Abstract
Deubiquitinating enzymes (DUBs) control the ubiquitination status of proteins in various cellular pathways. Regulation of the activity of DUBs, which is critically important to cellular homoeostasis, can be achieved at the level of gene expression, protein complex formation, or degradation. Here, we report that ubiquitination also directly regulates the activity of a DUB, ataxin-3, a polyglutamine disease protein implicated in protein quality control pathways. Ubiquitination enhances ubiquitin (Ub) chain cleavage by ataxin-3, but does not alter its preference for K63-linked Ub chains. In cells, ubiquitination of endogenous ataxin-3 increases when the proteasome is inhibited, when excess Ub is present, or when the unfolded protein response is induced, suggesting that the cellular functions of ataxin-3 in protein quality control are modulated through ubiquitination. Ataxin-3 is the first reported DUB in which ubiquitination directly regulates catalytic activity. We propose a new function for protein ubiquitination in regulating the activity of certain DUBs and perhaps other enzymes.
Keywords: ataxin-3, deubiquitinating enzyme, post-translational modification, spinocerebellar ataxia type 3, ubiquitin
Introduction
Post-translational modification of proteins by ubiquitin (Ub) regulates many cellular events. When conjugated to a protein, Ub targets it to specific subcellular compartments or to macromolecular complexes, such as the proteasome, achieving this essentially by altering protein–protein interactions. An 8 kDa globular protein, Ub, is covalently attached to proteins through the concerted action of three proteins (E1–E2–E3). The Ub-activating enzyme (E1) transfers Ub to an E2-conjugating enzyme. Through a Ub ligase (E3), Ub is ultimately transferred to a lysine (K) residue of either a target protein or another Ub, forming an isopeptide bond. Proteins can be mono-ubiquitinated (one Ub linked to a target protein), multi-mono-ubiquitinated (two or more single Ub linked to different lysine residues of a protein) or poly-ubiquitinated (one or more Ub chains linked to a protein). As Ub itself contains seven lysines, different chain linkages can be formed by the direct attachment of one Ub to another. The best studied among these are K48-linked chains, which target proteins for proteasomal degradation (Thrower et al, 2000). Other linkages, for example, K63-linked Ub, are implicated in other processes, including DNA repair, the NFκB pathway, Lewy body and aggresome formation, and autophagy (Arnason and Ellison, 1994; Hofmann and Pickart, 1999; Deng et al, 2000; Pickart and Fushman, 2004; Chen, 2005; Lim et al, 2005; Tan et al, 2007).
Protein ubiquitination is reversed by deubiquitinating enzymes (DUBs). Nearly 100 DUBs are encoded by the human genome (Nijman et al, 2005). Although relatively little is known about most DUBs, they clearly function as more than Ub recyclers. DUBs have important functions in numerous cellular pathways: from DNA transcription to protein degradation, from cell division to death. DUBs perform their functions by cleaving Ub from proteins, altering the length or type of Ub chains, or disassembling untethered Ub chains (reviewed by Amerik and Hochstrasser, 2004; Nijman et al, 2005; Ventii and Wilkinson, 2008). DUB activity can be regulated at the level of transcription, degradation, complex formation, or phosphorylation (Amerik and Hochstrasser, 2004; Nijman et al, 2005; Ventii and Wilkinson, 2008; Yao et al, 2008).
Several DUBs have been reported to be ubiquitinated (Shen et al, 2005; Wada and Kamitani, 2006; Fernandez-Montalvan et al, 2007; Meray and Lansbury, 2007; Todi et al, 2007a), but whether ubiquitination of DUBs directly regulates their activity has not been reported. The possibility that DUB activity could be regulated by ubiquitination is suggested by studies of other enzymes involved in Ub or Ub-like pathways. For example, the activity of cullin-based Ub ligases is enhanced through their modification by the Ub-like protein Nedd8 (Parry and Estelle, 2004; Duda et al, 2008). Ubiquitination of the E2 Cdc34 inhibits its activity (Scaglione et al, 2007). Substrate discrimination for Ubc9, the E2 for the Ub-like modifier SUMO, is regulated by its own SUMOylation (Knipscheer et al, 2008). Finally, SUMOylation of the DUB USP25 reduces its enzymatic activity by interfering with the ability of USP25 to interact with Ub (Meulmeester et al, 2008). Here, we report that the catalytic activity of a DUB, ataxin-3, is directly enhanced by ubiquitination.
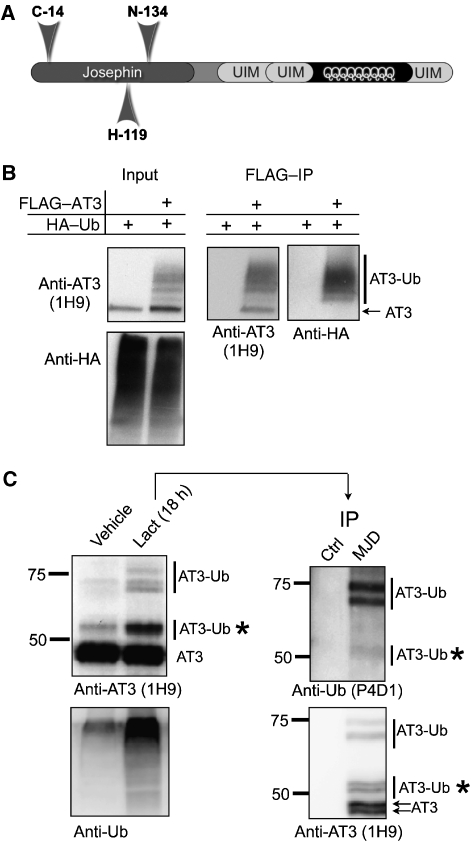
Ataxin-3 (AT3; Figure 1A) is a DUB implicated in Ub-dependent protein quality control (Wang et al, 2000, 2006; Burnett et al, 2003; Doss-Pepe et al, 2003; Chai et al, 2004; Mao et al, 2005; Nicastro et al, 2005; Warrick et al, 2005; Zhong and Pittman, 2006). An important feature of AT3 is its polyglutamine tract, which when expanded causes the neurodegenerative disorder Spinocerebellar Ataxia Type 3/Machado–Joseph disease (SCA3/MJD) (Kawaguchi et al, 1994; Stevanin et al, 1995a, 1995b). Through its multiple Ub-interacting motifs (UIMs; Figure 1A), AT3 binds both K48- and K63-linked Ub chains, yet preferentially cleaves K63-linked Ub chains in vitro, and is efficient at cleaving K63 linkages within short, mixed-linkage chains (Winborn et al, 2008). The UIMs are required both for this cleavage preference and for the ability to bind Ub chains (Winborn et al, 2008).
Figure 1.
Endogenous AT3 is ubiquitinated under basal conditions, and its ubiquitination is enhanced during proteasome inhibition. (A) Schematic of AT3 showing the N-terminal catalytic (Josephin) domain and the C-terminal Ub-interacting motifs (UIMs) flanking the polyglutamine tract. (B) AT3 is ubiquitinated in cells. FLAG–AT3 and HA–Ub were coexpressed in Cos7 cells. Lysates were subjected to stringent immunopurification with anti-FLAG antibody, then probed with anti-AT3 and anti-HA antibodies. Several high molecular weight ubiquitinated AT3 (AT3-Ub) bands are recognized by both anti-AT3 (1H9) and anti-HA antibodies. (C) Endogenous AT3 is ubiquitinated. Left: M17 cells were treated with or without the proteasome inhibitor lactacystin. Several higher molecular weight AT3-Ub species present in untreated cells are enriched when the proteasome is inhibited. Right: AT3 immunoprecipitated with polyclonal anti-AT3 antibody (MJD) from cells treated with lactacystin was probed with monoclonal anti-AT3 (1H9) and anti-Ub (P4D1) antibodies, confirming that the higher molecular weight forms of AT3 are ubiquitinated. Asterisks: mono-ubiquitinated AT3 exists in cells under basal conditions. AT3 doublets probably reflect allelic differences in the CAG/polyQ repeat in M17 cells. Ctrl: polyclonal, anti-HA antibody.
AT3 is a putative neuroprotective protein that functions in protein quality control pathways. AT3 rescues neurodegeneration caused by expanded polyQ proteins in Drosophila, doing so in an activity-dependent manner (Warrick et al, 2005). AT3 has also been implicated in ER-associated degradation (ERAD), where it assists in targeting proteins to the proteasome (Wang et al, 2006; Zhong and Pittman, 2006). When coexpressed with Ub, AT3 becomes ubiquitinated in cells (Shoesmith Berke et al, 2005; Todi et al, 2007a), but the functional consequences of this ubiquitination are unknown. Here, we show that ubiquitination of AT3 enhances cleavage of Ub chains by this DUB and that levels of ubiquitinated AT3 rise with certain cellular stressors. These results show protein ubiquitination as a direct enhancer of enzymatic activity, and suggest that the functions of AT3 in protein quality control are modulated through ubiquitination.
Results
Endogenous AT3 is ubiquitinated
FLAG–AT3 becomes ubiquitinated when coexpressed with Ub in mammalian cells (Figure 1B). To determine whether endogenous AT3 is also ubiquitinated, we conducted stringent denature/renature immunopurification of AT3 from cells (Figure 1C). In neural M17 cells, inhibition of the proteasome with lactacystin led to the accumulation of higher molecular weight AT3-immunoreactive bands (Figure 1C). Immunopurification confirmed that these bands are ubiquitinated forms of AT3 (AT3-Ub), including mono-ubiquitinated AT3 (Figure 1C). Ubiquitinated AT3 is present also under non-stressed conditions, although at lower levels. Thus, a fraction of AT3 is ubiquitinated in unperturbed cells under steady-state conditions, and proteasome inhibition increases this fraction.
Ubiquitinated AT3 cleaves Ub chains more quickly than unmodified AT3
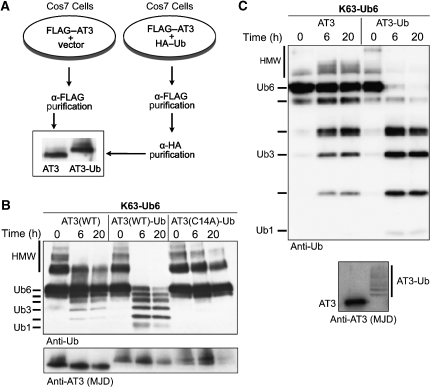
We hypothesized that AT3 ubiquitination might regulate its enzymatic activity. To investigate this, we purified unmodified or ubiquitinated AT3 from transfected cells and tested their activity in vitro (Figure 2A). Immunopurified AT3-Ub showed markedly increased DUB activity towards K63-linked hexa-Ub chains (K63-Ub6; Figure 2B). AT3-Ub also rapidly and quantitatively cleaved higher molecular weight Ub chain complexes (HMW; Figure 2B) that most likely represent longer polymers of Ub6 chains (Winborn et al, 2008). Mutating the catalytic cysteine at position 14 of AT3 to alanine renders the enzyme inactive (Burnett et al, 2003; Mao et al, 2005; Nicastro et al, 2005). Catalytically inactive, ubiquitinated AT3 (AT3(C14A)-Ub) showed no activity towards Ub chains (Figure 2B), indicating that the increased activity of AT3-Ub is not due to the spurious co-purification of another DUB. The major AT3-Ub species present in our immunopurifications is mono-ubiquitinated AT3, suggesting that a single conjugated Ub is sufficient to activate AT3 (Figure 2B).
Figure 2.
Ubiquitinated AT3 shows enhanced catalytic activity. (A) Diagram of preparation and immunoprecipitation of unmodified and ubiquitinated FLAG–AT3 from Cos7 cells transfected as indicated. (B) Ubiquitinated AT3 immunopurified from cells cleaves K63-Ub6 chains more rapidly than does unmodified AT3. AT3(WT), AT3(WT)-Ub, or catalytically inactive AT3-Ub (AT3(C14A)-Ub) (50 nM) were incubated with K63-Ub6 chains (250 nM). HMW: high molecular weight Ub species. Bottom: membrane was stripped and probed with anti-AT3 antibody. (C) AT3 ubiquitinated in vitro cleaves K63-Ub6 chains more quickly than does unmodified AT3. Anti-AT3 blot shows GST–AT3 species used in reactions.
To exclude the possibility that AT3-Ub isolated from cells co-purifies with modulators of its enzymatic activity, we performed similar reactions with recombinant AT3-Ub generated in vitro using the E2 UbcH5c and the E3 CHIP. Recombinant AT3-Ub also showed increased DUB activity (Figure 2C). Thus, ubiquitination enhances AT3 activity independent of potential cofactors/interactors or other types of post-translational modification.
Whereas AT3 does not cleave homotypic tetra-Ub (Ub4) chains efficiently, it does cleave mixed-linkage Ub4 chains (Winborn et al, 2008). Similar to unmodified AT3, AT3-Ub does not cleave K63-Ub4 chains but shows increased activity towards mixed-linkage Ub4 chains (Supplementary Figure 1). Therefore, ubiquitination enhances the activity of AT3 without altering its preference for K63-Ub6 or mixed-linkage Ub4 chains.
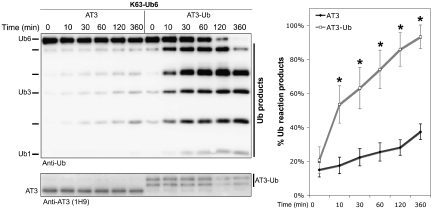
We next compared the kinetics of DUB activity of AT3-Ub with unmodified AT3. AT3-Ub prepared in vitro cleaved K63-Ub6 chains much more rapidly than unmodified AT3: reaction products were detectable within 10 min and went to completion by 6 h (Figure 3). The major reaction products were tetra-, tri-, and di-Ub (Figure 3) even in reactions extending for 20 h (Figure 2). The fact that both AT3 and AT3-Ub cleave longer chains into smaller chains rather than into mono-Ub (Figures 2 and 3) suggests that AT3 ubiquitination enhances activity towards K63-Ub6 chains without affecting the manner in which it cleaves.
Figure 3.
Accelerated cleavage of K63-Ub6 chains by AT3-Ub. Left: untagged AT3 or AT3-Ub (ubiquitinated in vitro) was incubated with K63-Ub6 chains for the indicated times. Anti-AT3 blot shows AT3 species used in reactions. Right: time-course of Ub reaction product appearance, determined by semi-quantification of western blots as shown on the left. Means±standard deviations (s.d.), N=5. Asterisks: statistically significant difference at P<0.001.
AT3-Ub prepared in vitro comprises a ladder of ubiquitinated species (Figures 2C and 3), suggesting that AT3 mono-ubiquitination may not be the only activating post-translational event. Therefore, we compared the activity of AT3-Ub prepared with wild-type UbcH5c, which can form poly-Ub chains, with AT3-Ub prepared with UbcH5c(S22R), which only mono-ubiquitinates substrates because it cannot extend Ub chains (Brzovic et al, 2006). AT3-Ub prepared either way was more active than unmodified AT3 (Supplementary Figure 2). Together, these data show that AT3 activity is enhanced by ubiquitination and that mono-ubiquitination of AT3 is sufficient for activation.
UIMs are not necessary for increased AT3 activity by ubiquitination
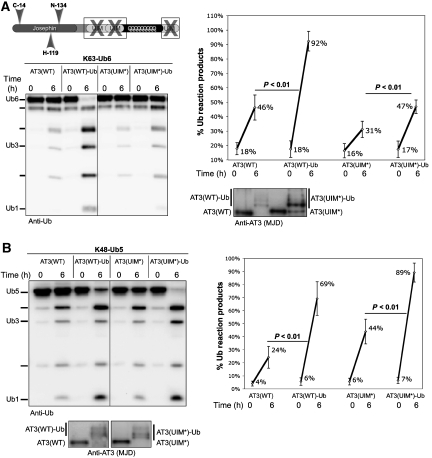
The UIMs of AT3 serve at least two functions: they mediate high-affinity binding to Ub chains and restrict the types of chains that can be cleaved by AT3. Whereas normal AT3 prefers cleaving K63 linkages, AT3 with mutated UIMs cleaves K63 and K48 linkages approximately equally well (Winborn et al, 2008). Considering this important function of UIMs in modulating the ability of AT3 to bind and cleave Ub chains, we hypothesized that an intramolecular interaction between UIMs and Ub conjugated to AT3 might underlie AT3 activation by ubiquitination. We investigated this possibility with AT3 mutated at conserved residues in each UIM (A → G/S → D; denoted as AT3(UIM*)). These mutations eliminate the ability of AT3 to bind Ub chains with high affinity (Supplementary Figure 3).
The extent of activation of AT3(UIM*)-Ub towards K63 chains was less robust than that observed with AT3(WT)-Ub (Figure 4A). In contrast, AT3(UIM*)-Ub showed strong enhancement of activity towards penta-Ub K48 chains (K48-Ub5; Figure 4B). Intriguingly, the pattern of activation towards K63 versus K48 chains is reversed for AT3(WT)-Ub and AT3(UIM*)-Ub: AT3(WT)-Ub shows greater enhancement of K63 chain cleavage, whereas AT3(UIM*)-Ub shows greater enhancement of K48 chain cleavage (Supplementary Figure 4). Thus, although the UIMs are required for preferential cleavage of homotypic K63 chains by AT3 (Winborn et al, 2008), they are not necessary for activation of AT3 by ubiquitination.
Figure 4.
The UIMs of AT3 are not necessary for activation by ubiquitination. (A) Ubiquitinated, UIM-mutated AT3 shows enhanced activity towards K63-Ub6 chains. Left: wild-type (AT3(WT)) or UIM-deficient AT3 (AT3(UIM*)) was ubiquitinated in vitro, then incubated with K63-Ub6 chains. Right: semi-quantitative representation of western blot results as on the left. Means±s.d.; N=6. (B) AT3(UIM*)-Ub cleaves K48-Ub5 chains more quickly than AT3(UIM*). Left: AT3 species were incubated with K48-Ub5 chains. Right: semi-quantitative representation of four independent experiments. Means±s.d. Anti-AT3 blots show GST–AT3 species used in reactions in (A) and (B).
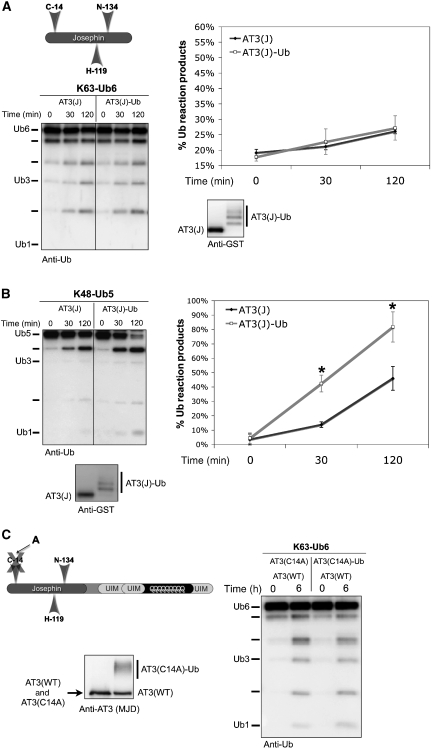
To examine which domains of AT3 are sufficient for ubiquitination-dependent activation, we studied the effect of ubiquitination on the activity of the isolated Josephin domain. The Josephin domain alone (denoted as AT3(J)) comprises amino acids 1–182 and excludes all UIMs. Although AT3(J)-Ub was not activated towards K63-Ub6 chains (Figure 5A), it cleaved K48-Ub5 chains more rapidly than unmodified Josephin domain (Figure 5B). These results support the conclusion that UIMs are not necessary for activation of AT3 by ubiquitination.
Figure 5.
Ubiquitination of the Josephin domain enhances cleavage of K48-linked Ub chains. (A) Ubiquitination of the Josephin domain does not enhance activity towards K63-Ub6 chains. Left: Josephin domain of AT3 (AT3(J)) was ubiquitinated in vitro and incubated with K63-Ub6 chains. Right: semi-quantitative representation of experiments conducted as on the left. Means±s.d.; N=3. (B) Left: ubiquitinated Josephin domain has enhanced activity towards K48-Ub5 chains. Right: semi-quantitative representation of experiments conducted as on the left. Means±s.d.; N=5. Asterisks: statistically significant difference at P<0.01. GST-tagged and untagged Josephin domain proteins yield similar results. (C) AT3 activation by ubiquitination does not occur in trans. AT3 species were prepared in vitro. Catalytically inactive AT3 (AT3(C14A)), either unmodified or ubiquitinated, was incubated with wild-type AT3 (AT3(WT)) and K63-Ub6 chains. Anti-AT3 blot shows GST–AT3 species used in reactions. Results representative of three independent experiments are shown.
AT3 activation by ubiquitination does not occur in trans
AT3 has been reported to self-associate in cells (Todi et al, 2007a) and in vitro (Ellisdon et al, 2006). It is thus possible that activation of AT3 by ubiquitination depends on homomeric interactions between AT3 proteins. To explore this possibility, we prepared unmodified and ubiquitinated forms of catalytically inactive AT3 (AT3(C14A)) to test in DUB assays with AT3(WT). AT3(C14A) or AT3(C14A)-Ub was mixed with unmodified, wild-type AT3 at equimolar concentrations, then incubated with K63-Ub6 chains. AT3(WT) activity towards K63-Ub6 was not enhanced by the presence of AT3(C14A)-Ub, indicating that activation most likely occurs in cis (Figure 5C). Moreover, as both AT3 and AT3-Ub fractionate identically by size-exclusion chromatography (Supplementary Figure 5), ubiquitination of AT3 does not appear to lead to a major change in AT3 tertiary structure. AT3 runs as an elliptical protein (Chow et al, 2006), enabling us to use size-exclusion chromatography to detect large conformational changes. This method, however, cannot detect conformational changes occurring at a domain or subdomain level.
Activity of pathogenic AT3 is also enhanced by ubiquitination
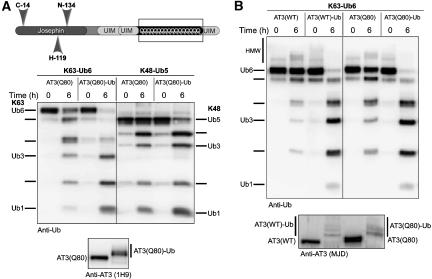
PolyQ expansion in AT3 causes the neurodegenerative disease SCA3/MJD. Disease protein context and the normal cellular functions of the implicated protein are important factors in the selective, progressive neurodegeneration seen in polyQ diseases (reviewed by Gatchel and Zoghbi, 2005; Todi et al, 2007b; Williams and Paulson, 2008). Therefore, understanding how AT3 function is affected by polyQ expansion may provide clues to pathogenesis. Expanded AT3 binds and cleaves Ub chains similarly to normal AT3 in vitro (Burnett et al, 2003; Chai et al, 2004; Winborn et al, 2008). In cells, however, expanded AT3 (AT3(Q80)) leads to accumulation of ubiquitinated proteins, probably through an indirect mechanism (Winborn et al, 2008). Given both increased AT3 activity when it is ubiquitinated and the fact that AT3(Q80) becomes ubiquitinated when coexpressed with Ub (Todi et al, 2007a; and data not shown), we investigated the ability of AT3(Q80)-Ub to cleave K63 and K48 chains. Expanded AT3-Ub prepared in vitro showed enhanced activity towards both K63 and K48 chains (Figure 6A). This enhancement did not differ from that of wild-type AT3-Ub (Figure 6B). Thus, polyQ expansion in AT3 does not appreciably alter activation by ubiquitination in vitro.
Figure 6.
Expanded (pathogenic) AT3 is also activated by ubiquitination. (A) Recombinant, expanded AT3 (AT3(Q80)) was ubiquitinated in vitro, then incubated with the indicated Ub chains. AT3(Q80)-Ub shows greater enhancement of activity towards K63-Ub6 chains. (B) Normal and expanded AT3 show similar enhancement in activity when ubiquitinated. Results in (A) and (B) are each representative of three independent experiments. Anti-AT3 blots show GST–AT3 species used in reactions.
Ubiquitination of AT3 increases with certain stressors
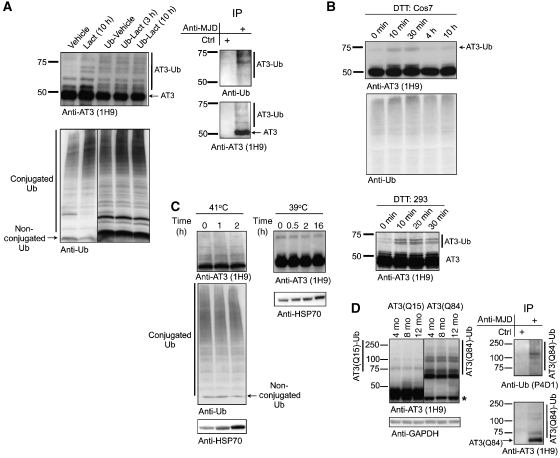
As AT3 is implicated in Ub-dependent protein quality control (Wang et al, 2000, 2006; Doss-Pepe et al, 2003; Chai et al, 2004; Warrick et al, 2005; Zhong and Pittman, 2006), we investigated whether perturbations in protein homoeostasis alter ubiquitination of endogenous AT3. In M17 (Figure 1C) and Cos7 cells (Figure 7A), levels of AT3-Ub increased during proteasome inhibition. Ub overexpression also led to higher levels of AT3-Ub (Figure 7A). Jointly inhibiting the proteasome and overexpressing Ub did not further increase AT3 ubiquitination, suggesting a saturation point for endogenous AT3 ubiquitination. Proteasome inhibition led to higher levels of conjugated Ub, whereas Ub overexpression increased levels of both conjugated and non-conjugated Ub (Figure 7A), suggesting that the ubiquitination status of AT3 functions as a feedback sensor of the overall levels of conjugated Ub in the cell.
Figure 7.
Levels of ubiquitinated endogenous AT3 are enhanced by certain stressors. (A) The fraction of endogenous AT3 ubiquitinated under basal conditions is enhanced by proteasome inhibition or by increasing Ub levels. Left: top blot shows Cos7 cells treated with lactacystin (10 μM; 10 h), or transfected with HA–Ub and treated as indicated. Bottom blot shows the same cell lysates loaded in 4–20% SDS–PAGE gel to probe for conjugated and non-conjugated Ub. All lanes are from the same exposure of the same blot. Right: AT3 was immunoprecipitated from Cos7 cells treated with lactacystin. Endogenous AT3-Ub bands are detected by both anti-AT3 and anti-Ub antibodies, confirming the identity of HMW AT3 species on the left as AT3-Ub. Representative results from four independent experiments are shown. (B) Induction of the unfolded protein response transiently leads to higher levels of ubiquitinated endogenous AT3. Treatment of Cos7 or 293 cells with the UPR inducer, DTT (5 μM), for the indicated times leads to the appearance of a higher molecular AT3 band consistent with ubiquitinated AT3. AT3 doublets in 293 cells most likely reflect allelic differences in CAG/polyQ repeat length. Representative results from at least three independent experiments are shown. (C) Heat shock does not alter the levels of ubiquitinated endogenous AT3. Cos7 cells were heat-shocked briefly (left) or for a prolonged time (right). Hsp70 levels confirmed induction of the heat shock response in treated cells. Representative results from at least three independent experiments are shown. Equal protein was loaded in (A–C). (D) Pathogenic AT3 is more heavily ubiquitinated than unexpanded AT3 in brain lysates from transgenic mice expressing normal (AT3(Q15)) or expanded (AT3(Q84)) AT3. Left: AT3(Q84) is more heavily ubiquitinated than AT3(Q15). Asterisk: endogenous AT3. AT3(Q15) mice express the protein more highly than the AT3(Q84) mice. Right: stringent immunopurification of 12-month-old AT3(Q84)-expressing brains shows that the HMW bands in left (AT3(Q84)-Ub) are ubiquitinated AT3.
Ubiquitination of endogenous AT3 also increased when the unfolded protein response (UPR) was induced by DTT, although DTT did not appreciably alter levels of conjugated Ub (Figure 7B). In contrast, acute or prolonged heat shock did not affect AT3 ubiquitination or Ub levels in Cos7 (Figure 7C), 293, or M17 cells (data not shown). Taken together, these data suggest a physiological function of AT3 ubiquitination during some, but not all, stressors.
Lastly, we investigated AT3 ubiquitination in brain lysates from transgenic mice expressing normal (AT3(Q15)) or pathogenic AT3 (AT3(Q84)) driven by the prion promoter (Cemal et al, 2002). Mice expressing AT3(Q84) show motor anomalies as early as 6 weeks of age (Cemal et al, 2002; and our unpublished data). AT3(Q84) was consistently more heavily ubiquitinated than AT3(Q15) in mice aged 4–12 months (Figure 7D). Importantly, this ubiquitinated AT3(Q84) is in the soluble fraction of brain lysates, indicating that it is not sequestered into inclusions. These data suggest that ubiquitination of expanded AT3 may be involved in SCA3/MJD pathogenesis.
Discussion
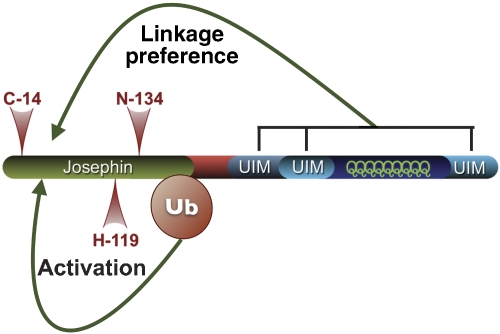
We have established that the catalytic activity of a disease-related DUB, AT3, is directly enhanced by ubiquitination. Although ubiquitination increases the enzymatic activity of AT3, it does not alter its preference for cleaving certain Ub linkages in vitro. The N-terminal half of AT3 containing the catalytic Josephin domain is sufficient for ubiquitination-dependent activation, although the C-terminal UIMs that are known to bind and restrict the types of Ub chains cleaved by AT3 are not necessary for this phenomenon (Figure 8). In mammalian cells, the fraction of endogenous AT3 that is ubiquitinated under normal conditions increases when the proteasome is inhibited, when excess conjugated Ub is present, or when the UPR is induced. Our results show that ubiquitination of a DUB can directly regulate its enzymatic properties, and suggest that activation of AT3 by this post-translational modification serves a function in the cellular response to certain stressors.
Figure 8.
Effects of protein domains on catalytic properties of AT3. Ubiquitination of the Josephin domain is sufficient for activation. Although the UIMs are not necessary for AT3 activation by ubiquitination, they confer Ub linkage preference to the catalytic domain.
Ubiquitination as a regulator of enzymatic activity
It makes biological sense that DUB activity would be regulated through feedback mechanisms tied to Ub-dependent pathways. Regulation of activity has been reported for some DUBs at various levels, including transcription (upregulation), phosphorylation (downregulation), interaction with single proteins (activation or inhibition), incorporation into complexes (upregulation or downregulation), or proteasomal degradation (inactivation) (reviewed by Nijman et al, 2005; Ventii and Wilkinson, 2008; Yao et al, 2008). In addition, USP1, a DUB that functions in DNA repair, inactivates itself through cleavage upon ultraviolet irradiation (Huang et al, 2006). Cleavage of USP7, which functions in apoptosis, is accomplished by caspase 3 (Vugmeyster et al, 2002). Here, we report for the first time that ubiquitination of a DUB directly regulates its activity.
Among the nearly 100 DUBs expressed in humans, at least six are known to be ubiquitinated: USP4, USP6, USP7, UCH-L1, AT3, and Josephin 1 (Shen et al, 2005; Wada and Kamitani, 2006; Fernandez-Montalvan et al, 2007; Meray and Lansbury, 2007; Todi et al, 2007a; and our unpublished data). At least five additional DUBs contain UIMs, a Ub-binding domain that often promotes ubiquitination of the host protein (Klapisz et al, 2002; Oldham et al, 2002; Polo et al, 2002; Shekhtman and Cowburn, 2002; Miller et al, 2004). Therefore, we anticipate that the ability of cells to control DUB function through ubiquitination will extend beyond AT3 to other DUBs. Modulation of DUB activity most likely will prove to be complex and tailored to the pathways in which the particular DUB participates.
Physiological function of ubiquitinated AT3
AT3 has been implicated in diverse cellular pathways of protein quality control. In cooperation with Valosin Containing Protein/p97, AT3 regulates ERAD substrate trafficking to the proteasome. In Drosophila, AT3 rescues neurodegeneration from expanded polyQ proteins in an activity- and proteasome-dependent manner (Wang et al, 2000, 2006; Doss-Pepe et al, 2003; Chai et al, 2004; Warrick et al, 2005; Zhong and Pittman, 2006). Building off these reports, we investigated changes in the ubiquitination levels of endogenous AT3 when the proteasome is impaired, when excess Ub is present, or when the UPR is induced. In each case, we observed an increase in levels of AT3-Ub, suggesting a physiological function of ubiquitinated AT3 during some stressors. AT3 binds both K48- and K63-linked Ub chains, but it preferentially cleaves K63-linked Ub chains, and is efficient at cleaving K63 linkages within mixed-linkage chains (Winborn et al, 2008). AT3 ubiquitination increases its activity, but does not alter the preference of AT3 for K63 linkages. Therefore, during certain types of stress, particularly those associated with perturbations in Ub chain homoeostasis, we propose that AT3 activity is upregulated through ubiquitination, thus helping to restore homoeostasis quickly. The fact that AT3-Ub levels do not increase during heat shock argues that this post-translational modification may not be part of a general response to proteotoxic stress.
We favour the view that increased levels of AT3-Ub when the proteasome is inhibited do not simply reflect AT3-Ub being a substrate for proteasomal degradation. When AT3-Ub is incubated in vitro with proteasome fractions, it is rapidly deubiquitinated by proteasome-associated DUBs, but the now unmodified AT3 is only slowly degraded (Todi et al, 2007a). This suggests that although AT3-Ub is readily deubiquitinated by proteasome-associated DUBs, it is not a favoured substrate for proteasomal degradation. Thus, it is possible that proteasome perturbation leads to accumulation of ubiquitinated proteins in cells, exceeding the capacity of proteasome-associated DUBs to rapidly deubiquitinate proteins, including AT3-Ub. The resultant increase in AT3-Ub (and correspondingly increased DUB activity) may help to restore Ub-dependent protein homoeostasis. Once homoeostasis is reestablished, AT3-Ub levels would be lowered by proteasome-associated DUBs.
Ubiquitination of AT3 could also regulate its functional associations with Ub ligases. AT3 interacts with at least three Ub ligases: E4B (Matsumoto et al, 2004), Hrd1 (Wang et al, 2006), and CHIP (KMS and HLP, manuscript in preparation). The interaction of AT3 with Ub ligases probably serves a function beyond merely targeting AT3 for degradation. Indeed, AT3 may exert an effect as a Ub chain editor within Ub ligase complexes. As AT3 is ubiquitinated by CHIP during ubiquitination cycles of model substrates (KMS and HLP, manuscript in preparation), we propose that AT3 ubiquitination serves in part to upregulate its DUB activity so as to facilitate Ub chain editing when Ub is conjugated to proteins.
The detrimental effects of expanded (i.e., pathogenic) AT3 on the nervous system were established before the cellular functions of this disease protein became recognized (Kawaguchi et al, 1994; Stevanin et al, 1995a, 1995b). As with other polyQ disease proteins, the mechanism by which pathogenic AT3 causes neurodegeneration is still poorly understood. What has become increasingly clear is the importance of protein context to polyQ disease pathogenesis. Studies of ataxin-1 (the disease protein in Spinocerebellar Ataxia Type 1) have shown that the relative abundance of specific ataxin-1 macromolecular complexes differs in normal and disease states (Lam et al, 2006; Bowman et al, 2007; Lim et al, 2008). How polyQ expansion affects protein–protein interactions for AT3 is still unknown. Thus far, expanded AT3 has behaved similarly to non-pathogenic AT3 in vitro (Burnett et al, 2003; Chai et al, 2004; Winborn et al, 2008). Likewise, in this study, polyQ expansion in AT3 did not alter activation by ubiquitination. We should stress, however, that although we do not notice differences in these in vitro assays, expanded AT3 could differ in its activity towards endogenous substrates. Indeed, pathogenic AT3 was shown to be less able than wild-type AT3 to reduce conjugated Ub levels in cells overexpressing Ub (Winborn et al, 2008).
We also observed increased ubiquitination of pathogenic AT3 in a mouse model of SCA3/MJD, suggesting that expanded AT3 ubiquitination is linked to pathogenesis in SCA3/MJD. Catalytically active AT3 is neuroprotective against toxic proteins in flies, and expanded AT3 retains this neuroprotective function (Warrick et al, 2005). As ubiquitinated AT3 has enhanced catalytic activity, it is possible that in SCA3/MJD mice, expanded AT3 ubiquitination contributes to neuronal protection. Future studies investigating whether AT3 ubiquitination is necessary for maintenance of cellular homoeostasis will address this and other possibilities.
AT3 regulation by ubiquitination
Our results indicate that ubiquitination of the isolated Josephin domain is sufficient to enhance its activity. Ubiquitination could enhance AT3 activity through at least two possibilities. First, if unmodified AT3 normally interacts with Ub chains in a manner that aligns chains so that the catalytic site has poor access to isopeptide bonds, AT3 ubiquitination could improve Ub chain presentation to the catalytic site. Ubiquitination could also directly alter the conformation of the catalytic site on the Josephin domain, effectively increasing its ability to cleave isopeptide bonds. The results of our size-exclusion chromatography, which argue against large conformational changes in AT3 upon ubiquitination, do not exclude smaller conformational changes occurring at a sub-domain level. The solution structure of the Josephin domain has been resolved (Mao et al, 2005; Nicastro et al, 2005). Therefore, identifying where AT3 is ubiquitinated in cells and in vitro, and which of its 15 lysines is important for activation, should offer structural insight into how AT3 is activated.
Although UIMs are dispensable for ubiquitination-dependent activation of AT3, they do confer upon AT3 a preference for K63 linkages. Unlike wild-type AT3, AT3 with mutated UIMs cleaves K48 and K63 linkages approximately equally well (Winborn et al, 2008). In the current studies, however, AT3-Ub with mutated UIMs showed more robust activation towards K48 than K63 linkages. Perhaps the catalytic site has an inherent preference for K48 linkages, which is altered by the UIMs in the full protein. In this case, ubiquitination of AT3 in the absence of UIMs would be expected to enhance its activity more significantly towards this preferred substrate. This model is supported by our results with the isolated Josephin domain, which when ubiquitinated showed activation towards K48- but not K63-linked Ub.
The enhanced activity of ubiquitinated Josephin domain is not as pronounced as that of ubiquitinated wild-type AT3 (e.g., compare Figures 3 and 5). Although we have ruled out the UIMs as necessary for ubiquitination-dependent activation of AT3, this by no means excludes their importance to the overall activity of AT3. Indeed, the marked activation of ubiquitinated wild-type AT3 may reflect the manner in which UIMs present isopeptide bonds to the catalytic site. It is also possible that full-length AT3 and the isolated Josephin domain are ubiquitinated at different lysines, with each having a different effect on activation. Knowing where AT3 is ubiquitinated and understanding the function of the UIMs in presenting Ub chains to its catalytic site should help our understanding of overall AT3 activity and its enhancement by ubiquitination.
Summary
The functional diversity of DUBs must be achieved through a combination of structural elements, modulatory protein domains, specific protein–protein interactions, and post-translational modifications. DUBs contain various protein motifs and interact with distinct partners that assign DUBs to specific pathways, confer substrate selectivity, and regulate enzymatic activity. We suggest that direct regulation of enzymatic activity by ubiquitination further refines the cellular functions of certain DUBs. In the case of AT3, enhancement of activity through ubiquitination may constitute a feed-forward regulatory process that helps to restore Ub-dependent homoeostasis. Ubiquitination could also serve as a direct enhancer of activity in other classes of enzymes.
Materials and methods
Constructs
Recombinant proteins are expressed in pGEX-4T1, pGEX-6P1, and pET28a vectors. FLAG-AT3 is expressed in pVETL (Todi et al, 2007a; Winborn et al, 2008). To inactivate UIMs, conserved alanine and serine residues of each UIM were mutated into glycine and aspartic acid residues, respectively (QuickChange Mutagenesis; Stratagene). Constructs for UbcH5c(WT) and UbcH5c(S22R) are Addgene plasmids 12643 and 12644.
Antibodies
The following antibodies were used: rabbit polyclonal anti-Ub (1:500; Dako); mouse monoclonal anti-Ub (1:500; P4D1; Santa Cruz Biotech); mouse monoclonal anti-AT3 (1:1000; 1H9; provided by Dr Yvan Trottier); rabbit polyclonal anti-MJD (1:40 000; Paulson et al, 1997); rabbit polyclonal anti-HSP70 (1:1000; StressGen); rabbit polyclonal anti-HA (1:500; Y11; Santa Cruz Biotech); goat polyclonal anti-GST (1:10 000; GE Healthcare); peroxidase-conjugated, goat anti-rabbit, goat anti-mouse, and rabbit anti-goat secondary antibodies (1:15 000; Jackson Immunoresearch).
Cellular treatments, transfections, and protein extraction
For proteasome inhibition, lactacystin (Boston Biochem) was used at a final concentration of 10–20 μM in regular cell media. For UPR induction, DTT (Sigma-Aldrich) was used at a final concentration of 5 μM in regular media. Heat shocking was conducted at 39 or 41°C. Cells were transfected using Lipofectamine-PLUS (Invitrogen). For western blotting, cells were lysed in 95°C Laemmli buffer with 100 mM DTT, boiled for 5 min, sonicated, centrifuged, and loaded on 4–20, or 10% SDS–PAGE gels.
Immunopurification and protein preparation
Stringent denature/renature AT3 immunopurification from cells was conducted as described previously (Todi et al, 2007a). Briefly, one 10 cm dish per experimental group was lysed in RIPA buffer (20 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholic acid, 1% Nonidet P-40, pH 7.4) with protease inhibitors (PI; Sigma-Aldrich), denatured for 30 min in 1% SDS, then renatured in 4.5% Triton X-100 for 30 min. Lysates were then incubated with antibody for 2 h at 4°C, rinsed 4 × with RIPA+PI and eluted with Laemmli buffer and boiled for a short time. Anti-AT3 (MJD) antibody used for IPs was cross-linked to protein A beads (Sigma-Aldrich) using 0.2 M triethanolamine.
Protein isolated from Cos7 cells for DUB reactions was collected by lysing cells in RIPA+PI. For unmodified AT3, cells were transfected with FLAG–AT3. Lysates from four 10 cm dishes per experiment were incubated in bead-bound anti-FLAG antibody for 2 h at 4°C, rinsed 5 × with RIPA+PI, 3 × with DUB reaction buffer (50 mM HEPES, 0.5 mM EDTA, 1 mM DTT, 0.1 mg/ml ovalbumin, pH 7.5), retained on beads, and used for reactions. We have not observed FLAG–AT3 ubiquitination in the absence of coexpressed Ub. To prepare AT3-Ub, cells were transfected with FLAG–AT3 and HA–Ub. FLAG–AT3 was isolated as above. AT3 was eluted using 3 × FLAG peptide (Sigma-Aldrich), incubated with bead-bound anti-HA antibody (2 h; Dako) to isolate AT3-Ub, rinsed 5 × with RIPA+PI and 3 × with DUB reaction buffer, and retained on beads for reactions. For quantification, a subset of the protein was eluted from beads using Laemmli buffer and quantified using serial dilutions and silver staining. Recombinant protein was prepared as described previously (Todi et al, 2007a; Winborn et al, 2008). Recombinant protein was eluted using reduced glutathione or PreScission Protease (GE Healthcare), and quantified using serial dilutions, Coomassie staining, and UV spectrophotometer (NanoDrop; Thermo Scientific).
Brain lysate preparation
Flash-frozen brains from YAC transgenic mice expressing AT3(Q15) or AT3(Q84) (Cemal et al, 2002) were homogenized in RIPA+PI, sonicated, and ultracentrifuged. For western blotting, supernatants were supplemented with 1% final SDS and 100 mM DTT, boiled, and loaded on 10% SDS–PAGE gels. Stringent immunopurification was conducted as outlined above.
Ubiquitination reactions
GST-bound AT3 was ubiquitinated in vitro using 1 μM CHIP (E3), 8 μM UbcH5c (E2), 0.16 μM E1 (Boston Biochem), 50 μM Ub, 4.5 μM MgCl2, and 4.5 μM ATP in kinase reaction buffer (50 mM TRIS, 50 mM KCl, 0.2 mM DTT, pH 7.5) for 2 h at 37°C. AT3 that was not ubiquitinated underwent the same treatment, without Ub or ATP/MgCl2. AT3 and AT3-Ub were purified using glutathione sepharose beads (GE Healthcare), and retained on beads, or eluted in DUB reaction buffer (PreScission Protease; GE Healthcare). Protein was quantified using Coomassie staining and UV spectrophotometer.
Deubiquitination reactions
All DUB reactions were continuous reactions. Protein was quantified before use with UV spectrophotometer and/or serial dilutions and silver or Coomassie staining. Ub chains (250 nM; Boston Biochem) were incubated with AT3 species (50–100 nM) in DUB reaction buffer at 37°C. Fractions were collected at the indicated times in 2% Laemmli, 100 mM DTT, and boiled for 1 min. Samples were loaded in 4–20, 10, or 15% SDS–PAGE gels.
Western blotting and quantification
Western blotting was conducted as described previously (Todi et al, 2007a; Winborn et al, 2008). Blots were imaged using autoradiography film (Kodak) or VersaDoc 5000 MP (Bio-Rad). For semi-quantification, images were collected exclusively using VersaDoc 5000 MP below-saturation levels, and quantified with Quantity One (Bio-Rad). Background was subtracted equally among lanes. Reaction products were added and divided by the sum of all bands (e.g., ∑(Ub5, Ub4, Ub3, Ub2, Ub1)/∑(Ub6, Ub5, Ub4, Ub3, Ub2, Ub1) for Ub6 chains). Two-tailed Student's t-test was used for statistical analyses.
Ub chain binding assay
Binding assays were conducted as described previously (Winborn et al, 2008). Briefly, GST–AT3 species (250 nM) were incubated with 250 nM Ub3-7 chains (Boston Biochem) for 30 min at 4°C to minimize proteolysis. Unbound supernatant fractions were removed and added to loading buffer. Beads were washed four times with buffer A (20 mM HEPES, 120 mM NaCl, 10% glycerol, 1% Triton X-100, pH 7.4) supplemented with 0.1% Nonidet P-40. Proteins were eluted with loading buffer.
Size exclusion chromatography
A volume of 100 μl of sample containing 2 μg of purified, recombinant AT3 or AT3-Ub were separated at 25°C on a Superdex 200 HR 10/30 column (Amersham) in U buffer (50 mM Tris (pH 7.5), 50 mM KCl, 0.2 mM DTT) with a flow rate of 1.0 ml/min and a fraction size of 1 ml. Gel filtration standards (Bio-Rad) were used to predict apparent molecular weight.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Ted Dawson for Ub plasmids, Dr Yvan Trottier for his generous gift of anti-AT3 (1H9) antibody, and Dr Rachel Klevit for UbcH5c(WT) and UbcH5c(S22R) plasmids. This research was supported in part by a Research Fellowship to SVT from the National Ataxia Foundation and by NIH/NINDS grant NS038712 to HLP.
References
- Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695: 189–207 [DOI] [PubMed] [Google Scholar]
- Arnason T, Ellison MJ (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol 14: 7876–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AB, Lam YC, Jafar-Nejad P, Chen HK, Richman R, Samaco RC, Fryer JD, Kahle JJ, Orr HT, Zoghbi HY (2007) Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet 39: 373–379 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 21: 873–880 [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet 12: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Cemal CK, Carroll CJ, Lawrence L, Lowrie MB, Ruddle P, Al-Mahdawi S, King RH, Pook MA, Huxley C, Chamberlain S (2002) YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet 11: 1075–1094 [DOI] [PubMed] [Google Scholar]
- Chai Y, Berke SS, Cohen RE, Paulson HL (2004) Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem 279: 3605–3611 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2005) Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7: 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow MK, Ellisdon AM, Cabrita LD, Bottomley SP (2006) Purification of polyglutamine proteins. Methods Enzymol 413: 1–19 [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K (2003) Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol 23: 6469–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA (2008) Structural insights into NEDD8 activation of Cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisdon AM, Thomas B, Bottomley SP (2006) The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J Biol Chem 281: 16888–16896 [DOI] [PubMed] [Google Scholar]
- Fernandez-Montalvan A, Bouwmeester T, Joberty G, Mader R, Mahnke M, Pierrat B, Schlaeppi JM, Worpenberg S, Gerhartz B (2007) Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J 274: 4256–4270 [DOI] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY (2005) Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet 6: 743–755 [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653 [DOI] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD (2006) Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8: 339–347 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, Kimura J, Narumiya S, Kakizuka A (1994) CAG expansions in a novel gene for Machado–Joseph disease at chromosome 14q32.1. Nat Genet 8: 221–228 [DOI] [PubMed] [Google Scholar]
- Klapisz E, Sorokina I, Lemeer S, Pijnenburg M, Verkleij AJ, van Bergen en Henegouwen PM (2002) A ubiquitin-interacting motif (UIM) is essential for Eps15 and Eps15R ubiquitination. J Biol Chem 277: 30746–30753 [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A (2008) Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 31: 371–382 [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, Zoghbi HY (2006) ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127: 1335–1347 [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY (2008) Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452: 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Dawson VL, Dawson TM (2005) Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson's and other conformational diseases? Neurobiol Aging 27: 524–529 [DOI] [PubMed] [Google Scholar]
- Mao Y, Senic-Matuglia F, Di Fiore PP, Polo S, Hodsdon ME, De Camilli P (2005) Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc Natl Acad Sci USA 102: 12700–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI (2004) Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J 23: 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meray RK, Lansbury PT Jr (2007) Reversible monoubiquitination regulates the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. J Biol Chem 282: 10567–10575 [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell 30: 610–619 [DOI] [PubMed] [Google Scholar]
- Miller SL, Malotky E, O'Bryan JP (2004) Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J Biol Chem 279: 33528–33537 [DOI] [PubMed] [Google Scholar]
- Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A (2005) The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci USA 102: 10493–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Oldham CE, Mohney RP, Miller SL, Hanes RN, O'Bryan JP (2002) The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr Biol 12: 1112–1116 [DOI] [PubMed] [Google Scholar]
- Parry G, Estelle M (2004) Regulation of cullin-based ubiquitin ligases by the Nedd8/RUB ubiquitin-like proteins. Semin Cell Dev Biol 15: 221–229 [DOI] [PubMed] [Google Scholar]
- Paulson HL, Das SS, Crino PB, Perez MK, Patel SC, Gotsdiner D, Fischbeck KH, Pittman RN (1997) Machado–Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann Neurol 41: 453–462 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Scaglione KM, Bansal PK, Deffenbaugh AE, Kiss A, Moore JM, Korolev S, Cocklin R, Goebl M, Kitagawa K, Skowyra D (2007) SCF E3-mediated autoubiquitination negatively regulates activity of Cdc34 E2 but plays a nonessential role in the catalytic cycle in vitro and in vivo. Mol Cell Biol 27: 5860–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhtman A, Cowburn D (2002) A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins. Biochem Biophys Res Commun 296: 1222–1227 [DOI] [PubMed] [Google Scholar]
- Shen C, Ye Y, Robertson SE, Lau AW, Mak DO, Chou MM (2005) Calcium/calmodulin regulates ubiquitination of the ubiquitin-specific protease TRE17/USP6. J Biol Chem 280: 35967–35973 [DOI] [PubMed] [Google Scholar]
- Shoesmith Berke SJ, Chai Y, Marrs GL, Wen H, Paulson HL (2005) Defining the role of ubiquitin interacting motifs in the polyglutamine disease protein, ataxin-3. J Biol Chem 280: 32026–32034 [DOI] [PubMed] [Google Scholar]
- Stevanin G, Cancel G, Didierjean O, Durr A, Abbas N, Cassa E, Feingold J, Agid Y, Brice A (1995a) Linkage disequilibrium at the Machado–Joseph disease/spinal cerebellar ataxia 3 locus: evidence for a common founder effect in French and Portuguese–Brazilian families as well as a second ancestral Portuguese–Azorean mutation. Am J Hum Genet 57: 1247–1250 [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Cassa E, Cancel G, Abbas N, Durr A, Jardim E, Agid Y, Sousa PS, Brice A (1995b) Characterisation of the unstable expanded CAG repeat in the MJD1 gene in four Brazilian families of Portuguese descent with Machado–Joseph disease. J Med Genet 32: 827–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, Gygi SP, Lee MK, Dawson VL, Dawson TM, Lim KL (2007) Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17: 431–439 [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Laco MN, Winborn BJ, Travis SM, Wen HM, Paulson HL (2007a) Cellular turnover of the polyglutamine disease protein ataxin-3 is regulated by its catalytic activity. J Biol Chem 282: 29348–29358 [DOI] [PubMed] [Google Scholar]
- Todi SV, Williams A, Paulson H (2007b) Polyglutamine repeat disorders, including Huntington's disease. In Molecular Neurology Waxman SG (ed), pp 257–276. London: Academic Press [Google Scholar]
- Ventii KH, Wilkinson KD (2008) Protein partners of deubiquitinating enzymes. Biochem J 414: 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vugmeyster Y, Borodovsky A, Maurice MM, Maehr R, Furman MH, Ploegh HL (2002) The ubiquitin-proteasome pathway in thymocyte apoptosis: caspase-dependent processing of the deubiquitinating enzyme USP7 (HAUSP). Mol Immunol 39: 431–441 [DOI] [PubMed] [Google Scholar]
- Wada K, Kamitani T (2006) UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem Biophys Res Commun 342: 253–258 [DOI] [PubMed] [Google Scholar]
- Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N (2000) Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet 9: 1795–1803 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y (2006) Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol 174: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM (2005) Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell 18: 37–48 [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL (2008) Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci 31: 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL (2008) The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits K63-linkages in mixed linkage ubiquitin chains. J Biol Chem 283: 26436–26443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW (2008) Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell 31: 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Pittman RN (2006) Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet 15: 2409–2420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information