Abstract
Phosphorylation of ribosomal protein L13a is essential for translational repression of inflammatory genes by the interferon (IFN)-gamma-activated inhibitor of translation (GAIT) complex. Here we show IFN-γ activates a kinase cascade in which death-associated protein kinase-1 (DAPK) activates zipper-interacting protein kinase (ZIPK), culminating in L13a phosphorylation on Ser77, L13a release from the ribosome, and translational silencing of GAIT element-bearing target mRNAs. Remarkably, both kinase mRNAs contain functional 3’UTR GAIT elements and thus the same inhibitory pathway activated by the kinases is co-opted to suppress their expression. Inhibition of DAPK and ZIPK facilitates cell restoration to the basal state and allows renewed induction of GAIT target transcripts by repeated stimulation. Thus, the DAPK-ZIPK-L13a axis forms a unique regulatory module that first represses, then re-permits inflammatory gene expression. We propose the module presents an important checkpoint in the macrophage “resolution of inflammation” program, and that pathway defects may contribute to chronic inflammatory disorders.
Introduction
Inflammatory gene expression is subject to control by an array of stimuli using diverse molecular mechanisms. Transcriptional regulation of macrophage inflammatory gene expression by cytokines is well-established, but much recent attention has focused on post-transcriptional mechanisms (Lindemann et al., 2005). In most cases, posttranscriptional regulation reduces gene expression, acting as a counteracting mechanism that limits the inflammatory response or resolves it after clearance of the initiating stimulus (Kracht and Saklatvala, 2002). Translational control mechanisms offer precise regulation of gene expression, economical use of resources (i.e., degradation of protein or mRNA is not required), and the possibility of rapid reversibility (Gebauer and Hentze, 2004; Mazumder et al., 2003b; Sonenberg and Hinnebusch, 2007). Global translational control regulates a majority of genes in response to extracellular stimuli, whereas transcript-selective translational control regulates expression of a specific gene subset. Transcript-selective translational control is generally mediated by the binding of a protein, protein complex, or microRNA to a defined structural element in either the 5’- or 3’-UTR of target mRNAs. Similar sequences and structural elements in multiple transcripts are recognized by the same RNA-binding protein(s) to enable co-regulation of translation, thereby constituting a post-transcriptional regulon (Keene, 2007; Lindemann et al., 2005).
Interferon (IFN)-γ induces formation of a heterotetrameric, IFN-gamma-activated inhibitor of translation (GAIT) complex in human monocytic cells (Mazumder et al., 2003a; Sampath et al., 2004). The complex binds a bipartite stem-loop element in the 3’-UTR of vascular endothelial growth factor (VEGF)-A mRNA, an angiogenic factor up-regulated in inflammation, and ceruloplasmin (Cp) mRNA, an acute-phase protein, and inhibits their translation (Ray and Fox, 2007; Sampath et al., 2003). The GAIT complex consists of ribosomal protein L13a, glutamyl-prolyl tRNA synthetase (EPRS), NS1-associated protein-1 (NSAP1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Phosphorylation of L13a has a critical role in GAIT system activation. Near-stoichiometric phosphorylation of L13a, occurring about 24 h after IFN-γ treatment, coincides with its release from the ribosome and recruitment into the cytosolic GAIT complex (Mazumder et al., 2003a). Phosphorylated L13a (phospho-L13a) is essential for conversion of the non-functional pre-GAIT complex consisting of EPRS and NSAP1 to the functional, RNA-binding GAIT complex (Jia et al., 2008; Sampath et al., 2004). Moreover, phospho-L13a is the actual translation inhibitor protein; after GAIT complex binding to target mRNA, phospho-L13a interacts specifically with the eIF3-binding site of eIF4G and suppresses translation by blocking recruitment of the 43S complex (Kapasi et al., 2007). These results strongly implicate L13a phosphorylation as the rate-limiting step in GAIT-mediated translational control; however, the specific phosphorylation event(s) responsible for L13a activation and the regulatory pathway are unknown.
Here, we identify the single site of L13a phosphorylation responsible for its release from ribosomes and for activation of the GAIT system in IFN-γ-treated monocytic cells. We determine death-associated protein kinase-1 (DAPK) and zipper-interacting protein kinase (ZIPK) as a kinase cascade responsible for delayed phosphorylation of L13a. Remarkably, the mRNAs of both kinases contain functional 3’-UTR GAIT elements and are themselves subject to translational repression by the same pathway they activate. Thus, DAPK, ZIPK, and L13a form a unique, RNA-based negative-feedback module that inhibits expression of a subset of late-onset inflammatory proteins, and then reverses the inhibition to restore the cell to the basal state and permit subsequent reactivation. The delayed activation of the GAIT system suggests it may be a critical checkpoint regulating late inflammatory gene expression.
Results
IFN-γ Induces L13a Phosphorylation at Ser77
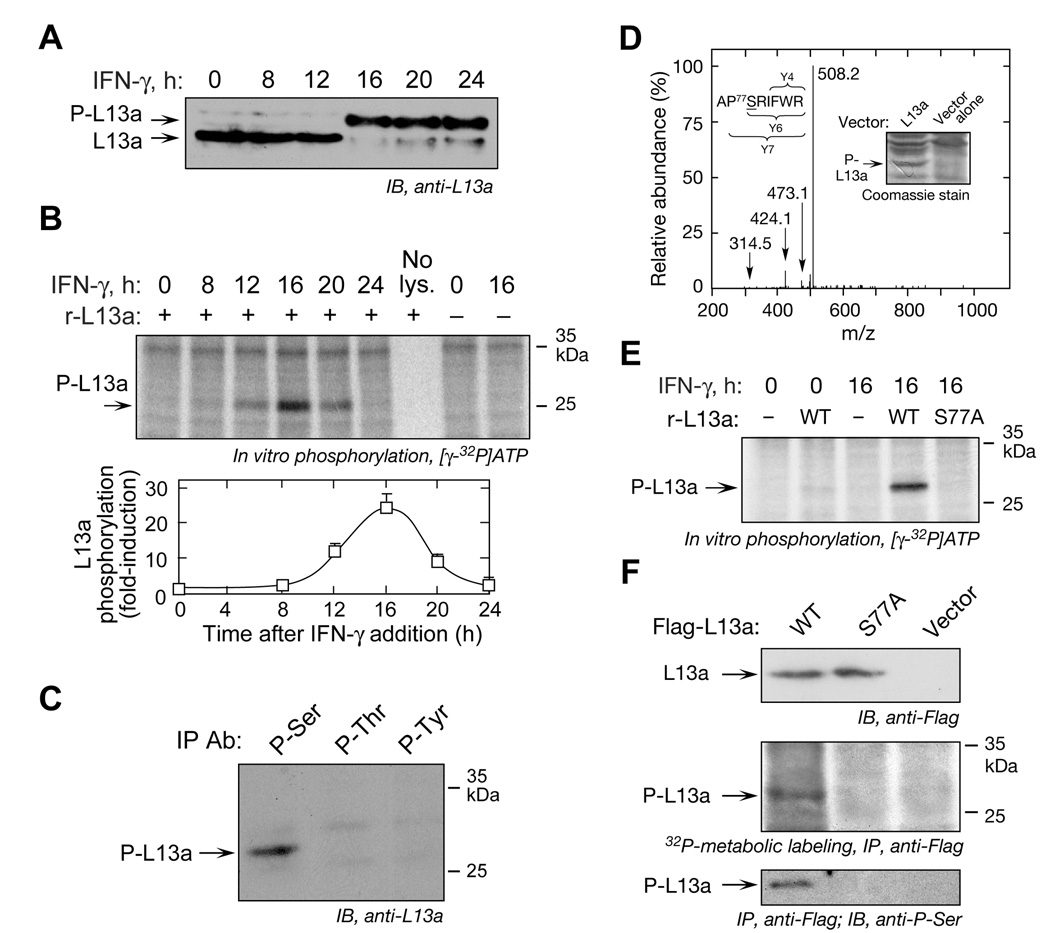
Nearly the entire cellular complement of L13a is phosphorylated 24 h after treatment of U937 cells with IFN-γ (Mazumder et al., 2003a). As an initial step in elucidating the L13a kinase, we established the time course of L13a phosphorylation and cellular L13a kinase activity. Cytosolic lysates were prepared after 8 h of IFN-γ treatment, and then at 4-h intervals up to 24 h. Phosphorylation, which reduces L13a mobility in SDS-PAGE, was induced between 12 and 16 h after IFN-γ treatment, and L13a remained phosphorylated for at least 24 h (Figure 1A). Kinase activity of cell lysates was determined by in vitro phosphorylation using purified, recombinant L13a as substrate. Kinase activity first appeared after 12 h and peaked at 16 h, consistent with the time course of L13a phosphorylation (Figure 1B). Phospho-specific antibodies were used to determine which L13a amino acid was modified. Immunoprecipitation of lysates from 24-h, IFN-γ-treated cells with antibodies against phospho-Ser, phospho-Thr, and phospho-Tyr, followed by immunoblot with anti-L13a, detected L13a phosphorylation on Ser only (Figure 1C). For mass spectrometric (MS) identification of the phosphorylation site, His-tagged human L13a was expressed in baculovirus-infected Hi-5 insect cells (Figure 1D, inset). Expressed L13a is phosphorylated and also functional as shown by inhibited translation of a GAIT element-bearing reporter RNA (Mazumder et al., 2003a). Gel-excised protein was subjected to tryptic digestion and 30 peptides, covering 74% of human L13a, were identified by LC-tandem MS (Figure 1D). The charge state of each ion set was used to calculate molecular mass of peptide fragments. Collision-induced dissociation (CID) spectra indicated 75APSRIFWR82 as the only detectable phosphopeptide, and Ser77 as the specific phosphorylation site.
Figure 1. IFN-γ Induces a Kinase that Phosphorylates L13a at Ser77.
(A) Delayed phosphorylation of L13a. Lysates from U937 cells treated with IFN-γ for up to 24 h were subjected to SDS-PAGE (12%) followed by immunoblot with anti-L13a antibody. The positions of phosphorylated (P-L13a) and non-phosphorylated L13a are indicated.
(B) Induction of L13a kinase activity in IFN-γ-treated cells. Lysates were used for in vitro phosphorylation assay using [γ-32P]ATP and recombinant L13a as substrate. Proteins were resolved by SDS-PAGE and subjected to autoradiography (top). Band intensities from three independent experiments were quantitated by densitometry and expressed as fold-induction compared to lysates from untreated cells (mean ± s.e.m., 3 independent experiments) (bottom).
(C) L13a is phosphorylated at Ser. Lysates from 24-h, IFN-γ-treated cells were immuno-precipitated with anti-phospho-Ser, phospho-Thr (Biodesign, Saco, ME) and phospho-Tyr (Santa Cruz) antibodies, followed by immunoblot analysis with anti-L13a antibody.
(D) Mass spectrometric analysis of phosphorylation site in L13a. Cell lysates from Hi-5 insect cells expressing His-tagged human L13a cDNA cloned into pFASTBAC1 vector, or empty vector, were resolved on SDS-PAGE and stained with Coomassie blue (inset). The phospho-L13a band was excised and the in-gel tryptic digest analyzed by mass spectrometry. The CID spectrum of the phosphopeptide APSRIFWR containing phosphorylated Ser77 revealed facile loss of H3PO4 to produce the doubly-charged base peak in the spectrum (m/z 508.2). The peptide sequence and site of phosphorylation were established by interpretation of low-abundance fragment ions, including labeled ions at m/z 473.1 and 424.1, doubly-charged ions corresponding to y7-H3PO4 and y6-H3PO4 ions, respectively.
(E) L13a is phosphorylated on Ser77 in vitro. His-tagged, wild-type (WT) and mutant (S77A) L13a were expressed in E. coli, purified by Ni-affinity chromatography, and used as substrate for in vitro phosphorylation with 0- or 16-h IFN-γ-treated cell lysates. Phosphorylation was visualized by SDS-PAGE and autoradiography.
(F) L13a is phosphorylated on Ser77 in vivo. Flag-tagged, wild-type and mutant (S77A) L13a were expressed in U937 cells. Expression was determined by immunoblot with anti-Flag antibody (top). Transfected cells were treated with IFN-γ for 20 h followed by a 4-h pulse of 32P-orthophosphate. Lysates were immunoprecipitated with anti-Flag antibody, and L13a phosphorylation determined by autoradiography (middle). Lysates from transfected cells were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-phospho-Ser antibody (bottom).
To confirm the phosphorylation site His-tagged, wild-type and S77A mutant L13a were expressed in E. coli and used as substrates for in vitro phosphorylation by lysates from IFN-γ-treated cells. Mutation at Ser77 completely prevented phosphorylation by the activated lysate (Figure 1E). Lysates from untreated cells did not phosphorylate L13a, showing dependence on an IFN-γ-stimulated kinase. In vivo phosphorylation of Ser77 was demonstrated in cells transfected with vectors expressing Flag-tagged wild-type or S77A L13a (Figure 1F, top). Transfected cells were treated with IFN-γ for 16 h and then labeled with a 4-h pulse of 32P-orthophosphate. Immunoprecipitation with anti-Flag antibody revealed 32P-labeling only in wild-type L13a (Figure 1F, middle). Immunoblot analysis of immunoprecipitated L13a protein with anti-phospho-Ser antibody confirmed the requirement of Ser77 for phosphorylation (Figure 1F, bottom). These studies indicate Ser77 is the sole L13a phosphorylation site in IFN-γ-treated U937 cells.
ZIPK Phosphorylates L13a in IFN-γ-stimulated cells
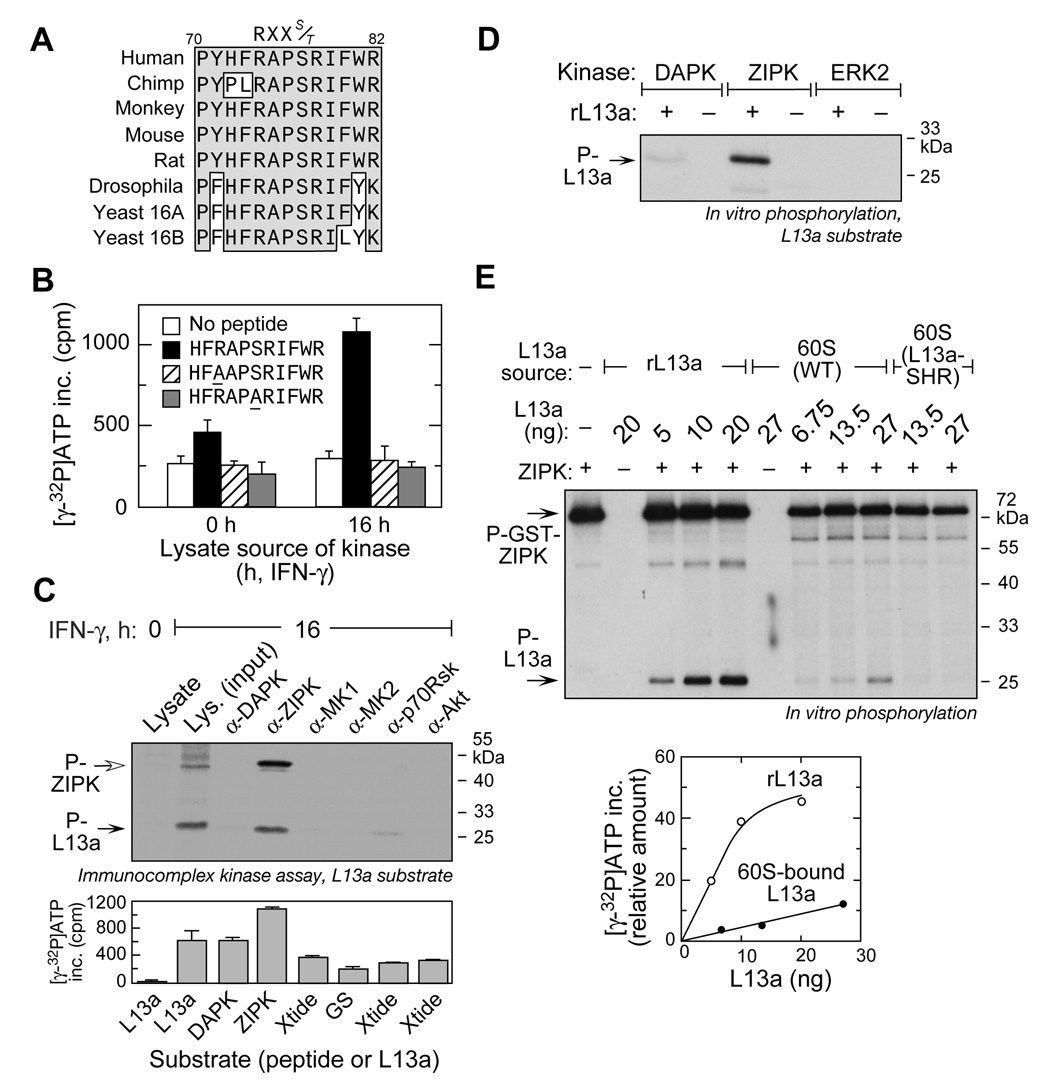
To identify the specific L13a kinase, we considered kinase recognition motifs surrounding Ser77. The consensus motif RXXS/T (Songyang et al., 1996; Ubersax and Ferrell, 2007) is conserved in L13a from multiple eukaryotic species including S. cerevisiae, Drosophila, rodents, and primates (Figure 2A). To validate the motif, 11-mer peptides carrying R74A and S77A mutations were subjected to in vitro phosphorylation. The lysate from IFN-γ-treated cells phosphorylated wild-type peptide, but neither mutant peptide, indicating the RXXS/T motif is essential for L13a phosphorylation (Figure 2B).
Figure 2. ZIPK Phosphorylates L13a in Vitro.
(A) Multiple sequence alignment of eukaryotic ribosomal protein L13a (and L16A/B paralogs in yeast) using ClustalW. Conserved residues surrounding the conserved R-X-X-S kinase recognition motif are boxed and shaded.
(B) The R-X-X-S/T motif is essential for L13a phosphorylation. Wild-type and mutant peptides (R74A and S77A) containing the L13a kinase recognition motif were subjected to in vitro phosphorylation using lysates from 0- and 16-h, IFN-γ-treated cells.
(C) ZIPK derived from U937 cells phosphorylates L13a. Candidate kinases were immunoprecipitated from lysate of 16-h, IFN-γ-treated cells, and the immunocomplex used to phosphorylate recombinant L13a. Auto-phosphorylated ZIPK (P-ZIPK) is indicated by an open arrow (top). Immunocomplex activity was tested by in vitro phosphorylation using appropriate substrate peptides (Xtide= crosstide, GS= glycogen synthase-derived peptide) (bottom).
(D) Recombinant ZIPK phosphorylates L13a. Active recombinant catalytic domain of DAPK, active full-length ZIPK, and active ERK2 were used to phosphorylate L13a by in vitro phosphorylation.
(E) ZIPK phosphorylates 60S-bound L13a. Recombinant ZIPK was used for in vitro phosphorylation of recombinant L13a (rL13a) and 60S ribosomal subunit isolated from either wild-type or L13a-knockdown (L13a-SHR) U937 cells (top). L13a in 60S subunit was quantitated by western blot and comparison to purified recombinant L13a (not shown). Phosphorylated L13a was quantitated by densitometry and plotted against L13a amount (bottom).
The RXXS/T motif is recognized by multiple basophilic Ser/Thr kinases, including several regulated by IFN-γ (Deiss et al., 1995; Songyang et al., 1996). Candidate kinases MAP kinase-activated protein kinase 1 and 2 (MK1/2), p70Rsk, Akt, DAPK, and ZIPK were investigated by immunocomplex kinase assay. The kinases were immunoprecipitated from lysates of IFN-γ-treated cells, and tested for in vitro phosphorylation of recombinant L13a substrate. Only the ZIPK-immunocomplex robustly phosphorylated L13a (Figure 2C, top). The 52-kDa phosphoprotein is consistent with ZIPK autophosphorylation, which increases its catalytic activity (Graves et al., 2005). In a control experiment, activity of immunoprecipitated kinases was shown by in vitro phosphorylation of kinase-specific substrate peptides (Figure 2C, bottom). Purified, active recombinant kinases were tested for in vitro phosphorylation of L13a. ZIPK efficiently phosphorylated recombinant L13a (Figure 2D). However, DAPK, the prototypic member of the same kinase family (Bialik and Kimchi, 2006), exhibited only marginal phosphorylation activity, and the negative control ERK2 was inactive. To test whether ZIPK can utilize ribosome-bound L13a as substrate, the 60S ribosomal subunit isolated from U937 cells was subjected to in vitro phosphorylation. Dose-responsive phosphorylation of L13a was observed, and confirmed by its absence in U937 cells in which L13a was knocked down by shRNA (Figure 2E, top). L13a is the only 60S protein visibly phosphorylated by ZIPK indicating marked specificity. However, the efficiency of phosphorylation of 60S-bound L13a was about an order-of-magnitude less than free L13a (Figure 2E, bottom). Together, these studies indicate ZIPK is the IFN-γ-activated kinase that phosphorylates L13a.
DAPK and ZIPK Form a Kinase Cascade Required for L13a Phosphorylation
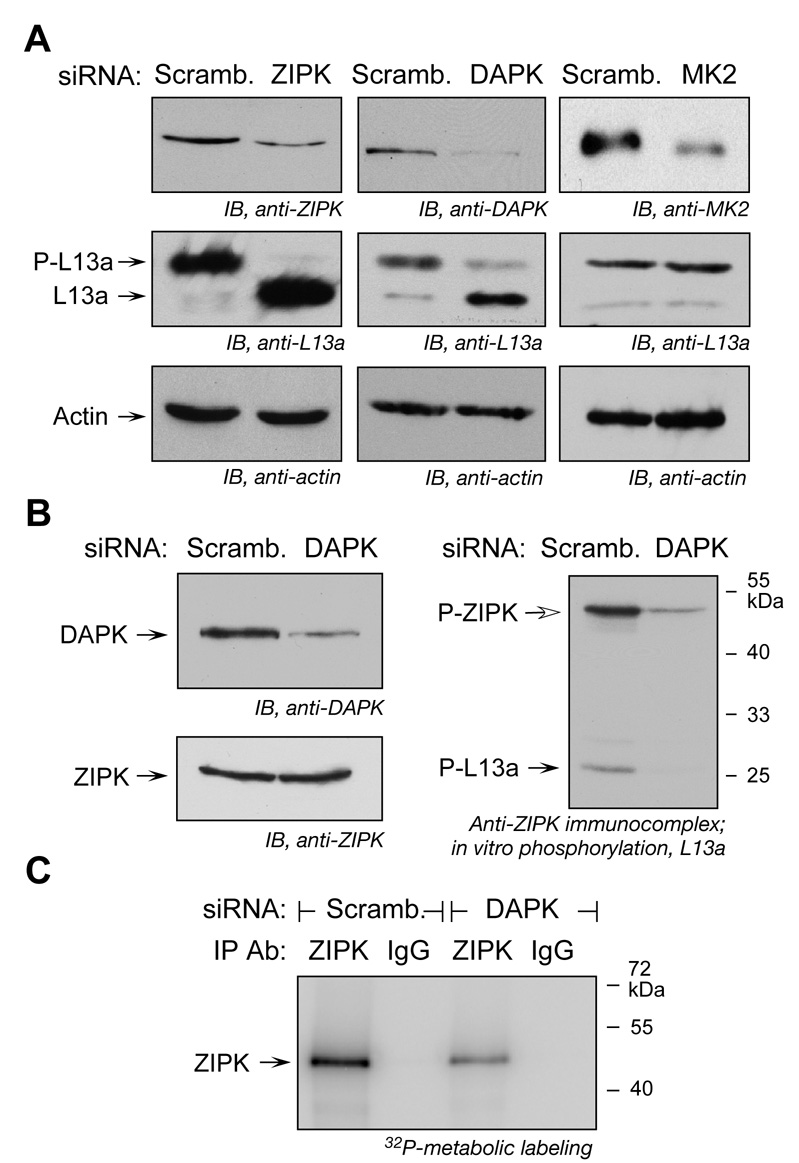
To determine the in vivo role of candidate kinases, cells were transfected with siRNAs targeted against ZIPK, DAPK, and MK2. All siRNAs substantially reduced expression of their target kinases (Figure 3A, top row). IFN-γ-induced phosphorylation of L13a was almost completely blocked by siRNA targeted against ZIPK (Figure 3A, middle row). siRNA targeted against DAPK also inhibited L13a phosphorylation suggesting that the kinases form a cascade in which DAPK is upstream of ZIPK. Knock-down of MK2 was ineffective. These results provide in vivo evidence for a DAPK-ZIPK cascade in which ZIPK is the proximal kinase that phosphorylates L13a.
Figure 3. DAPK and ZIPK form a Kinase Cascade required for L13a Phosphorylation.
(A) siRNA-mediated knock-down of ZIPK or DAPK inhibits L13a phosphorylation. U937 cells were transfected with siRNAs against ZIPK, DAPK, and MK2. Scrambled siRNAs were used as controls. After 24 h, knock-down efficiency was determined by immunoblot analysis of cell lysates (top). Transfected cells were treated with IFN-γ for 16 h, and L13a and phospho-L13a (P-L13a) detected by immunoblot (middle). Immunoblots with anti-actin served as loading controls (bottom).
(B) DAPK is required for ZIPK activation and L13a phosphorylation. U937 cells were transfected with DAPK (or scrambled) siRNA. After recovery, cells were incubated with IFN-γ for 16 h and cell lysates were immunoblotted with anti-DAPK and anti-ZIPK antibodies (left). The same lysates were immunoprecipitated with anti-ZIPK and the immunocomplex was used to phosphorylate recombinant L13a in vitro (right).
(C) DAPK knockdown reduces ZIPK activity. U937 cells were transfected with DAPK (or scrambled) siRNA. After recovery, cells were incubated with IFN-γ for 12 h and metabolically labeled with 32P-orthophosphoric acid for an additional 4 h. Cytosolic extracts were immunoprecipitated with anti-ZIPK antibody or mouse IgG, and ZIPK phosphorylation visualized by SDS-PAGE and autoradiography.
To rigorously establish the requirement of DAPK for ZIPK activation and L13a phosphorylation, ZIPK function was determined in DAPK-depleted cells. DAPK siRNA significantly reduced the amount of DAPK protein without affecting ZIPK protein expression (Figure 3B, left). The cells were treated with IFN-γ and the immunocomplex precipitated with anti-ZIPK antibody was used in an in vitro phosphorylation assay with recombinant L13a substrate. The ZIPK-immunocomplex from cells treated with scrambled siRNA caused phosphorylation of L13a, as well as autophosphorylation of ZIPK (Figure 3B, right). In contrast, the ZIPK-immunocomplex from DAPK-depleted cells failed to phosphorylate L13a, and a marked reduction in ZIPK autophosphorylation was seen. To confirm this observation in vivo, cells were transfected with DAPK siRNA and then incubated with IFN-γ for 12 h. Metabolic labeling with 32P-orthophosphate for 4 h and immunoprecipitation with anti-ZIPK antibody showed decreased ZIPK phosphorylation in DAPK knock-down cells, confirming the important role of DAPK in ZIPK activation (Figure 3C).
DAPK-ZIPK Cascade is Required for Activation of L13a and the GAIT Complex
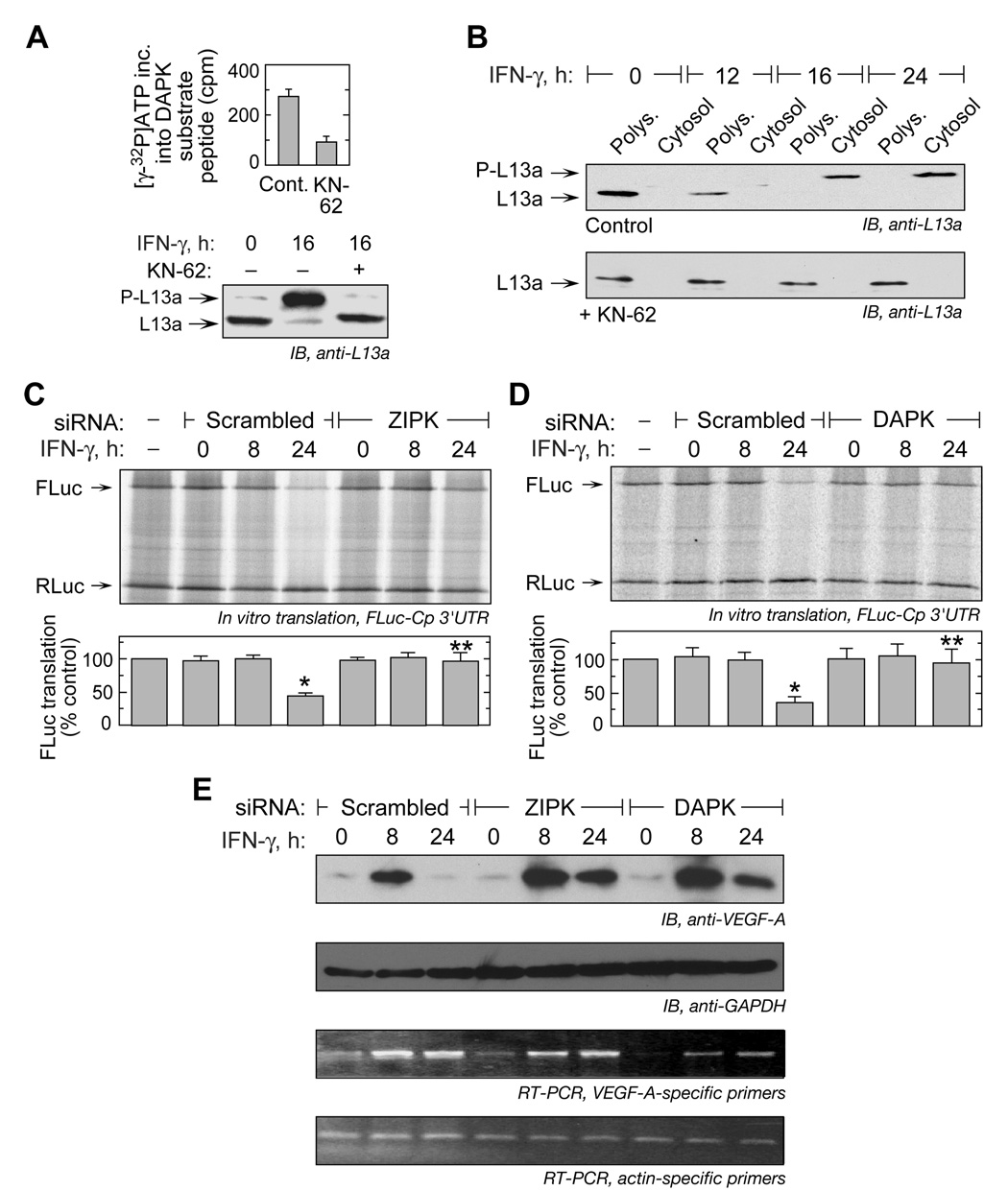
To determine the role of the DAPK-ZIPK cascade in release of L13a from the ribosome, we took advantage of the essential calmodulin-binding domain in DAPK (not present in ZIPK) to inhibit its activity (Bialik and Kimchi, 2006). U937 cells were treated with IFN-γ for 8 h, and then for another 8 h with KN-62, a selective, cell-permeable inhibitor of calcium/calmodulin-dependent protein kinases. The lysates were immunoprecipitated with anti-DAPK antibody and immunocomplex kinase activity was tested using a DAPK-specific substrate peptide (Tocris Bioscience, Ellisville, MO). KN-62 markedly reduced phosphorylation activity of the immunocomplex (Figure 4A, left), and almost completely inhibited L13a phosphorylation in IFN-γ treated cells (Figure 4A, right). To determine L13a release from the ribosome, lysates from cells treated with IFN-γ were fractionated by ultracentrifugation on a 20% sucrose cushion, and L13a was detected in cytosolic and polysomal fractions by immunoblot analysis (Mazumder et al., 2003a). In control cells not treated with KN-62, L13a remained ribosome-bound for at least 12 h, but after 16 and 24 h, nearly the entire complement of L13a was observed in the cytosolic fraction in the phosphorylated form (Figure 4B, top). However, in KN-62-treated cells, L13a remained ribosome-bound in the unphosphorylated form for the entire 24-h period, indicating a strict requirement for L13a phosphorylation by the DAPK-ZIPK cascade for ribosome release (Figure 4B, bottom).
Figure 4. DAPK-ZIPK Cascade is Required for L13a Release from the Ribosome and GAIT-mediated Translational Silencing Function.
(A) KN-62 inhibits DAPK activity and L13a phosphorylation. U937 cells were treated with IFN-γ for 8 h and then with 5 µM KN-62 (or DMSO) for an additional 8 h. DAPK was immunoprecipitated from cell lysates, and the immunocomplex was used for in vitro phosphorylation of DAPK-specific substrate peptide (left). Lysates were immunoblotted to detect L13a and phospho-L13a (P-L13a) (right).
(B) Inhibition of L13a phosphorylation blocks its release from ribosomes. U937 cells were treated with IFN-γ and DMSO control (above) or KN-62 (below), and polysome (Polys.) and cytosol fractions isolated by ultracentrifugation on a 20% (w/v) sucrose cushion in the presence of cycloheximide. Fractions were analyzed by immunoblot with anti-L13a.
(C) siRNA-mediated knock-down of ZIPK suppresses translational silencing. U937 cells were transfected with siRNA (or scrambled) against ZIPK, and treated with IFN-γ for up to 24 h. Cell lysates were added to in vitro translation reactions of capped, FLuc-Cp-3’ UTR-poly(A) RNA reporter in RRL (top). Capped, RLuc RNA lacking a 3’UTR was co-translated as a control. Fluc expression was normalized by Rluc, both determined by densitometry, and expressed as % of control (mean ± s.e.m., 3 experiments) (bottom). The significant (*) and non-significant (**) differences between 24-h and control lysates are indicated (p < 0.05, two-tailed t-test).
(D) siRNA-mediated knock-down of DAPK inhibits translational silencing. U937 cells were transfected with siRNA (or scrambled control) against DAPK, and in vitro translation determined as in (C).
(E) DAPK and ZIPK knock-down restores VEGF-A expression. U937 cells were transfected with DAPK, ZIPK, or scrambled siRNAs. Cells were treated with IFN-γ for up to 24 h. Cytosolic lysates were immunoblotted with anti-VEGF-A or GAPDH antibodies (top 2 panels). RT-PCR analysis of total cellular RNA was done using primers specific for VEGF-A or β-actin (bottom 2 panels).
The role of the DAPK-ZIPK cascade on transcript-selective translational silencing was investigated by in vitro translation. U937 cells were transfected with siRNAs targeted to ZIPK or DAPK and then incubated with IFN-γ for 8 or 24 h. Translation of a capped, poly(A)-tailed reporter RNA containing firefly luciferase (FLuc) ORF upstream of the GAIT element-bearing Cp 3’-UTR was determined in a rabbit reticulocyte lysate (RRL). Lysates from cells treated with IFN-γ for 24 h efficiently inhibited Fluc reporter translation in the presence of scrambled ZIPK or DAPK siRNA (Figure 4C,D). However, siRNA targeted against either kinase substantially overcame the translation-inhibition, indicating a non-functional GAIT complex. As shown previously, lysates from untreated cells or cells treated with IFN-γ for 8 h did not silence GAIT element-bearing reporter translation under any condition, because L13a is not phosphorylated at this time. Capped, renilla luciferase (RLuc) RNA lacking the Cp 3’-UTR was used as a control template and translation was unaffected by any treatment. To investigate the role of the kinases in regulating GAIT target expression, VEGF-A was measured in lysates from cells treated with ZIPK or DAPK siRNA. Only a minor reduction in VEGF-A expression was observed 24 h after IFN-γ treatment of cells treated with either siRNA (Figure 4E). Thus, L13a phosphorylation by the DAPK-ZIPK cascade is essential for GAIT-mediated translational silencing.
DAPK and ZIPK are Targets of the GAIT-mediated Translational Silencing Pathway
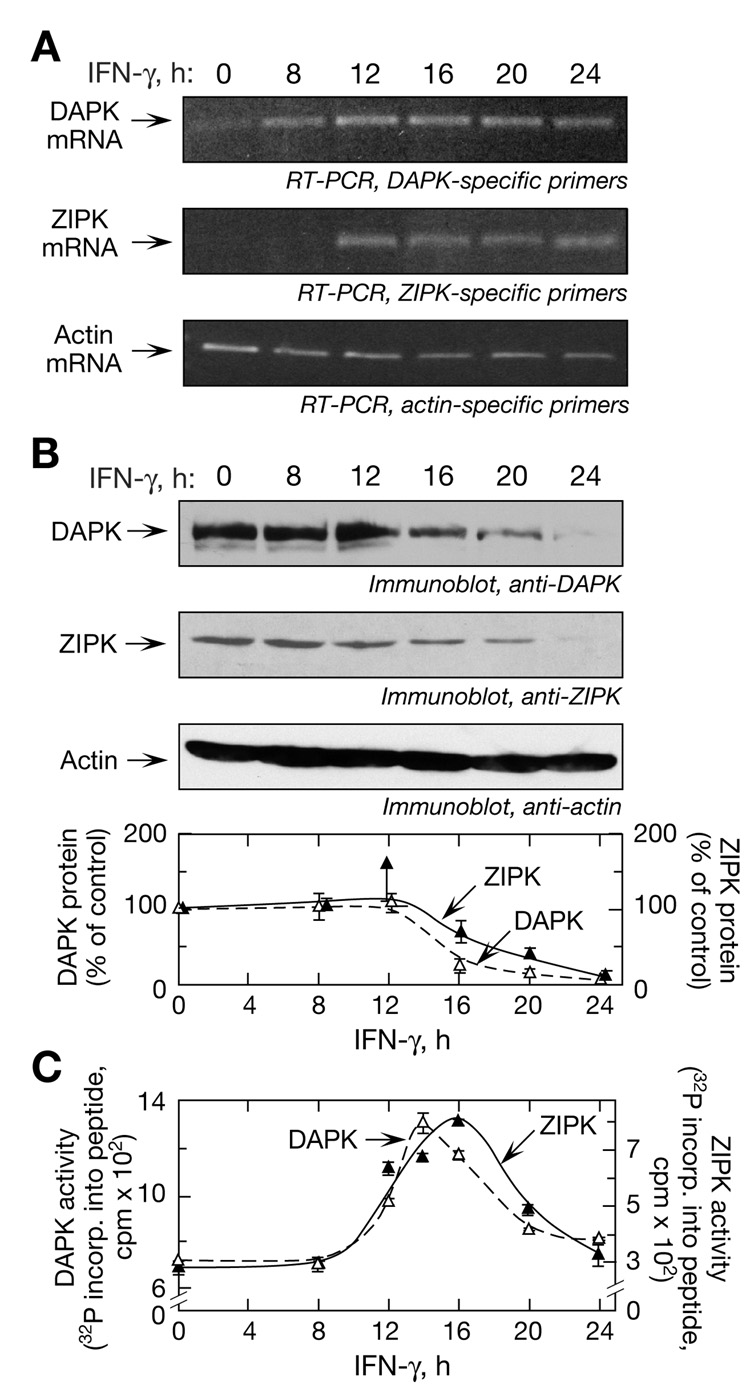
We determined whether the temporal expression of the kinases was consistent with the observed delay in L13a phosphorylation. IFN-γ induces DAPK mRNA in HeLa cells, but virtually nothing is known about regulation of ZIPK expression (Bialik and Kimchi, 2006; Deiss et al., 1995). In unstimulated cells, DAPK mRNA was expressed at a low level and ZIPK mRNA was undetectable (Figure 5A). Both transcripts were induced by IFN-γ, with maximal expression at 12 h persisting until at least 24 h. The protein levels of both kinases were constant for the first 12 h; however, they declined precipitously and were undetectable at 24 h despite robust expression of DAPK and ZIPK mRNA at this time (Figure 5B). Actin mRNA and protein expression were unaltered (Figure 5A,B). DAPK and ZIPK activities were measured by in vitro phosphorylation using kinase-specific substrate peptides. Maximal induction of DAPK activity was observed after 14 h of IFN-γ treatment, with ZIPK lagging by about 2 h (Figure 5C). The time of maximal induction of ZIPK activity corresponded to L13a phosphorylation (Figure 1A). The marked induction of kinase activities observed after 8 h despite essentially unchanged kinase levels suggests a higher specific activity of IFN-γ-induced, newly synthesized kinases compared to constitutively present kinases, consistent with the important role of posttranslational modification in activation of both kinases (Chen et al., 2005; Shani et al., 2004). After attaining their maxima, DAPK and ZIPK activities gradually declined, consistent with the decline in protein levels. In view of the high-level expression of DAPK and ZIPK mRNA that persists until at least 24 h, these results indicate that posttranscriptional regulation is responsible for reduced kinase expression and activity.
Figure 5. Regulation of DAPK and ZIPK Expression and Activity in Response to IFN-γ.
(A) Induction of DAPK and ZIPK mRNA. U937 cells were treated with IFN-γ for up to 24 h and RT-PCR analysis of total cellular RNA was done using primers targeted against DAPK (top panel), ZIPK (middle panel), and actin (bottom panel).
(B) Temporal expression of DAPK and ZIPK protein. Cell lysates described in (A) were immunoblotted with antibodies against DAPK (top panel), ZIPK (2nd panel), and actin (3rd panel). DAPK (△) and ZIPK (▲) expression was quantitated by densitometry and expressed as % of control (mean ± s.e.m.) (bottom panel).
(C) Determination of DAPK and ZIPK activities. Cell lysates were used for in vitro phosphorylation of DAPK- (△) and ZIPK- (▲) specific substrate peptides (mean ± s.e.m.).
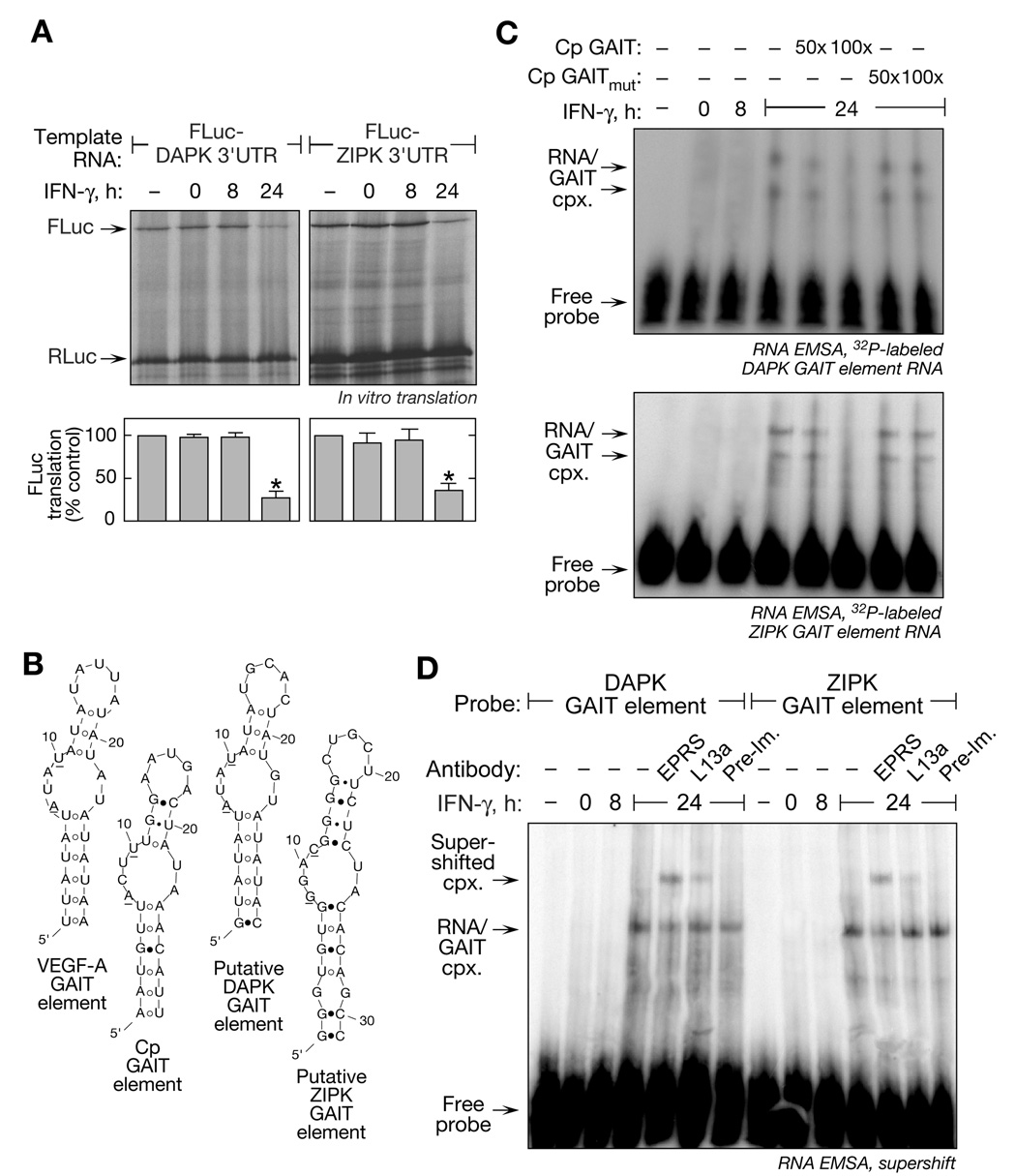
Remarkably, both DAPK and ZIPK mRNAs are potential GAIT targets. A GAIT element in the DAPK 3’-UTR was predicted by PatSearch, a pattern-matching algorithm (Grillo et al., 2003; Ray and Fox, 2007). Similarly, ZIPK mRNA was tentatively identified as a GAIT target by RNA-immunoprecipitation. GAIT mRNPs were isolated by immunoprecipitation with antibody against EPRS (a GAIT complex component) from lysates of 24-h, IFN-γ-treated cells. Quantitative RT-PCR with DAPK- and ZIPK-specific primers revealed specific association of DAPK and ZIPK mRNA with the GAIT complex as shown by 510- and 147-fold enrichment, respectively, compared to an immunoprecipitate with pre-immune rabbit IgG (Supplementary Figure 1). We tested whether the kinase mRNA 3’UTRs conferred translational silencing to heterologous reporter transcripts. FLuc reporters bearing full-length 3’UTRs from both kinases were subjected to in vitro translation in RRL. Translation of Fluc upstream of either the DAPK or ZIPK 3’UTR was markedly inhibited by lysates from 24-h, IFN-γ treated cells, indicating both 3’UTRs confer translational silencing activity (Figure 6A).
Figure 6. DAPK and ZIPK are Post-Transcriptionally Regulated.
(A) DAPK and ZIPK mRNA 3’UTRs mediate translational inhibition of reporters. Capped, FLuc-DAPK 3’UTR(1–1225)-A30 RNA and FLuc-ZIPK 3’UTR(50–645)-A30 RNA was translated in RRL in presence of cytosolic extracts from U937 cells treated with IFN-γ for up to 24 h (top). Capped, RLuc RNA lacking the GAIT element was co-translated in each reaction as control. Fluc expression was normalized by Rluc expression and expressed as % of control reaction without lysate (mean ± s.e.m.); significant differences (p < 0.05, two-tailed t-test) between 24-h and control lysates are indicated (*) (bottom panel).
(B) Secondary structures of GAIT elements. The predicted secondary structures of VEGF-A (3’UTR nt 358–386), Cp (nt 78–106), putative DAPK (nt 1141–1169) and ZIPK (nt 174–206) GAIT elements (by Mfold). Base-pairing between A7:U23 and U8:G22 was disallowed for DAPK RNA, and between A10:U25 for ZIPK RNA.
(C) The GAIT complex binds DAPK and ZIPK GAIT elements. Radiolabeled, GAIT element riboprobes were incubated with cytosolic extracts from U937 cells treated with IFN-γ for up to 24 h. RNA-protein complexes were resolved by non-denaturing 5% PAGE. RNA-protein complexes were competed with 50- and 100-fold excess of unlabeled Cp GAIT element or mutant Cp GAIT (U10C) element RNA.
(D) EMSA-supershift analysis of GAIT complex binding to DAPK and ZIPK GAIT elements. Cytosolic extracts from IFN-γ-treated U937 cells were incubated with anti-EPRS, anti-L13a and pre-immune IgG and then added to radiolabeled, GAIT element riboprobes. RNA-protein complexes were resolved by non-denaturing 5% PAGE and visualized by autoradiography.
The secondary structures of putative DAPK and ZIPK GAIT elements, as predicted by the Mfold RNA-folding algorithm (Jaeger et al., 1989), were compared to the known Cp and VEGF-A GAIT elements (Ray and Fox, 2007; Sampath et al., 2003). A putative GAIT element in the DAPK 3’UTR was predicted by PatSearch (Grillo et al., 2003). Mfold confirmed a secondary structure identical to the Cp and VEGF-A GAIT elements, including the conserved A and U in the proximal bulge (Figure 6B). Likewise, Mfold predicted the ZIPK 3’UTR to contain a stem-loop with an internal bulge-loop similar, but not identical, to the consensus structural element. Function of the putative GAIT elements was determined by RNA electrophoretic mobility shift assay. Radiolabeled DAPK (Figure 6C, top) and ZIPK (Figure 6C, bottom) RNA probes bound proteins in lysates from 24-h, but not 0- or 8-h, IFN-γ-treated cells. Binding specificity was shown by effective competition with unlabeled Cp GAIT element RNA, but not with a mutant Cp GAIT RNA that does not bind the GAIT complex (Sampath et al., 2003). Binding of the GAIT complex to DAPK and ZIPK GAIT elements was verified by supershift analysis of the RNA EMSA using anti-EPRS and anti-L13a antibodies (Figure 6D). Therefore, both DAPK and ZIPK mRNAs are specific targets of the GAIT translational silencing pathway that is activated by their encoded kinases.
GAIT-Mediated Translational Silencing of DAPK and ZIPK form a Negative Feedback Circuit
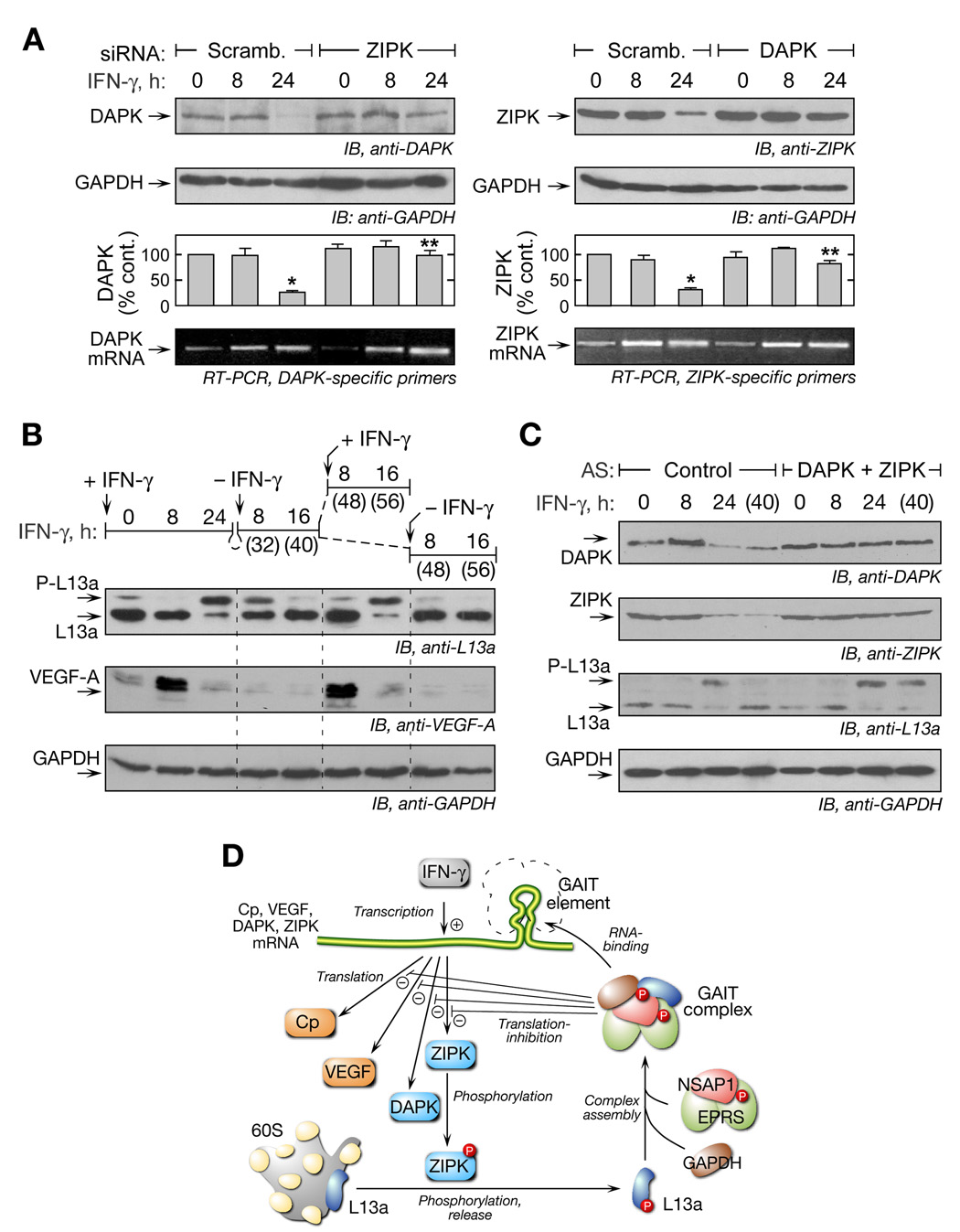
The presence of functional GAIT elements in DAPK and ZIPK mRNAs suggests they form, with L13a, a negative-feedback circuit in which the kinases activate a pathway that ultimately inhibits their own expression. To test this mechanism we determined whether inhibiting expression of one kinase prevents translational silencing of the other. Cells were transfected with siRNA against ZIPK (or with scrambled siRNA) and then treated with IFN-γ. The repression of DAPK expression after 24 h was not observed in ZIPK-depleted cells (Figure 7A, left). Likewise, DAPK depletion prevented the repression of ZIPK, confirming the negative-feedback circuit (Figure 7A, right). Similar results were observed using KN-62 to inhibit DAPK activity (Supplementary Figure S2).
Figure 7. Negative-feedback Regulation of GAIT System Allows Cell to Return to Basal State.
(A) Knockdown of ZIPK prevents translational silencing of DAPK and vice versa. U937 cells were transfected with siRNA directed against ZIPK (or scrambled). After recovery, cells were treated with IFN-γ up to 24 h and lysates immunoblotted with anti-DAPK and anti-GAPDH antibodies (left, top 2 panels). Similarly, cells were transfected with siRNA against DAPK, and lysates were probed with anti-ZIPK and anti-GAPDH antibodies (right, top 2 panel). Significant (*) and non-significant (**) differences between 24-h and control lysates are indicated (p < 0.001, two-tailed t-test). RT-PCR analysis of total cellular RNA was performed with primers targeted against DAPK and ZIPK (bottom).
(B) Time course of L13a phosphorylation and VEGF-A translation after recurrent IFN-γ treatment. U937 cells were treated with IFN-γ initially for up to 24 h (0–24 h). Cells were then washed and resuspended in fresh medium without IFN-γ for another 8 or 16 h (t = 32, 40 h). At that time, cells were washed again and incubated with IFN-γ or medium for an additional 8 or 16 h (t = 48, 56 h). Lysates were immunoblotted with anti-L13a (top), anti-VEGF-A (middle), and anti-GAPDH (bottom) antibodies.
(C) RNA oligomers complementary to DAPK and ZIPK GAIT elements disrupt negative-feedback regulation. U937 cells were transfected with DAPK and ZIPK (or control) morpholino antisense oligomers (10 µM). Cells were treated with IFN-γ for up to 24 h, then washed and resuspended in medium without IFN-γ for another 16 h (40 h). Lysates were immunoblotted with anti-DAPK, anti-ZIPK, anti-L13a and anti-GAPDH antibodies (panels 1–4, respectively).
(D) Schematic illustrating negative-feedback regulation in the GAIT system. IFN-γ causes transcriptional induction of inflammatory genes including Cp, VEGF, DAPK, and ZIPK. The kinases then activate the GAIT complex that ultimately silences translation of GAIT element-bearing genes, including both kinases, thereby inactivating the GAIT system.
The consequences of negative-feedback regulation of the DAPK-ZIPK-L13a axis was examined by determining the kinetics of GAIT pathway activation and inactivation. Phosphorylation of L13a, which is near complete at 24 h, exhibited a gradual decline, returning to the initial level about 16 h after removal of the stimulus (Figure 7B, top). When the cells were re-stimulated with IFN-γ, renewed induction of L13a phosphorylation was observed after 16 h (but was absent in untreated cells). To establish that the L13a phosphorylation state reflects translation-regulation, VEGF-A expression was measured (Figure 7B, middle) as well as GAPDH as control (Figure 7B, bottom). VEGF-A was induced after 8 h of IFN-γ treatment and suppressed after 24 h, as shown previously (Ray and Fox, 2007). Re-stimulation with IFN-γ induced a second round of VEGF-A expression after 8 h and subsequent suppression.
To confirm that negative-feedback regulation of the kinases is required for L13a return to the non-phosphorylated state, morpholino oligomers antisense to the DAPK and ZIPK GAIT elements were used to block GAIT complex-mediated silencing of the kinases. Both kinases were targeted simultaneously because the activities of both are required for L13a phosphorylation. Cells were incubated with IFN-γ for 24 h and then permitted to recover for 16 h in medium without IFN-γ. DAPK and ZIPK protein expression declined at 24 h and remained low at 40 h in cells treated with a control oligomer (Figure 7C). L13a was primarily in the phosphorylated state at 24 h, but non-phosphorylated at 40 h. In contrast, in cells treated with the antisense morpholinos, DAPK and ZIPK expression remained high even after 40 h. Importantly, L13a remained phosphorylated after 40 h showing that disruption of GAIT complex binding to the kinase GAIT elements prevents negative-feedback regulation of L13a and extends the lifetime of phospho-L13a. Thus, the GAIT-mediated negative-feedback pathway is responsible for return of L13a to the non-phosphorylated, inactive form, and for restoration of the cell to the basal state to allow subsequent, stimulus-dependent induction of target mRNA translation.
Discussion
Protein phosphorylation plays a particularly important role in translational control mechanisms. Phosphorylation of components of the translation machinery, e.g., eIF2α, eIF4B, eIF4E, eIF4G, or eEF2, is common to mechanisms regulating global translation (Dever, 2002; Raught et al., 2004), whereas phosphorylation of components of RNA-binding complexes, e.g., maskin, hnRNP K, and EPRS, is common to transcript-selective translational control (Barnard et al., 2005; Ostareck-Lederer et al., 2002; Sampath et al., 2004). We have elucidated the kinase-dependent pathway that activates GAIT-mediated translational silencing. The delayed activation of the DAPK and ZIPK cascade clarifies the molecular basis underlying the 16-h delay before the onset of translational repression following IFN-γ treatment. The experiments reveal ribosomal protein L13a as a novel target of ZIPK. Importantly, the regulation of Cp and VEGF-A expression in response to IFN-γ ascribe a novel role to the DAPK and ZIPK cascade in the inflammatory response of myeloid cells; previous studies focused primarily on the role of these kinases in induction of apoptosis and muscle contraction (Inbal et al., 1997; Shani et al., 2004). With the addition of DAPK and ZIPK to the previously identified targets VEGF-A and Cp, it is apparent that the GAIT system constitutes a significant RNA regulon, and it is the first case which is self-limiting in nature (Keene, 2007; Keene and Tenenbaum, 2002). Negative translational regulation of the kinases inactivates the GAIT pathway and restores the cell to the basal state to allow reactivation of the IFN-γ-induced gene expression program by recurrent stimuli. This negative-feedback system defines a unique “network motif” (Milo et al., 2002) in which activation of the system is mediated by phosphorylation of an effector protein, i.e., L13a, whereas negative regulation occurs by a distinct process, namely, translational repression of the kinases (Figure 7D).
Ribosomal Protein L13a is a Novel ZIPK Substrate
ZIPK is a mammalian-specific member of a family of serine/threonine kinases implicated in cell death and associated functions (Bialik and Kimchi, 2006; Inbal et al., 1997; Shani et al., 2004). Its N-terminal kinase domain shares significant sequence similarity with DAPK, the prototypic member of the family, and with DAPK-related protein-1 (Bialik and Kimchi, 2006). ZIPK is distinguished from other family members by the presence of C-terminal nuclear localization sequences and a leucine zipper, and the absence of calmodulin-binding or death domains (Bialik and Kimchi, 2006). Overexpression of ZIPK induces morphological changes consistent with apoptosis including cell rounding with shrinkage, formation of autophagic vesicles, and chromatin condensation, DNA fragmentation and ultimately, apoptosis by a caspase-3-independent mechanism (Bialik and Kimchi, 2006; Kawai et al., 1998; Mills et al., 1998). ZIPK was originally identified by its leucine zipper-mediated interaction with ATF4, a transcription factor implicated in cell growth and development (Kawai et al., 1998; Masuoka and Townes, 2002). Myosin light chain may be an important ZIPK substrate, and phosphorylation of smooth muscle myosin phosphatase and its inhibitor, CPI-17, synergistically maintain myosin in its phosphorylated state (Endo et al., 2004; MacDonald et al., 2001). Myosin phosphorylation by ZIPK has been linked to cell movement and smooth muscle contraction, and may contribute to apoptosis, e.g., by induction of membrane blebbing (Haystead, 2005; Murata-Hori et al., 2001). Other ZIPK targets have been reported including ZIPK itself (Kögel et al., 1998), histone H3 (Kögel et al., 1998), p21WAF1 (Burch et al., 2004), and STAT3 (Sato et al., 2005). Several substrates are nuclear factors, and indeed, overexpressed ZIPK is localized primarily in the nucleus in unstimulated COS-7 and REF52.2 cells (Kawai et al., 1998; Kögel et al., 1999).
Our results indicate an unexpected expansion of the function of this IFN-inducible kinase cascade, and establish ribosomal protein L13a as a novel ZIPK target. The cytosolic localization of L13a and its parent complex, the large ribosomal subunit, suggest the kinase is cytosolic in monocytic U937 cells, and cytosolic retention may be influenced by IFN-γ. Autophosphorylation of ZIPK at Thr180, Thr225, and Thr265 are essential for its catalytic activity (Graves et al., 2005). ZIPK is also autophosphorylated at Thr299 which induces cytosolic retention. Interestingly, Thr299 is also a target of DAPK (Shani et al., 2004). Our results show that both DAPK and ZIPK mRNAs are markedly induced by IFN-γ, accompanied by a small induction of the proteins at 12 h. We speculate that the presence of both newly synthesized kinases in the cytoplasm facilitates phosphorylation of ZIPK by DAPK leading to cytosolic retention and subsequent activation of the former.
Role of DAPK-ZIPK Cascade and GAIT System in Inflammation and Disease
Unique features of this anti-inflammatory mechanism distinguish it from established signaling pathways. Most obvious is the delayed nature of the inhibition of gene expression caused by the 12–16 h lag between IFN-γ addition and induction of DAPK and ZIPK activities. The lag in appearance of DAPK mRNA may be due to delayed transcriptional induction of via C/EBP-β (Gade et al., 2008) (and ZIPK via an unknown pathway), and there may be delayed post-transcriptional modification of one or both kinases (Chen et al., 2005; Shani et al., 2004). Delayed activation of the GAIT pathway can facilitate suppression of mRNAs that avoid early transcriptional blocks, for example immediate-early mRNAs that are highly stable or transcripts induced with delayed kinetics. The mechanism of inhibition by the GAIT system is entirely distinct from mechanisms described for known inflammation-suppression pathways. In the latter case, specific components of the transcriptional activation pathways are targeted, i.e., receptor-activated kinases (e.g., JAKs) and transcription factors (e.g., STATs), whereas the GAIT complex directly targets a regulon of inflammatory transcripts. Moreover, the established anti-inflammatory mechanisms involve inactivation of targets by dephosphorylation (e.g., phosphatase inactivation of JAK and MAPK) (Liu et al., 2007), by binding proteins (e.g., JAK inhibition by suppressors of cytokine signaling-1) (Starr et al., 1997), or by proteasomal degradation (e.g., STAT1) (Kim and Maniatis, 1996). In contrast, the GAIT system inhibits translation by binding a structural element in the noncoding region of the target mRNA. A potential benefit of the GAIT mechanism is the ability to silence mRNAs induced by multiple transcriptional pathways, e.g., JAK-STAT, ERK-C/EBP-β, and others. Unexpectedly, the mRNAs encoding DAPK and ZIPK, the inducers of the translational silencing pathway, are themselves targets of the pathway, thereby describing a novel mechanism for kinase inactivation and a pathway for returning the cell to the basal state. The GAIT system is a unique network module that may exemplify a principle of parsimonious pathway evolution in which the principal function of an existing system, in this case translational silencing of a regulon of inflammatory transcripts, is co-opted to down-regulate the initial stimulus.
This unique regulatory system can potentially fine-tune inflammatory gene expression in the presence of persistent inflammatory stimuli, and also contribute to inflammation-resolution after the stimulus is eliminated. It is tempting to speculate that genetic or environmental defects in the assembly or activation of the GAIT complex could influence the extent or duration of inflammatory responses. In this regard, a genome-wide scan for genes associated with late-onset Alzheimer’s disease identified two intronic, single nucleotide polymorphisms in DAPK (Li et al., 2006). Inflammation is an important contributor to Alzheimer’s disease, and DAPK mRNA is highly expressed in the adult hippocampus and cortex, the most severely affected brain regions. Prolonged expression of VEGF and other inflammatory genes recently has been recognized as an important contributor to malignant tumor progression (Ferrara and avis-Smyth, 1997). Interestingly, mutation of the gene encoding GAPDH, a component of the GAIT complex, is also associated with Alzheimer’s disease (Li et al., 2004). Failure to silence inflammatory gene expression by tumor-associated macrophages may be particularly critical because they regulate the angiogenic switch that drives the transition from small, dormant, avascular tumors to large, rapidly growing, vascularized tumors (Lin et al., 2006). DAPK has been implicated in occurrence and progression of several cancers. Hypermethylation of the DAPK promoter, accompanied by diminished transcription and mRNA expression, is strongly associated with non-small-cell lung cancer, Barrett’s esophageal adenocarcinoma, and chronic lymphocytic leukemia (Kim et al., 2001; Kuester et al., 2007; Raval et al., 2007). The mechanism by which diminished DAPK contributes to carcinogenesis is not known. However, recent studies indicate inflammation is an important contributor to tumorigenesis (Coussens and Werb, 2002) and the anti-inflammatory function of DAPK may contribute to its established tumor suppressive activity (Inbal et al., 1997; Kim et al., 2001). Similarly, diminished DAPK may contribute to carcinogenesis by over-expression of inflammatory proteins.
EXPERIMENTAL PROCEDURES
Cell Culture
Human U937 monocytic cells (CRL 1593.2; American Type Culture Collection, Rockville, MD) were cultured in RPMI 1640 containing 10% fetal bovine serum. The cells (1 × 107) were treated with IFN-γ (500 units/ml, R&D Systems, Minnneapolis, MN) as described (Mazumder et al., 2003). Cytosolic lysates were prepared in Phosphosafe extraction buffer (Novagen, Darmstadt) containing protease inhibitor cocktail (Roche, Mannheim, Germany).
Plasmid Construction and Protein Expression
Full-length wild type and S77A mutant L13a was obtained by the PCR amplification of cDNA from U937 cells and cloned into pRSET B vector (Invitrogen, Carlsbad, CA). S77A mutation was introduced using mega primer approach by two-step PCR of L13a cDNA. For expression, both plasmids were transformed into E. coli BL21(DE3)pLysS (Novagen). Expressed, His-tagged proteins were purified using Ni-affinity chromatography (Pierce, Rockford, IL). For expression in U937 cells, N-terminal Flag tagged-wild type and S77A mutant L13a were cloned in pcDNA3 vector under a CMV promoter (Invitrogen).
Mass Spectrometric Analysis of Phosphorylation Sites
His-tagged, phospho-L13a (cDNA cloned in pFastBac vector) was expressed in baculovirus-infected insect cells (Mazumder et al, 2003). The proteins were resolved by SDS-PAGE and the L13a band was excised, digested with trypsin in-gel, and subjected to LC-tandem MS (Proteomics Facility, Lerner Research Institute, Cleveland Clinic). The peptide mapping and phosphopeptide analyses were performed using reversed-phase capillary HPLC on an LCQ-Deca ion trap MS system (ThermoFinnigan, San Jose, CA). The data were acquired by the data dependent multitask capability mode, recording the full scan mass spectra and the CID spectra. All CID spectra were used to search the sequence of human ribosomal protein L13a (NCBI accession no. 6912634). The phosphopeptide was identified by a characteristic neutral loss of 49 Da, corresponding to the loss of H3PO4 from doubly-charged molecular ion.
Cell Transfection
U937 cells were transfected using Amaxa nucleofection technology (Koeln, Germany). Cells were suspended in 100 µl of cell line nucleofector solution V with 2 µg of plasmid DNA or 250 nM siRNAs (Dharmacon, Chicago, IL). The mix was transfected according to the manufacturer’s protocol. Cells were immediately transferred to pre-warmed in OPTIMEM media for 6 h, and then to RPMI 1640 containing 10% fetal bovine serum for 18 h. Morpholino antisense oligomers against DAPK (3’UTR nt 1145–1169) and ZIPK GAIT elements (3’UTR nt 179–203) were synthesized by Gene Tools (Philomath, OR), and transfected into U937 cells using protocol described above.
Metabolic Labeling
24 h after transfection with wild type or S77A mutant L13a-expressing plasmid DNA, cells were treated with IFN-γ for 20 h and then labeled with a 4-h pulse of 500 µCi [32P]orthophosphate (Perkin-Elmer, Boston, MA) in phosphate free medium. Labeled cells were lysed in phosphosafe buffer and expressed Flag-L13a proteins were immunoprecipitated with anti-Flag antibody-conjugated agarose (Sigma) in detergent-containing RIPA buffer. Proteins in the immunoprecipitates were resolved by 12% SDS-PAGE and bands were visualized by autoradiography.
In Vitro Kinase Assays
Equal quantity of lysates from control or IFN-γ treated cells and recombinant wild-type or mutant (S77A) His-tagged L13a substrate protein were pre-incubated for 5 min in kinase assay buffer (50 mM Tris-HCl, pH 7.6), 1 mM dithiotheitol, 10 mM MgCl2, and 1 mM CaCl2), followed by incubation in presence of [γ-32P]ATP (5 µCi, Perkin-Elmer) for 20 min, and terminated using SDS gel-loading dye followed by SDS-PAGE and autoradiography. In the peptide phosphorylation assay synthetic peptides (100 µM) were in vitro phosphorylated in 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM CaCl2, and 5 µCi [γ-32P]ATP. Equal volumes were spotted onto P81-phosphocellulose squares, which were washed and measured by scintillation counting. In the immunocomplex kinase assay, the target kinase was immunoprecipitated from lysates of 16-h, IFN-γ-treated U937 cells. The immunoprecipitated kinase was resuspended in kinase assay buffer and used to phosphorylate either purified L13a or peptides by in vitro phosphorylation.
In Vitro Translation
Capped, poly(A)-tailed template RNAs were translated for 90 min in RRL (Promega, Madison. WI) in the presence of a methionine-free amino acid mixture and translation-grade [35S]methionine. Cytosolic extract (500 ng of protein) from untreated or IFN-γ-treated U937 cells were added to translation reactions. Reactions were resolved by SDS-PAGE (10% polyacrylamide) and visualized by phosphorimaging. Template RNAs (Jia et al., 2008) and the GAIT complex (not shown) remain intact and functional for the duration of the in vitro translation assay.
RNA EMSA
[α-32P]UTP-labeled DAPK or ZIPK GAIT element RNA was incubated with cytosolic extracts from U937 cells incubated with IFN-γ for up to 24 h. RNA-protein complexes were resolved by native gel electrophoresis and visualized by phosphorimaging. For competition experiments, lysates were incubated with 50 and 100-fold excess of unlabeled Cp wild-type or mutant GAIT RNAs before binding with the radiolabeled DAPK or ZIPK GAIT element RNAs. For supershift assays, antibodies against EPRS and L13a were pre-incubated with cytosolic extract for 15 min at 4°C before incubation with radiolabeled GAIT element probes.
Ribosome Fractionation
For isolation of 60S ribosomal subunits wild-type and L13a-knockdown U937 cells (Ray and Fox, 2007) were lysed in sucrose gradient buffer and cytosolic extracts were obtained by centrifugation at 10,000 × g for 20 min. The extract was overlayed on a 10–50% (w/v) sucrose gradient and centrifuged for 4 h at 100,000 × g. Fractions containing 60S ribosomal subunits were collected, dialysed, and resuspended in kinase assay buffer. For isolation of total ribosomes, the cytosolic extract was overlayed on a 20% (w/v) sucrose cushion and centrifuged at 150,000 × g for 2 h. The ribosome-containing pellet and the non-ribosomal supernatant were collected separately and immunoblotted with anti-L13a antibody.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants HL29582, HL67725, and HL76491 (to P.L.F.), and by an AHA Postdoctoral Fellowship, Ohio Valley Affiliate (to A.A). We appreciate helpful discussions with Barsanjit Mazumder, Cleveland State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barnard DC, Cao Q, Richter JD. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell. Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialik S, Kimchi A. The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- Burch LR, Scott M, Pohler E, Meek D, Hupp T. Phage-peptide display identifies the interferon-responsive, death-activated protein kinase family as a novel modifier of MDM2 and p21WAF1. J. Mol. Biol. 2004;337:115–128. doi: 10.1016/j.jmb.2003.10.081. [DOI] [PubMed] [Google Scholar]
- Chen CH, Wang WJ, Kuo JC, Tsai HC, Lin JR, Chang ZF, Chen RH. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J. 2005;24:294–304. doi: 10.1038/sj.emboj.7600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the γ interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Endo A, Surks HK, Mochizuki S, Mochizuki N, Mendelsohn ME. Identification and characterization of zipper-interacting protein kinase as the unique vascular smooth muscle myosin phosphatase-associated kinase. J. Biol. Chem. 2004;279:42055–42061. doi: 10.1074/jbc.M403676200. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr. Rev. 1997;18:4–15. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. A critical role for transcription factor C/EBP-β in regulating the expression of death-associated protein kinase-1. Mol. Cell. Biol. 2008;28:2528–2548. doi: 10.1128/MCB.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Winkfield KM, Haystead TA. Regulation of zipper-interacting protein kinase activity in vitro and in vivo by multisite phosphorylation. J. Biol. Chem. 2005;280:9363–9374. doi: 10.1074/jbc.M412538200. [DOI] [PubMed] [Google Scholar]
- Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haystead TA. ZIP kinase, a key regulator of myosin protein phosphatase 1. Cell. Signal. 2005;17:1313–1322. doi: 10.1016/j.cellsig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Inbal B, Cohen O, Polak-Charcon S, Kopolovic J, Vadai E, Eisenbach L, Kimchi A. DAP kinase links the control of apoptosis to metastasis. Nature. 1997;390:180–184. doi: 10.1038/36599. [DOI] [PubMed] [Google Scholar]
- Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi P, Chaudhuri S, Vyas K, Baus D, Komar AA, Fox PL, Merrick WC, Mazumder B. L13a blocks 48S assembly: Role of a general initiation factor in mRNA-specific translational control. Mol. Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol. Cell. Biol. 1998;18:1642–1651. doi: 10.1128/mcb.18.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Kim DH, Nelson HH, Wiencke JK, Christiani DC, Wain JC, Mark EJ, Kelsey KT. Promoter methylation of DAP-kinase: association with advanced stage in non-small cell lung cancer. Oncogene. 2001;20:1765–1770. doi: 10.1038/sj.onc.1204302. [DOI] [PubMed] [Google Scholar]
- Kim TK, Maniatis T. Regulation of interferon-γ-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- Kögel D, Bierbaum H, Preuss U, Scheidtmann KH. C-terminal truncation of Dlk/ZIP kinase leads to abrogation of nuclear transport and high apoptotic activity. Oncogene. 1999;18:7212–7218. doi: 10.1038/sj.onc.1203169. [DOI] [PubMed] [Google Scholar]
- Kögel D, Plottner O, Landsberg G, Christian S, Scheidtmann KH. Cloning and characterization of Dlk, a novel serine/threonine kinase that is tightly associated with chromatin and phosphorylates core histones. Oncogene. 1998;17:2645–2654. doi: 10.1038/sj.onc.1202204. [DOI] [PubMed] [Google Scholar]
- Kracht M, Saklatvala J. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine. 2002;20:91–106. doi: 10.1006/cyto.2002.0895. [DOI] [PubMed] [Google Scholar]
- Kuester D, Dar AA, Moskaluk CC, Krueger S, Meyer F, Hartig R, Stolte M, Malfertheiner P, Lippert H, Roessner A, et al. Early involvement of death-associated protein kinase promoter hypermethylation in the carcinogenesis of Barrett's esophageal adenocarcinoma and its association with clinical progression. Neoplasia. 2007;9:236–245. doi: 10.1593/neo.06802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Grupe A, Rowland C, Nowotny P, Kauwe JS, Smemo S, Hinrichs A, Tacey K, Toombs TA, Kwok S, et al. DAPK1 variants are associated with Alzheimer's disease and allele-specific expression. Hum. Mol. Genet. 2006;15:2506–2568. doi: 10.1093/hmg/ddl178. [DOI] [PubMed] [Google Scholar]
- Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, Tacey K, Doil L, van Luchene R, Garcia V, et al. Association of late-onset Alzheimer's disease with genetic variation in multiple members of the GAPD gene family. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, Qian H, Xue XN, Pollard JW. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66:11238–11246. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- Lindemann SW, Weyrich AS, Zimmerman GA. Signaling to translational control pathways: diversity in gene regulation in inflammatory and vascular cells. Trends Cardiovasc. Med. 2005;15:9–17. doi: 10.1016/j.tcm.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shepherd EG, Nelin LD. MAPK phosphatases–regulating the immune response. Nat. Rev. Immunol. 2007;7:202–212. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- MacDonald JA, Borman MA, Muranyi A, Somlyo AV, Hartshorne DJ, Haystead TA. Identification of the endogenous smooth muscle myosin phosphatase-associated kinase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2419–2424. doi: 10.1073/pnas.041331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka HC, Townes TM. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood. 2002;99:736–745. doi: 10.1182/blood.v99.3.736. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto P, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003a;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem. Sci. 2003b;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Mills JC, Stone NL, Erhardt J, Pittman RN. Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J. Cell Biol. 1998;140:627–636. doi: 10.1083/jcb.140.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Murata-Hori M, Fukuta Y, Ueda K, Iwasaki T, Hosoya H. HeLa ZIP kinase induces diphosphorylation of myosin II regulatory light chain and reorganization of actin filaments in nonmuscle cells. Oncogene. 2001;20:8175–8183. doi: 10.1038/sj.onc.1205055. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen SS, Hackanson B, Grever MR, Lucas DM, Matkovic JJ, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Fox PL. A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 2007;26:3360–3372. doi: 10.1038/sj.emboj.7601774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Fox PL. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3' untranslated region. Mol. Cell. Biol. 2003;23:1509–1519. doi: 10.1128/MCB.23.5.1509-1519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Sato N, Kawai T, Sugiyama K, Muromoto R, Imoto S, Sekine Y, Ishida M, Akira S, Matsuda T. Physical and functional interactions between STAT3 and ZIP kinase. Int. Immunol. 2005;17:1543–1552. doi: 10.1093/intimm/dxh331. [DOI] [PubMed] [Google Scholar]
- Shani G, Marash L, Gozuacik D, Bialik S, Teitelbaum L, Shohat G, Kimchi A. Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol. Cell. Biol. 2004;24:8611–8626. doi: 10.1128/MCB.24.19.8611-8626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: Primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.