impediment to αC reorientation by disrupting the L856 hydrophobic network helps to overcome the second impediment posed by the inhibitory E643-R834 salt bridge in the T882A mutant, where the latter cannot be disrupted by autophosphorylation. T882 phosphorylation still contributes to Gcn2 activation when the L856 hydrophobic network is impaired, which is fully consistent with the idea that the hydrophobic network and E643-R834 salt bridge both contribute to impeding the reorientation of αC to the active conformation.
It is highly interesting that L856 corresponds to a key residue in the epidermal growth factor receptor (EGFR), L858, which appears to help buttress an outward displacement of αC in the inactive form of this kinase (38). Moreover, mutating L858 to Arg activates EGFR in vitro, and this mutation is found frequently in a subset of lung cancers (40, 41). In the case of inactive EGFR, L858 resides within a helical turn in the activation loop that seems to force αC into the incorrect position, and L858 makes hydrophobic contacts with other residues in the KD interior. Presumably, the charged side chain introduced by the activating substitution L858R is incompatible with these hydrophobic interactions and destabilizes the helical structure needed to displace αC. However, most of the hydrophobic residues interacting with L858 in inactive EGFR, located in β2, β3, β5, and the activation loop (22), are distinct from those in the hydrophobic pocket surrounding L856 in inactive Gcn2. Unlike in Gcn2, residues in αC itself do not participate in the EGFR hydrophobic pocket, and two Leu residues in the activation loop helical turn in EGFR (L861 and L862) are not conserved, and do not participate, in the hydrophobic pocket of Gcn2. Hence, while the equivalent Leu residue plays a key role in both Gcn2 and EGFR latency, the hydrophobic pocket that supports its inhibitory function is constituted quite differently in the two kinases.
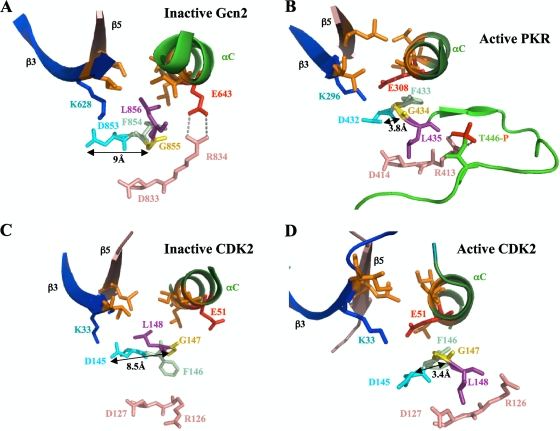
Azam et al. (2) recently proposed an alternative mechanism for the role of L858 in maintaining the latency of EGFR through destabilizing the “hydrophobic spine,” which links the N and C lobes in the active conformation of EGFR and numerous other protein kinases (21). The equivalent Leu residues in ABL and SRC also seem to participate in dismantling the hydrophobic spines in the inactive forms of these kinases (2). In particular, L403 in inactive ABL interacts with the Phe residue of the DFG motif (F401) in the “DFG-out” conformation, where F401 is flipped out of the active site. Furthermore, the DFG Asp is not hydrogen bonded to the DFG Gly, so that inactive ABL lacks an interaction thought to be critical for proper positioning of the DFG Asp for catalysis (21). It is unclear whether a similar mechanism applies to the role of L856 in Gcn2 latency. Although the DFG Asp and Gly are too far apart (∼9 Å) to hydrogen bond in the inactive Gcn2 KD (D853 and G855; Fig. 8A), we found that the hydrophobic spine appears to be intact in inactive Gcn2 (25).
FIG. 8.
Different orientations of the DFG + 1 Leu residue correlate with distinct positions of the DFG Asp in inactive versus active forms of eIF2α kinases and CDK2. In the structures of the inactive forms of Gcn2 (A) and CDK2 (C), interaction of the DFG + 1 Leu residue (L856 or L148, respectively, shown in purple) with hydrophobic residues in β3, β5, and αC (all in orange) correlates with an incorrect position of the Asp (in cyan) of the DFG motif, wherein its ∼9-Å separation from the DFG Gly residue (shown in yellow) precludes H bonding. The proposed disruption of the hydrophobic network during kinase activation of PKR (B) (considered a model for activated Gcn2) and CDK2 (D) might help to achieve the correct position of the DFG Asp and its H bonding to the DFG Gly residue.
In the PKR KD, all but one of the residues corresponding to the L856 hydrophobic network in Gcn2 are perfectly conserved or (in the case of Gcn2 I630) replaced by a different bulky hydrophobic amino acid. Thus, a hydrophobic network involving the equivalent of L856 (L435) might contribute to the latency of PKR in its monomeric state in the manner described here for Gcn2. In the crystal structure of active PKR, L435 is oriented away from the putative hydrophobic pocket (6) (Fig. 8B), adopting essentially the same position as the corresponding “DFG + 1” residue observed in the active conformations of many different kinases (21). Moreover, the DFG Asp and Gly are close enough (3.8 Å) to hydrogen bond in active PKR (D432 and G434; Fig. 8B). In activated Gcn2, it seems likely that L856 will isomerize to the outward position observed for L435 in active PKR, eliminating the impediment to αC reorientation and achieving the stereotypical active conformation of the DFG + 1 motif (21). As shown in Fig. 8C, the inactive form of kinase CDK2 (33) shows the unfavorable positions of the Asp (D145) and + 1 (L148) residues of the DFG + 1 motif, similar to those present in (inactive) Gcn2 (cf. Fig. 8A and C), whereas the active form of CDK2 (3) displays the favorable conformation of the DFG + 1 motif (cf. Fig. 8B and D) (21). Interestingly, we realized that inactive CDK2 also contains a hydrophobic network involving L148 that is highly similar to that identified here in Gcn2, with residues I35, I52, and L55 corresponding to I630, V644, and L647 in Gcn2 (cf. Fig. 8A and C), which might contribute to the latency of this critical kinase as well.
Activation of Gcn2 in amino acid-starved cells requires the binding of uncharged tRNA to the HisRS-like domain, dependent on motif 2. If the postulated reorientation of αC is a crucial step in the activation pathway, then decreasing the impediment to αC reorientation by disrupting the L856 hydrophobic network should reduce the requirement for motif 2. Our results confirm this prediction by showing that the L856A and I640A L647A activating mutations restore kinase function to the m2 mutant. As would be expected if αC reorientation is not the only conformational change during kinase activation, the m2 substitutions, in turn, reduce the functions of the L856A and I640A L647A proteins. We propose that stimulatory input from uncharged tRNA and the HisRS domain is still required to remodel the hinge and expose the ATP-binding site even after a key impediment to αC reorientation has been eliminated by disrupting the L856 hydrophobic network. The lethality of the Hyper mutations is likewise suppressed by the m2 mutation (29), which can now be explained by proposing that tRNA-dependent reorientation of αC is still necessary for full kinase activation even when hinge rigidity and obstruction of the ATP-binding pocket has been diminished by the Hyper substitutions. This inference is supported by the crystal structure of the R794G KD, in which the hinge is remodeled but αC remains in the inactive orientation, presumably because T882 is unphosphorylated (25).
The results presented here also provide strong evidence that adopting the PKR mode of parallel KD dimerization (6) is a regulated step in the Gcn2 activation pathway. Whereas inactive Gcn2 displays a distinct, antiparallel mode of dimerization (25), the functional importance of Gcn2 residues (R594-D598) corresponding to the crucial salt bridge at the PKR parallel interface argues that Gcn2 assumes the PKR mode of dimerization in the active state (11). Supporting this conclusion, Gcn2 is constitutively activated by substitutions (L650F and K850R) corresponding to the same substitutions in PKR that stimulate dimerization and kinase function in the absence of the extrinsic (double-stranded RNA binding) dimerization domain (10). The fact that these mutations activate Gcn2 in nonstarved cells suggests that stabilizing the parallel dimer interface normally occurs in WT Gcn2 upon amino acid starvation. Supporting this interpretation, the L650F K850R double mutation restores significant kinase activity to the m2 mutant, indicating that stabilization of the parallel dimer interface decreases the requirement for strong tRNA binding in Gcn2 activation.
The L650F and K850R substitutions at the predicted dimer interface also restore function to the T882A autophosphorylation mutant. One way to interpret this result is to propose that autophosphorylation promotes parallel dimerization, so that the requirement for T882 is diminished by mutations that strengthen the parallel dimer interface. Indeed, there is evidence that autophosphorylation is a prerequisite for PKR KD dimerization (10). A second, not mutually exclusive, explanation is that parallel dimerization and autophosphorylation both promote the same activating conformational transition in the KD, so that enhancing dimerization lessens the importance of autophosphorylation in achieving the active conformation. If T882 autophosphorylation promotes reorientation of αC, as argued above, then this second interpretation would imply that αC reorientation is also stimulated by parallel dimerization. In fact, this conclusion fits perfectly with our finding that L856A and I640A L647A, which facilitate αC reorientation, restore activity to the dimerization-defective mutant R594D lacking the critical salt bridge at the parallel dimer interface. Therefore, it seems very likely that autophosphorylation stabilizes reorientation of αC indirectly by enhancing parallel dimerization in addition to dissolving the inhibitory salt bridge E643-R834 by the charge neutralization mechanism mentioned above (Fig. 1B and C).
Our proposal that parallel dimerization facilitates αC reorientation is consonant with the fact that αC in its proper location constitutes a portion of the parallel dimer interface in active PKR (6). Thus, it is easy to imagine that parallel dimerization helps to push αC into the correct orientation. Indeed, a related mechanism is supported for EGFR, in which asymmetric KD dimerization achieves αC reorientation, wherein the C lobe of one protomer interacts with the N lobe of the second protomer at an interface containing αC in its inwardly rotated, active conformation (41). This is also analogous to a role played by cyclin in cyclin-dependent kinase, wherein inwardly rotated αC is present at the cyclin-KD interface (19).
The role of the hinge region in Gcn2 latency represents another layer of control not described for other kinases. The Hyper activating mutations, which alter the hinge conformation and expose the ATP-binding site (25), elicit a strong recovery of activity in the face of defects in tRNA binding (m2 substitutions), parallel dimerization (R594D), or autophosphorylation (T882A). These findings imply that, in WT Gcn2, tRNA binding, autophosphorylation, and parallel dimerization all combine to stabilize the favorable hinge conformation mimicked by the Hyper substitutions. We argued above that these events also promote the reorientation of αC. Although the R794G KD crystal structure displays an altered hinge conformation in the presence of the incorrect orientation of αC (25), it is possible that αC reorientation actually facilitates hinge remodeling. In fact, we envision that these various conformational transitions are mutually reinforcing to provoke a concerted switch from inactive to active conformation on tRNA binding and that the ensuing autophosphorylation of T882 locks in the fully activated state.
Integrating our current findings with previous results allows us to propose the hypothetical model for Gcn2 activation shown in Fig. 9. In nonstarved cells with low levels of uncharged tRNA, we propose that the Gcn2 KD exists in equilibrium between two inactive states. In state A, the KDs of the full-length Gcn2 dimer are monomeric and contain an improperly rotated αC and rigid hinge, incompatible with ATP binding or catalysis. (The KDs might undergo antiparallel dimerization in this state; however, this possibility is disfavored by our finding that destabilizing substitutions at the antiparallel dimer interface do not activate Gcn2.) State B contains a parallel KD dimer with properly oriented αC that still binds ATP weakly and is defective for catalysis, owing to hinge rigidity. Binding of uncharged tRNA reduces hinge rigidity via interaction with the HisRS domain, establishing a new equilibrium between the monomeric (state C) and parallel (state D) dimer forms, which both bind ATP more tightly and possess the requisite interlobe flexibility needed for catalysis. Stochastic transition from the monomeric (state C) to the parallel (state D) mode of dimerization elicits rapid autophosphorylation of T882 in state D to produce a highly stable, fully activated conformation (state E). According to this model, the Hyper substitutions shift the equilibrium from states A and B to C and D, even at the low-level tRNA occupancies of the HisRS domain in nonstarved cells. Given that the Hyper substitutions suppress the deleterious effects of alterations at the parallel dimer interface, they also appear to shift the equilibrium toward the parallel dimer in the conversion of state C to state D to enhance the formation of fully activated Gcn2 (state E). Disrupting the hydrophobic network by L856A strongly shifts the equilibrium from state A to state B in nonstarved cells, so that even the low occupancy of bound tRNA under nonstarvation conditions provokes substantial state B to D conversion and, with ensuing autophosphorylation, fully activated Gcn2 (state E). States A and C in our model correspond to the crystal structures of the inactive forms of the WT and R794G KDs determined previously (25). In the future, it would be interesting to determine whether the KDs of the L856A mutant and L856A R794G double mutant crystallize in states B and E, respectively, with parallel dimer interfaces and αC oriented in the active conformation, and (for state E) autophosphorylation of T882.
FIG. 9.
Hypothetical model for multistep activation of Gcn2 by uncharged tRNA. The mode of KD dimerization and disposition of αC and key residues controlling ATP binding and catalysis are indicated schematically for different states in the proposed activation pathway for Gcn2. In all states, we envision that Gcn2 is dimerized through the self-interaction of the C-term (not shown), and the model depicts only the disposition of the KDs within the full-length dimer. (A and B) At the low levels of uncharged tRNA in nonstarved cells, Gcn2 exists in equilibrium between two inactive conformations, with that shown in panel A being highly favored; it contains nondimerized KDs in which ATP binding is hindered by a closed conformation of the N and C lobes and by N793, which forms a flap over the ATP-binding pocket, and catalysis is blocked by hinge rigidity, the incorrect rotation of αC, and the attendant E643-R834 inhibitory salt bridge. (B) The KDs adopt the parallel dimer conformation in which αC is properly oriented, but catalysis and ATP binding are still hindered by hinge rigidity, the closed conformation of N and C lobes, and the N793 flap. (C to E) Binding of tRNA to the HisRS domain occurs in starved cells and shifts the equilibrium to the three different states, of which the fully activated form (E) is highly favored. In all three of these states, tRNA binding to the HisRS domain has derigidified the hinge, removing the flap over the ATP-binding site and increasing interlobe flexibility to expand the separation between the N and C lobes. (C) This state is inactive, owing to the absence of parallel dimerization of the KDs and incorrect rotation of αC. (D) This state is active, containing the proper conformation of αC and the crucial K628-E643 salt bridge, and is capable of autophosphorylation to produce the fully functional kinase locked into the active conformation (E).
Acknowledgments
We are indebted to Anil K. Padyana for suggesting the likely importance of L856 in Gcn2 latency and to Frank Sicheri for helpful suggestions.
This work was supported by the Intramural Research Program of the NIH.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Adams, J., P. Huang, and D. Patrick. 2002. A strategy for the design of multiplex inhibitors for kinase-mediated signaling in angiogenesis. Curr. Opin. Chem. Biol. 6486-492. [DOI] [PubMed] [Google Scholar]
- 2.Azam, M., M. A. Seeliger, N. S. Gray, J. Kuriyan, and G. Q. Daley. 2008. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat. Struct. Mol. Biol. 151109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, N. R., M. E. Noble, J. A. Endicott, and L. N. Johnson. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1438-443. [DOI] [PubMed] [Google Scholar]
- 4.Cai, R., and B. R. Williams. 1998. Mutations in the double-stranded RNA-activated protein kinase insert region that uncouple catalysis from eIF2alpha binding. J. Biol. Chem. 27311274-11280. [DOI] [PubMed] [Google Scholar]
- 5.Costa-Mattioli, M., D. Gobert, H. Harding, B. Herdy, M. Azzi, M. Bruno, M. Bidinosti, C. Ben Mamou, E. Marcinkiewicz, M. Yoshida, H. Imataka, A. C. Cuello, N. Seidah, W. Sossin, J. C. Lacaille, D. Ron, K. Nader, and N. Sonenberg. 2005. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 4361166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dar, A. C., T. E. Dever, and F. Sicheri. 2005. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122887-900. [DOI] [PubMed] [Google Scholar]
- 7.Dever, T. E., J. J. Chen, G. N. Barber, A. M. Cigan, L. Feng, T. F. Donahue, I. M. London, M. G. Katze, and A. G. Hinnebusch. 1993. Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 in the GCN4 translational control mechanism of yeast. Proc. Natl. Acad. Sci. USA 904616-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dever, T. E., A. C. Dar, and F. Sicheri. 2007. The elF2α kinases, p. 319-344. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. D. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68585-596. [DOI] [PubMed] [Google Scholar]
- 10.Dey, M., C. Cao, A. C. Dar, T. Tamura, K. Ozato, F. Sicheri, and T. E. Dever. 2005. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122901-913. [DOI] [PubMed] [Google Scholar]
- 11.Dey, M., C. Cao, F. Sicheri, and T. E. Dever. 2007. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J. Biol. Chem. 2826653-6660. [DOI] [PubMed] [Google Scholar]
- 12.Dong, J., H. Qiu, M. Garcia-Barrio, J. Anderson, and A. G. Hinnebusch. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6269-279. [DOI] [PubMed] [Google Scholar]
- 13.Gellman, S. H. 1991. On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry 306633-6636. [DOI] [PubMed] [Google Scholar]
- 14.Guo, F., and D. R. Cavener. 2007. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 5103-114. [DOI] [PubMed] [Google Scholar]
- 15.Hao, S., J. W. Sharp, C. M. Ross-Inta, B. J. McDaniel, T. G. Anthony, R. C. Wek, D. R. Cavener, B. C. McGrath, J. B. Rudell, T. J. Koehnle, and D. W. Gietzen. 2005. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 3071776-1778. [DOI] [PubMed] [Google Scholar]
- 16.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 61099-1108. [DOI] [PubMed] [Google Scholar]
- 17.Hartman, J. L. T., B. Garvik, and L. Hartwell. 2001. Principles for the buffering of genetic variation. Science 2911001-1004. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch, A. G. 1996. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2, p. 199-244. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Jeffrey, P. D., A. A. Russo, K. Polyak, E. Gibbs, J. Hurwitz, J. Massague, and N. P. Pavletich. 1995. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376313-320. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, L. N., M. E. M. Noble, and D. J. Owen. 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85149-158. [DOI] [PubMed] [Google Scholar]
- 21.Kornev, A. P., N. M. Haste, S. S. Taylor, and L. F. Eyck. 2006. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 10317783-17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, A., E. T. Petri, B. Halmos, and T. J. Boggon. 2008. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J. Clin. Oncol. 261742-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovell, S. C., J. M. Word, J. S. Richardson, and D. C. Richardson. 2000. The penultimate rotamer library. Proteins 40389-408. [PubMed] [Google Scholar]
- 24.Narasimhan, J., K. A. Staschke, and R. C. Wek. 2004. Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J. Biol. Chem. 27922820-22832. [DOI] [PubMed] [Google Scholar]
- 25.Padyana, A. K., H. Qiu, A. Roll-Mecak, A. G. Hinnebusch, and S. K. Burley. 2005. Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2alpha protein kinase GCN2. J. Biol. Chem. 28029289-29299. [DOI] [PubMed] [Google Scholar]
- 26.Pavitt, G. D., W. Yang, and A. G. Hinnebusch. 1997. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 171298-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu, H., J. Dong, C. Hu, C. S. Francklyn, and A. G. Hinnebusch. 2001. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J. 201425-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu, H., M. T. Garcia-Barrio, and A. G. Hinnebusch. 1998. Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell. Biol. 182697-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, H., C. Hu, J. Dong, and A. G. Hinnebusch. 2002. Mutations that bypass tRNA binding activate the intrinsically defective kinase domain in GCN2. Genes Dev. 161271-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez, M., R. C. Wek, C. R. Vazquez de Aldana, B. M. Jackson, B. Freeman, and A. G. Hinnebusch. 1992. Mutations activating the yeast eIF-2α kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 125801-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, G. A., and G. Schatz. 1982. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J. Biol. Chem. 25713056-13061. [PubMed] [Google Scholar]
- 32.Romano, P. R., M. T. Garcia-Barrio, X. Zhang, Q. Wang, D. R. Taylor, F. Zhang, C. Herring, M. B. Mathews, J. Qin, and A. G. Hinnebusch. 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol. 182282-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulze-Gahmen, U., H. L. De Bondt, and S. H. Kim. 1996. High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design. J. Med. Chem. 394540-4546. [DOI] [PubMed] [Google Scholar]
- 34.Vattem, K. M., and R. C. Wek. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 10111269-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wek, R. C., B. M. Jackson, and A. G. Hinnebusch. 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. USA 864579-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wek, R. C., M. Ramirez, B. M. Jackson, and A. G. Hinnebusch. 1990. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol. Cell. Biol. 102820-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 154497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood, E. R., A. T. Truesdale, O. B. McDonald, D. Yuan, A. Hassell, S. H. Dickerson, B. Ellis, C. Pennisi, E. Horne, K. Lackey, K. J. Alligood, D. W. Rusnak, T. M. Gilmer, and L. Shewchuk. 2004. A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 646652-6659. [DOI] [PubMed] [Google Scholar]
- 39.Yon, J., and M. Fried. 1989. Precise gene fusion by PCR. Nucleic Acids Res. 174895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun, C. H., T. J. Boggon, Y. Li, M. S. Woo, H. Greulich, M. Meyerson, and M. J. Eck. 2007. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, X., J. Gureasko, K. Shen, P. A. Cole, and J. Kuriyan. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 1251137-1149. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, D., L. R. Palam, L. Jiang, J. Narasimhan, K. A. Staschke, and R. C. Wek. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 2837064-7073. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, S., A. Y. Sobolev, and R. C. Wek. 1996. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J. Biol. Chem. 27124989-24994. [DOI] [PubMed] [Google Scholar]