Abstract
To develop transgenic lines for conditional expression of desired genes in rats, we generated several lines of the transgenic rats carrying the tetracycline-controlled transactivator (tTA) gene. Using a vigorous, ubiquitous promoter to drive the tTA transgene, we obtained widespread expression of tTA in various tissues. Expression of tTA was sufficient to strongly activate its reporter gene, but was below the toxicity threshold. We examined the dynamics of Doxycycline (Dox)-regulated gene expression in transgenic rats. In the two transmittable lines, tTA-mediated activation of the reporter gene was fully subject to regulation by Dox. Dox dose-dependently suppressed tTA-activated gene expression. The washout time for the effects of Dox was dose-dependent. We tested a complex regime of Dox administration to determine the optimal effectiveness and washout duration. Dox was administered at a high dose (500 μg/ml in drinking water) for two days to reach the effective concentration, and then was given at a low dose (20 μg/ml) to maintain effectiveness. This regimen of Dox administration can achieve a quick switch between ON and OFF statuses of tTA-activated gene expression. In addition, administration of Dox to pregnant rats fully suppressed postnatal tTA-activated gene expression in their offspring. Sufficient levels of Dox are present in mother's milk to produce maximal efficacy in nursing neonates. Administration of Dox to pregnant or nursing rats can provide a continual suppression of tTA-dependent gene expression during embryonic and postnatal development. The tTA transgenic rat allows for inducible and reversible gene expression in the rat; this important tool will be valuable in the development of genetic rat models of human diseases.
Keywords: Rats, transgenic, tetracycline-controlled transactivator, tTA, Doxycycline, Leucine Rich Repeat Kinase 2, LRRK2
1. Introduction
Rodents are an ideal laboratory animal model for functional genetic studies, because breeding cycles are short and because methods of gene manipulation have been developed in laboratory mice and rats 1-5. Compared to mice, rats have a longer history of use in physiological and pharmacologic studies 6, 7. Therefore, a wealth of experimental data on rats can be supplemented with genetic analysis 1. Rats show advantage over mice in the ease of microsurgery 8, 9, in multiple sampling, and in behavior analyses. Additionally, the rats are more affordable for breeding than larger mammalian animals 1, 6, 7. However, mice have been preferred by geneticists for decades because totipotent embryonic stem (ES) cells have been initially developed in mice rather than in other mammalian animals 10. The situation is going to change as totipotent ES cells are developed in rats 11, 12. Moreover, a need for transgenic rats has been pursued because rats often serve as a superior model of human disease, compared to mice 1, 2, 6, 7, 13.
Rats often recapitulate human disease phenotypes more accurately than the mice, as exemplified by rats transgenic for the rennin-2 gene, the cholesterylester transfer gene, or the huntingtin gene 14-16. Rats model type 1 diabetes better than mice in both the pathogenesis of disease and the response to treatment. As suggested by epidemiological studies, type 1 diabetes is caused by non-genetic environmental factors that operate in a genetically susceptible host to initiate a destructive immune process. A viral infection readily induces diabetes in several rat models 17, but fails to induce diabetes in mice 18. Clinical studies suggest that neither parenteral nor oral insulin can prevent or delay diabetes onset in human patients 19. Both parenteral and oral administration of insulin prevents diabetes in mouse models 20, but fail to do so in rats. Data from the rat model, but not the mouse model, of diabetes is consistent with clinical studies. Such differences may be caused by species-specific factors, a hypothesis that is supported by analysis of the rat genome 21. The rat genome conserves genes that are involved in immunity, metabolic detoxification, reproduction, chemo-sensation, and human diseases 22, 23. Increasing evidence indicates that rats recapitulate human diseases more accurately than mice. To increase the diversity of animal models for human diseases, it is compelling to expand the availability of genetically manipulated rats.
To model human diseases in animals, inducible mutations or overexpression of desired genes is necessary when the genetic manipulation causes embryonic or early postnatal lethality 24, 25. As such, multiple systems have been developed for conditional gene manipulations. The Tet-off system has readily been used in transgenic mice 26-29. Compared to thousands of transgenic mice reported, the availability of transgenic rats is limited to about 100 transgenic lines characterized 1, 30-32. As transgenic technology advances in rats 1, 2, the number of transgenic rats is increasing at an accelerated speed 1, 33, 34. To express genes of interest in a conditional pattern in rats, we developed transgenic rats carrying and expressing tetracycline-controlled transactivator (tTA) under the control of a ubiquitous promoter. The tTA transgenic rats expressed the transgene tTA in many tissues. The expression levels of the tTA transgene were sufficient to vigorously and reversibly activate reporter gene under control of the Tet-responsive promoter. These tTA transgenic rat lines will be useful in the conditional expression of human disease genes in the rats.
2. Materials and Methods
Construction of transgenic DNA and creation of transgenic rats
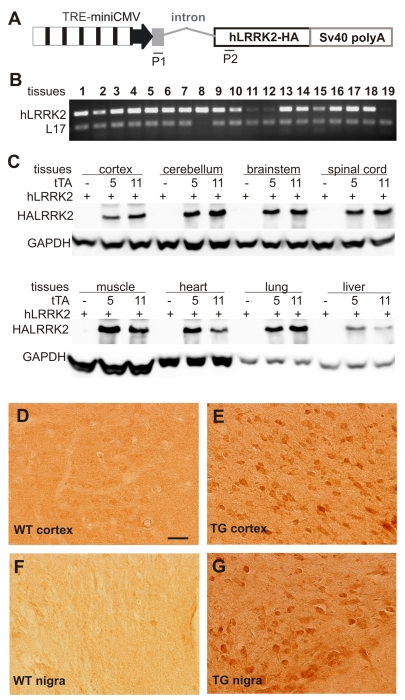
A Tet-off system was chosen to produce inducible and reversible gene expression in transgenic rats. This system is readily used in transgenic mice 26-29. The Tet-off system is comprised of two elements: a tetracycline-controlled transactivator (tTA) and a tTA-dependent promoter. The tTA-dependent promoter is a hybrid promoter that is constructed by juxtaposing multiple tetracycline-responsive elements (TRE) upstream of a minimal cytomegalovirus promoter (TREminiCMV). When a gene is placed under control of the TREminiCMV promoter, transcription of the gene depends on activation by the transcriptional activator tTA. The tTA activates its reporter gene in the absence of tetracycline or tetracycline derivatives such as Dox (Dox), but is inactivated in the presence of Dox. For robust gene expression, a strong hybrid promoter, CAG (cytomegalovirus enhancer fused to chicken beta actin promoter), was used to drive a tTA gene that was extracted from the vector pTet-off (Clontech) 35, 36. The transgenic construct CAG-tTA consisted of the CAG promoter, the noncoding exon along with the first intron of the beta actin gene, tTA cDNA, and three repeats of the SV40 polyadenylation signal (Figure 1A). The human Leucine Rich Repeat Kinase 2 (LRRK2) gene tagged with hemaglutinin (HA) was driven by the TRE promoter and used as the reporter gene for tTA (Figure 2A).
Figure 1.
Widespread expression of the tTA transgene in tTA transgenic rats. A, Schematic shows the structure of the tTA transgene. Two pairs of primers were designed for regular PCR to determine the genotypes of transgenic rats (P2 + P3) or for reverse-transcriptional PCR (RT-PCR) to detect expression of the transgene tTA (P1 + P3). B, Results of representative PCR with the primers P2 and P3 show three founder rats (lines 4, 5, and 11). C+, nontransgenic rat DNA mixed with the transgene tTA; C-, nontransgenic rat DNA. C, RT-PCR with primers P1 and P3 detected a wide expression of the transgene tTA in various tissues: OB, olfactory bulb; hippo, hippocampus; CB, cerebellum; BS, brainstem; SC, spinal cord; SM, skeletal muscle; and other tissues as indicated. RT-PCR for the rat L17 gene serves as an internal control for equal loading of cDNA pools. RT-PCR was run for 25 cycles to show the difference in transgene expression between the two expressing lines. D, Western blotting showed that expression of the tTA transgene was not detected at protein level in the two transmittable lines. C+, HEK293 cells were transiently transfected with the tTA transgene construct and the cell lysate was used as the positive control.
Figure 2.
A reporter gene driven by a tTA-dependent promoter is transcriptionally activated by tTA and robustly expressed in various tissues. A, Schematic shows the structure of the reporter gene for probing tTA transcriptional activity. The human LRRK2 gene (hLRRK2), tagged by hemaglutinin (HA), was driven by a hybrid promoter comprising a minimal CMV promoter of 110 bp and seven repeats of tetracycline-responsive elements. The noncoding exon 1 and the first intron of human ubiquitin C gene were placed between the hybrid promoter and the human LRRK2 gene to enhance gene expression. A pair of primers (P1, P2) was designed to magnify the mature cDNA of the transgene, giving a band of 240 base-pairs. B, Representative RT-PCR shows that the reporter gene was widely expressed in various tissues of a double transgenic rat carrying both the CAGtTA and the TREhLRRK2 transgenes. Genomic DNA-eliminated cDNA pools were extracted from the following tissues: 1, olfactory bulb; 2, cortex; 3, striatum; 4, hippocampus; 5, cerebellum; 6, brainstem; 7, spinal cord; 8, skeletal muscle; 9, heart; 10, lung; 11, liver; 12, spleen; 13, kidney; 14, stomach; 15, intestine; 16, colon; 17, testis; 18, ovary; 19, skin. RT-PCR for rat L17 gene serves as an internal control for equal loading of cDNA pools. C, Western blot detected widespread expression of the reporter gene hLRRK2 upon activation by the tTA transgene in transgenic lines 5 or 11. D-G, Immunoreactivity to HA tag is detected only in the tissues of the CAGtTA/TREhLRRK2-HA double transgenic rat (E, G), not in the tissues (D, F) of nontransgenic rats (WT).
Both the CAGtTA and the TREhLRRK2-HA transgenic constructs were linearized by restriction digestion and purified from an agarose gel. The transgene DNA was injected into the pronucleus of fertilized single-cell embryos of Sprague-Dawley (SD) rats. Surviving eggs were transferred into pseudopregnant female rats. Transgenic founder rats were identified by PCR-magnifying part of the transgenes with the following primers: 5'-AGCCTCTGCTAACCATGTTC-3' (forward) and 5'-AACCTTCGATTCCGACCTCA-3' (reverse) for the tTA transgene, and 5'-CTGTTGATCGTCTTGGACTC-3' (forward) and 5'-CCCAATCATTTCCAACATCC-3' (reverse) for the report gene hLRRK2. Copy number of the transgenes was determined by quantitative PCR. Transgenes were maintained on the genomic background of SD rats. Each line of the tTA transgenic founders was crossed with an expressing line of the hLRRK2-HA transgenic rats to produce double transgenic offspring (F1), in which the transcriptional activity of tTA was examined. Animal use followed NIH guidelines. The animal use protocol was approved by the Institutional Animal Care and Use Committees (IACUC) at Thomas Jefferson University.
Doxycycline treatment
To suppress tTA-activated gene expression, doxycycline hydrochloride (Dox; a Sigma-Aldrich) was continually supplied in drinking water at desired concentrations. Dox-containing water was supplied in a light-protected bottle and was renewed every three days. Rats were given regular food and water supplied with or without Dox of the indicated concentrations.
Quantitative PCR
Copy number of the transgene was estimated by quantitative PCR. Genomic DNA was extracted from tail biopsy and was adjusted to a constant concentration of 100 ng/μl. A pair of primers was selected specifically for the tTA transgene: 5'-AGCCTCTGCTAACCATGTTC-3' (forward) and 5'-AACCTTCGATTCCGACCTCA-3' (reverse). In parallel, the rat tyrosine hydroxylase (TH; a known single-copy gene) gene was used as an internal control for PCR quantification and was magnified with the primers 5'-CTCAAGAATCCTGTCACCAG-3' (forward) and 5'-ACTGCCTTTCAGGGTATGTC-3' (reverse). Rat genomic DNA was estimated to contain 3 x 109 base-pairs. The tTA transgene DNA was dissolved in genomic DNA of nontransgenic rats to obtain a series of standards from one to ten copies per genome. The resulting standard curve of tTA gene copy number was used for estimation of tTA transgene copy number in individual transgenic lines.
The mRNA level of the reporter gene human LRRK2 was also estimated with quantitative PCR. Total RNA was isolated from rat tissue with Trizol reagent (Sigma) and was further purified by digesting genomic DNA on a column (RNA easy kit and RNase-free DNase: Qiagen). One microgram of the purified RNA from each sample was reversely transcribed to cDNA with oligo-dT primer (RT kit, Invitrogen). Relative levels of hLRRK2 mRNA in individual samples were estimated by quantitative PCR with the following gene-specific primers: 5'-CAGCCTGACTATCAACTTCT-3' (forward) and 5'-TTGTTTGTGGATCGCTGTGA-3' (reverse) for the reporter gene hLRRK2, and 5'-TGGTTCGCTACTCCCTTGAC-3' (forward) and 5'-CTTGATGGCCTGGGCAGTT-3' (reverse) for rat L17 gene. The mRNA level of the L17 gene was used as an internal control for PCR quantification.
For quantitative PCR, the gene-specific primers were used at a concentration of 500 nM. Gene doses were measured by quantitative PCR with SYBR green kit per manufacturer's instruction (Qiagen). Cycling conditions were 15 min at 95°C followed by 40 cycles of 15 seconds at 94°C, 30 seconds at 60°C, and 20 seconds at 72°C. Aliquots of the amplified products were separated on 3% agarose gels to ensure amplification of the specific products at the predicted length. The threshold cycle number (Ct) for an examined gene was normalized to the Ct for the individual internal genes: TH gene for estimation of tTA transgene copy and L17 gene for estimation of reporter gene mRNA. The relative mRNA level of the reporter gene hLRRK2 was determined and expressed as a ratio of the mRNA level in the Dox-treated rats to that in the Dox-untreated rats, as described previously 37.
Western blotting
Animal tissues were homogenized in phosphate buffer (pH 7.4) supplied with protease inhibitors (Sigma). Tissue debris, unbroken cells, and cell nuclei were removed by centrifugation at 16,000 × g for 10 minutes. Protein content in the cleared lysate was determined by BCA assay (BioRad). Since protein product of the reporter gene human LRRK2 was about 280 kDa, the LRRK2 protein was barely detected in completely denatured sample 38. Protein sample of certain amounts was mixed with 2% SDS-containing loading dye and was partially denatured by incubating at 60°C for 5 minutes. Proteins were separated on 4-20% gradient SDS-PAGE and blotted onto GeneScreen Plus membrane (Perkin Elmer). Immunoreactivity to the epitope HA was detected by incubating the membrane with rabbit polyclonal antibody to HA (Rockland Inc., Pennsylvania). Immunoreactivity to GAPDH was also detected by incubating the membrane with a mouse monoclonal antibody to GAPDH (Abcam) and used as a control for equal loading. The immunoreactivity signal was developed with Super Signal kit (Pierce).
Immunohistochemistry
Rats were transcardially perfused with 4% paraformaldehyde in 0.1 M ice-cooled phosphate buffer. Their brains were dissected and further fixed in the same fixative for additional 24 hours. The dissected tissues were dehydrated in 30% sucrose and cut into serial coronal sections (30 μm thick) on a Cryostat. For detection of human LRRK2 protein, brain sections were incubated sequentially with a rabbit polyclonal antibody to the epitope HA (1:200; Rockland), with biotinylated goat anti-rabbit IgG (1:500; Vector Laboratories), and with peroxidase-conjugated avidin-biotin complex (ABC kit; Vector Laboratories). After thorough washing, bound antibodies were visualized by addition of diaminobenzidine (Vector Laboratories). Immunostained cells were observed under a Nikon microscope and were documented with a Nikon digital camera.
Rotarod test
Rats were first trained to stay on the rod of a rotarod (Med Associates) at a constant speed of 5 rpm, once a day for 5 days. Following the training, rats were tested on the Rotarod with an accelerating speed of 0.2 rpm/s, starting at 5 rpm. During a testing day, each rat was given three successive trials with a 20-minute interval. The latency to fall from the rod was recorded for each trial. The average latency was calculated for each testing day.
Statistical analysis
Data are expressed as means + SD. The statistical significance was tested with analysis of variance (ANOVA) followed by Tukey's post hoc test to compare group means. In all analyses, the null hypothesis was rejected at a level of 0.05.
3. Results
Copy-related expression of the tTA transgene in CAG-tTA transgenic rats
Production of transgenic rats appears more difficult than creation of transgenic mice because the pronuclei of rat zygotes are less regular and less uniform than those of mouse zygotes. Furthermore, the plasma and pronuclear membranes of rat zygotes are more plastic and sticky than those of mouse zygotes. About 31-75% of rat zygotes are viable after microinjection and are ready for implantation into pseudopregnant females 2, 30. We injected 540 single-cell embryos of SD rats with the CAGtTA transgene DNA at a concentration of 2ng/µl (microinjection buffer: Invitrogen) and transplanted 386 viable embryos into 12 pseudopregnant SD female rats (Figure 1A). From 68 pups, we identified three founder rats carrying the CAGtTA transgene (Figure 1B). Two of the three transgenic founders transmitted the transgene (Figure 1B: lines 5 and 11); however, founder 4 was infertile. We determined the copy number of the transgene in individual lines by quantitative PCR and detected 2 copies of the CAGtTA transgene in the line 5, 8 copies in the line 11, and 25 copies in the founder 4. The transgene was integrated at a single location in the genomes of the lines 5 and 11 because the copy number of the CAGtTA transgene was constant across generations. In addition, we tried to breed tTA transgenic lines to obtain homozygotes and succeeded with line 5 but not with line 11 (0 homozygotes in 30 transgenic offspring). No difference was observed in gross health between the homozygote and the hemizygote tTA rats.
To detect transgene expression, we first tried immunoblotting with commercially available antibodies that were raised with peptides corresponding to the tTA protein. Unfortunately, we could not detect tTA protein by western blotting in the two transmittable lines (Figure 1). In previous studies, low expression of tTA transgene has been shown sufficient to vigorously activate a tTA-dependent reporter gene 26, 39. We designed a pair of primers spanning the intron of tTA transgene, and thus, we could identify mature tTA mRNA by RT-PCR (Figure 1A). We detected expression of the tTA transgene in various tissues of the two transmittable lines (Figure 1C). In many tissues, the tTA transgene was expressed at higher levels in the 8-copy line (line-11) than in the 2-copy line (line-5), suggesting a pattern of transgene copy-related expression.
Vigorous activation of tTA-dependent reporter gene in the double transgenic rats
To test whether limited levels of tTA gene expression were sufficient to activate its target genes in vivo, we crossed the tTA transgenic lines with a tTA-responsive reporter line that carried human LRRK2 gene under control of the TREminiCMV promoter (Figure 2A). By RT-PCR, we detected a widespread expression of the reporter gene in many tissues examined (Figure 2B). Since the reporter gene was tagged by hemaglutinin (HA), expression of the reporter gene could be detected by immunoblotting or immunohistochemistry for HA antigen. HA immunoreactivity was detected only when both the tTA and the reporter genes were presented in the transgenic rats, but was not detected when only the reporter transgene was present (Figure 2C). These results indicate that expression of the reporter gene depended on tTA activation and that leakage of the reporter gene was negligible. Further, western blotting detected strong HA immunoreactivity in the double transgenic rats carrying the 8-copy tTA transgene, but detected relatively weak HA immunoreactivity in the rats carrying 2-copy tTA transgene (Figure 2C). Expression levels of the reporter gene corresponded with those of the tTA transgene. In addition, immunohistochemistry with an HA antibody detected widespread expression of the reporter gene in many tissues of double-transgenic animals, but no expression in tissues of animals carrying the reporter transgene alone (Figure 2D-G). Consistent with the previous findings in transgenic mice 26, 39, our results suggest that limited levels of tTA transgene expression were sufficient to vigorously activate its reporter gene in vivo.
Suppression of tTA-activated gene expression by Dox
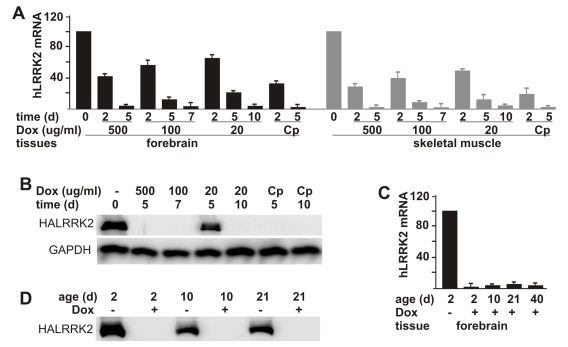
One advantage of the tetracycline-regulatory system is that transgene expression can be suppressed temporally. To test whether the tTA-activated gene was responsive to Dox, we first treated adult rats carrying both the tTA and the reporter transgenes. When administered at a constant concentration in drinking water, Dox effectively suppressed expression of the tTA-dependent reporter gene and reached its maximal effect in a dose-dependent pattern (Figure 3A, B). By day 5, Dox administered at 500 μg/ml reduced mRNA and protein levels of the reporter gene below detection threshold. In contrast, lower doses of Dox took a longer time to reach its maximal effects: 100 μg/ml and 20 μg/ml of Dox reached the maximal effect by day 7 and by day 10, respectively. To improve drug efficiency, we tested a complex regime of Dox administration: Dox administered at 500 μg/ml for 2 days and then at 20 μg/ml for the remainder of the experiment. The complex (Cp) regime reached maximal effect by day 5 and achieved a comparable result to the highest dosage for Dox (Figure 3A, B).
Figure 3.
Dox-dependent suppression of tTA-activated gene expression in transgenic rats. A, Dox turned off a tTA-dependent report gene at a dose-dependent speed. Adult female rats doubly transgenic for CAGtTA (line 11) and TREhLRRK2 were given Dox in drinking water at the indicated concentrations (Cp: 500 μg/ml for 2 days and then 20 μg/ml for the duration of the experiment). At the indicated time (d: day) after Dox treatment, rats were sacrificed for tissue collection. The mRNA levels of the report gene were measured by quantitative RT-PCR and expressed as a percentage of levels in the Dox-untreated rats. Data are means + SD (n = 3). B, Consistent with the RT-PCR results shown in A, western blot detected no expression of the reporter gene in the forebrain after Dox was administered for certain time (d: day). Brain tissues were taken from the same rats used for the RT-PCR shown in A. Immunoreactivity to GAPDH was also measured as a control for equal loading. C, Milk-secreted Dox continually suppressed tTA-dependent gene expression in neonatal rats. Pregnant rats were given Dox (20 μg/ml) in drinking water from the time of mating onward. The pups were sacrificed at indicated ages (d: day). Their forebrain was removed from the skull and dissected for extraction of RNA and protein. The mRNA levels of the reporter gene were measured by quantitative RT-PCR and expressed as a percentage of levels in the Dox-untreated rats. Data are means + SD (n = 4). Protein levels of the reporter gene were measured by western blot (D). For detection of HA-tagged human LRRK2 protein, 50 µg of total protein from each sample was loaded.
To study the functions of embryo-lethal genes in adult animals, we should avoid expression of genes at the embryonic and postnatal stages. Therefore, we tested the gene-suppression efficiency of milk-secreted Dox in postnatal rats. Expression of the reporter gene was effectively suppressed in postnatal rats from 2 days of age through weaning when their mother was given Dox (20 μg/ml) in drinking water since the date of mating (Figure 3C, D). Our results show that expression of the tTA-dependent transgene was subject to a tight and constant suppression by Dox in both adult and postnatal rats.
Reversibility of tTA-dependent gene expression after Dox withdrawal
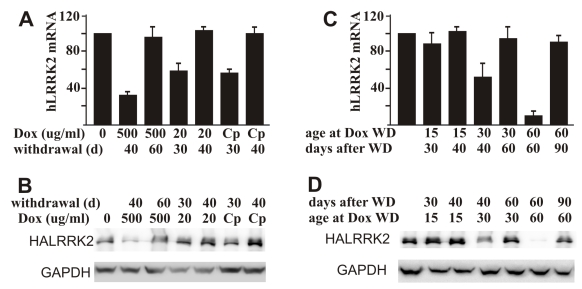
Compared to other gene regulatory systems, such as the Cre-lox system, a big advantage of the Tet regulatory system is that gene suppression can be released by Dox withdrawal. We first tested gene restoration in adult rats that were treated with Dox for 20 days. Expression of the reporter gene was efficiently restored in all the rats treated with three regimes of Dox (Figure 4A, B). Sixty days were required to remove the remnant effect of Dox administered at 500 μg/ml, but only 40 days were required to eliminate the remnant effect of Dox administered at 20 μg/ml (Figure 4A, B). Restoration time of gene expression is inversely relative to the dosage of Dox administered. We then tested recovery of gene expression in the rats inherently treated with Dox. We observed that gene recovery time depended on the age at Dox withdrawal. Periods of 40, 60, and 90 days were needed to recover gene expression in the rats depleted of Dox at ages of 15, 30, and 60 days, respectively (Figure 4C, D). Rats grow faster at early ages than at later ages (Figure 5A). Early withdrawal of Dox may require less time to remove the remnant effect of Dox due to a dilution effect from physical body expansion.
Figure 4.
Restoration of tTA-dependent gene expression depends on the dosage of Dox administered. A, B: Residual effect of Dox temporally administered depends on the dose of Dox. Female rats at age 40 days were given Dox in drinking water at indicated concentrations (Cp: 500 μg/ml for 2 days and then 20 μg/ml for the rest time) for 20 consecutive days. Rats were sacrificed at indicated days (d: day) after Dox withdrawal. Restoration of gene expression in the forebrain was examined by quantitative RT-PCR for human LRRK2 mRNA (A) and by western blotting for HA immunoreactivity (B). C, D: Residual effects of Dox depend on the duration of Dox administration. Mating rats were given Dox in drinking water at 20 μg/ml. Dox was withdrawn (WD: withdrawal) at the indicated ages of newborn rats. Forebrains of neonates were dissected at the indicated times after Dox withdrawal. Expression of the reporter gene was examined by quantitative RT-PCR for human LRRK2 mRNA (C) and by western blotting for HA immunoreactivity (D). Immunoreactivity to GAPDH served as a control for equal loading (50 μg of total protein per lane). Data are means + SD (n = 3-4).
Figure 5.
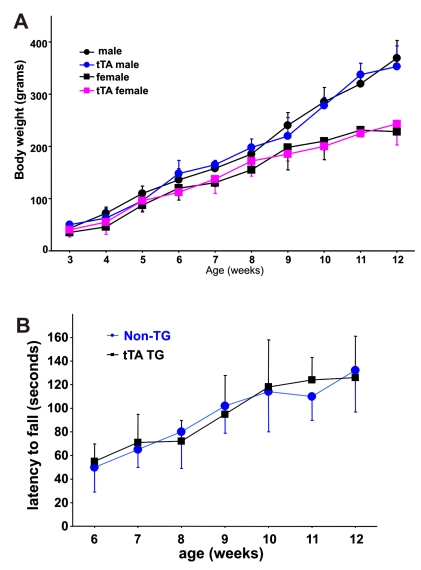
The tTA transgene does not affect rat growth or psychomotor function. A, The body weight of each rat was measured weekly after genotypes were determined. B, Psychomotor function of male rats was tested with a Rotarod weekly after a 5-day training period. Performance on the Rotarod was not different between the tTA transgenic rats and their nontransgenic littermates. Data are means + SD (n = 5-8).
No effect of the tTA transgene on animal growth or motor function
In transgenic studies, gene insertion may cause unwanted phenotypes due to interruption of other genes at the insertion site. In addition, overexpression of the transcriptional factor tTA may cause side effects 39. We examined tTA transgenic rats for overall health, but failed to observe any abnormal phenotypes. No differences were observed in the size of liver, heart, kidney, and brain between the tTA transgenic and the nontransgenic rats (data not shown). Compared to nontransgenic littermates, the tTA transgenic rats gained body weight at comparable speeds during postnatal development (Figure 5A). We monitored motor function using Rotarod test and observed no difference in psychomotor function between tTA transgenic and nontransgenic littermates (Figure 5B). Insertion and expression of the tTA transgene did not affect the growth or psychomotor function of rats.
4. Discussion
To develop a transgenic line for conditional expression of desired genes in rats, we used the tetracycline regulatory system and generated several lines of the transgenic rats carrying the tTA transgene. Using a vigorous and ubiquitous promoter to drive the tTA transgene, we obtained widespread expression of tTA in the central nerve system, skeletal muscle, heart, lung, and in most abdominal organs. Expression of the tTA was sufficient to vigorously activate its reporter gene, but was below the threshold to produce toxicity to the rats. These tTA transgenic rats are a model of inducible and reversible gene expression in the rat, and could be valuable to the development of genetic rat models for human diseases.
The Tet-regulatory system is widely used to achieve inducible and reversible expression of desired genes in vivo. To date, a great number of transgenic mice have been generated to express tTA (Tet-off) or rtTA (Tet-on) transactivators in ubiquitous or tissue-specific patterns 26, 27, 39. While few lines of transgenic rats have been created to express tTA in cell-specific patterns 29, 40, 41, a single line of tTA or rtTA transgenic rat is not available for ubiquitous activation of reporter gene in various cell types. To obtain ubiquitous expression of the tTA transgene in rats, we chose the ubiquitous promoter CAG, a hybrid promoter that has been constructed by fusing the cytomegalovirus enhancer with the chicken beta actin promoter. CAG has been shown to produce ubiquitous and robust gene expression in transgenic mice 35, 42, 43. Although CAG is a vigorous promoter, expression of tTA protein was not detectable by immunoblotting in the two transmittable lines (data not shown). We used the original bacterial codon for tTA. Sanbe et al. reported that bacterial codon usage bias leads to alternative splicing of tTA mRNA and results in a low abundance of full-length tTA mRNA 39. As a transcriptional activator, tTA at limited levels should be sufficient to activate its target gene. Excessive expression of tTA protein may cause toxicity to the cells because high abundance of exogenous transactivator may interfere with cellular transcription activity.
Indeed, enhanced expression of tTA using mammalian codon causes toxicity in transgenic mice 39, 44. To balance ubiquitous with adequate gene expression, we chose a ubiquitous promoter as the driver, but used the original bacterial codon for tTA. Consistent with previous finding in transgenic mice 26, 35, 39, 42, 43, our results in transgenic rats demonstrated that expression of tTA with bacterial codon was under the detection threshold by western blotting, but was sufficient to vigorously activate its target gene in various tissues. Insertion or expression of the tTA transgene may interfere with endogenous genes and cause toxicity to the animals. If the toxicity of the tTA transgene is evident, the toxicity should be reflected as an effect on growth, overall health, or organ development. In the two transmittable lines of transgenic tTA rats, no detectable abnormality was observed. These results suggest that insertion of the tTA transgene did not result in a deleterious dominant mutation and that expression of the tTA transgene did not obviously interfere with endogenous genes. These transgenic rats express tTA within a safe dosage and at sufficient levels to activate the target gene.
The dynamics of Dox-regulated gene expression were thoroughly examined for the first time in transgenic rats. We not only examined suppression of tTA-activated gene expression by Dox, but also examined reversibility of tTA-dependent gene expression after Dox withdrawal. Both the suppression and the reversal of gene expression were tested in adult and neonatal rats. In the two transmittable lines, tTA-mediated activation of the reporter gene was fully subjected to the regulation by Dox. In transgenic mice, Dox dose-dependently suppresses tTA-activated gene expression and washout of Dox remnant effect also corresponds to Dox dosage used 26, 39. Similarly, Dox reached its maximal effect of gene suppression at speeds proportional to its dose. High dose of Dox (500 μg/ml in drinking water) took a short time to fully suppress tTA-activated gene expression, but required a much long time to remove its remnant effect. In our studies, a duration of 5 days appeared to be the minimal time for complete gene suppression by orally administered Dox. We tested a complex regime of Dox administration to quickly reach the maximal effect but to successfully avoid extended duration of Dox cleanout. Dox was used at a high dose (500 μg/ml in drinking water) for 2 days to reach its effective concentration quickly and then used at a low dose (20 μg/ml) to maintain its effect constantly. This improved regime of Dox administration can help achieve a quick switch of tTA-activated gene expression between ON and OFF status. In addition, parental administration of Dox fully suppressed tTA-activated gene expression in offspring rats. Milk-secreted Dox was sufficient to reach its effective concentration in neonatal rats. Parental administration of Dox can help provide a constant suppression of tTA-dependent gene expression during embryonic and postnatal development. We also tested the tTA transgenic lines with another reporter gene and observed similar results (data not shown).
Although laboratory mice are preferred for functional genetics, laboratory rats show significant advantages as model animals in pharmacological and toxicological studies, in behavioral analysis, in microsurgery such as cell transplantation, and in studies on neurological diseases. In modeling some human diseases, animal species may play an important role in phenotypic expression. For example, no substantial loss of dopaminergic neurons is observed in human alpha-synuclein transgenic mice 45, 46, but progressive loss of the neurons is induced by transient expression of human alpha-synuclein in rats 47. Animal species may not be a sole factor contributing to the variation of phenotypic expression in alpha-synuclein models. In some genetic models for human disease, developmental compensation may dilute phenotypic expression when the gene is constitutively mutated. Such compensatory changes are increasingly observed in genetic models 48, 49. Disrupting one gene of the JNK family causes compensatory changes in the other related genes 49. Studies on Jnk2 knockout mice with the gene disrupted from germline suggest a negative regulatory role in cell proliferation for the protein kinase Jnk2 50. In contrast, studies of Jnk2 inhibition in adult mice suggest Jnk2 as a positive regulator for cell proliferation 49. These distinct phenotypes may result from developmental compensation by upregulated expression of the related genes 49. Compared with the human, the rodent has a short lifetime. Developmental compensation may buffer the detrimental effects of gene mutation and thus attenuate or even prevent the phenotypes of gene mutation from developing in a rodent's lifetime. Selective expression of disease genes in adult animal may increase the chance to reproduce the phenotypes of human diseases in the animals. Recent advances in transgenic RNAi indicates that RNAi-mediated gene silencing can reproduce phenotypes of the gene knockout 37, 51, 52. Transgenic RNAi is a convenient approach to achieve hypomorphic phenotypes in the transgenic rats. To maintain RNAi transgenic lines, conditional expression of the RNAi transgene is necessary when the hypomorphic phenotype is lethal or infertile. In the aforementioned applications, the tTA transgenic rats would be a valuable tool for accomplishment of these goals.
Acknowledgments
We thank Dr. Miyazaki at Osaka University Medical School and Dr. Nagy at the University Of Toronto for CAG constructs. We thank Dr. Saunders and Dr. Filipiak at the Michigan University for teaching us the technique to create transgenic rats.
This work was supported by NIH/National Center for Research Resources [R21RR024586 to Xu Gang Xia] and by NIH/National Institute of Environmental Health Sciences [R01ES016760 to Xu Gang Xia].
References
- 1.Tesson L, Cozzi J et al. Transgenic modifications of the rat genome. Transgenic Res. 2005;14(5):531–46. doi: 10.1007/s11248-005-5077-z. [DOI] [PubMed] [Google Scholar]
- 2.Filipiak WE, Saunders TL. Advances in transgenic rat produc-tion. Transgenic Res. 2006;15(6):673–86. doi: 10.1007/s11248-006-9002-x. [DOI] [PubMed] [Google Scholar]
- 3.Aiba A, Nakao H. Conditional mutant mice using tetracy-cline-controlled gene expression system in the brain. Neurosci Res. 2007;58(2):113–7. doi: 10.1016/j.neures.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Fisch SG. Animal models and human neuropsychiatric disor-ders. Behav Genet. 2007;37(1):1–10. doi: 10.1007/s10519-006-9117-0. [DOI] [PubMed] [Google Scholar]
- 5.Gaveriaux-Ruff C, Kieffer LB. Conditional gene targeting in the mouse nervous system: Insights into brain function and dis-eases. Pharmacol Ther. 2007;113(3):619–34. doi: 10.1016/j.pharmthera.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Lull EM, Freeman MW, Vrana EK, Mash CD. Correlating hu-man and animal studies of cocaine abuse and gene expression. Ann N Y Acad Sci. 2008;1141:58–75. doi: 10.1196/annals.1441.013. [DOI] [PubMed] [Google Scholar]
- 7.Bouwknecht AJ, Paylor R. Pitfalls in the interpretation of genetic and pharmacological effects on anxiety-like behaviour in rodents. Behav Pharmacol. 2008;19(5-6):385–402. doi: 10.1097/FBP.0b013e32830c3658. [DOI] [PubMed] [Google Scholar]
- 8.Cao Q, Xu et al. Functional recovery in traumatic spinal cord injury after transplantation of multineurotrophin-expressing glial-restricted precursor cells. J Neurosci. 2005;25(30):6947–57. doi: 10.1523/JNEUROSCI.1065-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang PY, Shields et al. Use of magnetic stimulation to elicit motor evoked potentials, somatosensory evoked potentials, and H-reflexes in non-sedated rodents. J Neurosci Methods. 2007;165(1):9–17. doi: 10.1016/j.jneumeth.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26(3):137–48. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 11.Buehr M, Meek et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135(7):1287–98. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Li P, Tong et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135(7):1299–310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng HJ, She et al. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294(1):G39–49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 14.Mullins JJ, Peters J, Ganten D. Fulminant hypertension in transgenic rats harbouring the mouse Ren-2 gene. Nature. 1990;344(6266):541–4. doi: 10.1038/344541a0. [DOI] [PubMed] [Google Scholar]
- 15.Herrera VL, Makrides SC, Xie HX, Adari H, Krauss RM, Ryan US, Ruiz-Opazo N. Spontaneous combined hyperlipidemia, coronary heart disease and decreased survival in Dahl salt-sensitive hypertensive rats transgenic for human choles-teryl ester transfer protein. Nat Med. 1999;5(12):1383–9. doi: 10.1038/70956. [DOI] [PubMed] [Google Scholar]
- 16.von Horsten S, Schmitt I, Nguyen HP et al. Transgenic rat model of Huntington's disease. Hum Mol Genet. 2003;12(6):617–24. doi: 10.1093/hmg/ddg075. [DOI] [PubMed] [Google Scholar]
- 17.Mordes JP, Bortell R, Blankenhorn EP, Rossini AA, Greiner DL. Rat models of type 1 diabetes: genetics, environment, and autoimmunity. Ilar J. 2004;45(3):278–91. doi: 10.1093/ilar.45.3.278. [DOI] [PubMed] [Google Scholar]
- 18.Atkinson MA, Leiter EH. The NOD mouse model of type 1 diabetes: as good as it gets? Nat Med. 1999;5(6):601–4. doi: 10.1038/9442. [DOI] [PubMed] [Google Scholar]
- 19.Chaillous L, Lefevre et al. Oral insulin administration and residual beta-cell function in recent-onset type 1 diabetes: a multicentre randomised controlled trial. Diabete Insuline Orale group. Lancet. 2000;356(9229):545–9. doi: 10.1016/s0140-6736(00)02579-4. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson AM, Maclaren KN, Luchetta R. Insulitis and dia-betes in NOD mice reduced by prophylactic insulin therapy. Diabetes. 1990;39(8):933–7. doi: 10.2337/diab.39.8.933. [DOI] [PubMed] [Google Scholar]
- 21.Mullins JL, Mullins JJ. Insights from the rat genome se-quence. Genome Biol. 2004;5(5):221. doi: 10.1186/gb-2004-5-5-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs RA, Weinstock GM, Metzker ML et al. Genome sequence of the Brown Norway rat yields insights into mammalian evo-lution. Nature. 2004;428(6982):493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 23.Canzian F. Phylogenetics of the laboratory rat Rattus norvegicus. Genome Res. 1997;7(3):262–7. doi: 10.1101/gr.7.3.262. [DOI] [PubMed] [Google Scholar]
- 24.Kuan YC, Yang et al. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain de-velopment. Neuron. 1999;22(4):667–76. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 25.Fon AE, Pothos et al. Vesicular transport regulates monoam-ine storage and release but is not essential for amphetamine ac-tion. Neuron. 1997;19(6):1271–83. doi: 10.1016/s0896-6273(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 26.Hong KH, Chong et al. Inducible and reversible Clock gene expression in brain using the tTA system for the study of cir-cadian behavior. PLoS Genet. 2007;3(2):e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Z, Zheng T, Lee CG, Homer RJ, Elias JA. Tetracy-cline-controlled transcriptional regulation systems: advances and application in transgenic animal modeling. Semin Cell Dev Biol. 2002;13(2):121–8. doi: 10.1016/s1084-9521(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 28.Stark KL, Gross C et al. A novel conditional knockout strategy applied to serotonin receptors. Handb Exp Pharmacol. 2007;178:347–63. doi: 10.1007/978-3-540-35109-2_14. [DOI] [PubMed] [Google Scholar]
- 29.Braudeau C, Bouchet D, Toquet C et al. Generation of heme oxygenase-1-transgenic rats. Exp Biol Med (Maywood) 2003;228(5):466–71. doi: 10.1177/15353702-0322805-07. [DOI] [PubMed] [Google Scholar]
- 30.Tesson L, Cozzi J, Menoret S, Remy S, Usal C, Fraichard A, Anegon I. Transgenic modifications of the rat genome. Transgenic Res. 2005;14(5):531–46. doi: 10.1007/s11248-005-5077-z. [DOI] [PubMed] [Google Scholar]
- 31.van den Brandt J, Wang D, Kwon SH, Heinkelein M, Reichardt HM. Lentivirally generated eGFP-transgenic rats allow efficient cell tracking in vivo. Genesis. 2004;39(2):94–9. doi: 10.1002/gene.20037. [DOI] [PubMed] [Google Scholar]
- 32.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes de-livered by lentiviral vectors. Science. 2002;295(5556):868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 33.Heng YM, Detloff JP, Albin LR. Rodent genetic models of Huntington disease. Neurobiol Dis. 2008;32(1):1–9. doi: 10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Cozzi J, Fraichard A, Thiam K. Use of genetically modified rat models for translational medicine. Drug Discov Today. 2008;13(11-12):488–94. doi: 10.1016/j.drudis.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 36.Novak A, Guo et al. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28(3-4):147–55. [PubMed] [Google Scholar]
- 37.Zhou H, Falkenburger BH, Schulz JB, Tieu K, Xu Z, Xia XG. Silencing of the Pink1 Gene Expression by Conditional RNAi Does Not Induce Dopaminergic Neuron Death in Mice. Int J Biol Sci. 2007;3(4):242–50. doi: 10.7150/ijbs.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biskup S, Moore et al. Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci. 2007;8:102. doi: 10.1186/1471-2202-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanbe A, Gulick et al. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92(6):609–16. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 40.Barton DM, Dunlop et al. Modified GFAP promoter auto-regulates tet-activator expression for increased transacti-vation and reduced tTA-associated toxicity. Brain Res Mol Brain Res. 2002;101(1-2):71–81. doi: 10.1016/s0169-328x(02)00170-5. [DOI] [PubMed] [Google Scholar]
- 41.Tesson L, Charreau et al. Endothelial expression of Fas ligand in transgenic rats under the temporal control of a tetracy-cline-inducible system. Transplant Proc. 1999;31(3):1533–4. doi: 10.1016/s0041-1345(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 42.Kubo J, Yamanouchi K, Naito K, Tojo H. Expression of the gene of interest fused to the EGFP-expressing gene in trans-genic mice derived from selected transgenic embryos. J Exp Zool. 2002;293(7):712–8. doi: 10.1002/jez.10163. [DOI] [PubMed] [Google Scholar]
- 43.Vintersten K, Monetti et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult ani-mals. Genesis. 2004;40(4):241–6. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- 44.Morimoto M, Kopan R. rtTA toxicity limits the usefulness of the SP-C-rtTA transgenic mouse. Dev Biol. 2009;325(1):171–8. doi: 10.1016/j.ydbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein RL, King MA, Hamby ME, Meyer EM. Dopaminergic cell loss induced by human A30P alpha-synuclein gene transfer to the rat substantia nigra. Hum Gene Ther. 2002;13(5):605–12. doi: 10.1089/10430340252837206. [DOI] [PubMed] [Google Scholar]
- 46.Maingay M, Romero-Ramos M, Kirik D. Viral vector medi-ated overexpression of human alpha-synuclein in the ni-grostriatal dopaminergic neurons: a new model for Parkinson's disease. CNS Spectr. 2005;10(3):235–44. doi: 10.1017/s1092852900010075. [DOI] [PubMed] [Google Scholar]
- 47.Hashimoto M, Rockenstein E, Masliah E. Transgenic models of alpha-synuclein pathology: past, present, and future. Ann N Y Acad Sci. 2003;991:171–88. [PubMed] [Google Scholar]
- 48.Sage J, Miller et al. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424(6945):223–8. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 49.Jaeschke A, Karasarides et al. JNK2 is a positive regulator of the cJun transcription factor. Mol Cell. 2006;23(6):899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 50.Sabapathy K, Hochedlinger K, Nam SY, Bauer A, Karin M, Wagner EF. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol Cell. 2004;15(5):713–25. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 51.Xia GX, Zhou et al. Pol II-expressed shRNA knocks down Sod2 gene expression and causes phenotypes of the gene knockout in mice. PLoS Genet. 2006;2(1):27. doi: 10.1371/journal.pgen.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dickins AR, McJunkin et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39(7):914–21. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]