Abstract
T-cell precursors remain developmentally plastic for multiple cell generations after entering the thymus, preserving access to developmental alternatives of macrophage, dendritic cell, and even mast-cell fates. The underlying regulatory basis of this plasticity is that early T-cell differentiation depends on transcription factors which can also promote alternative developmental programs. Interfactor competition, together with environmental signals, keep these diversions under control. Here the pathways leading to several lineage alternatives for early pro-T cells are reviewed, with close focus on the mechanisms of action of three vital factors, GATA-3, PU.1, and Notch-Delta signals, whose counterbalance appears to be essential for T-cell specification.
Keywords: T cell development, Lineage commitment, Mast cells, Dendritic cells, Transcription factor
1. MODELS FOR HEMATOPOIETIC LINEAGE CHOICE
Hematopoietic stem cells can develop into any of at least ten different kinds of effector cells, and these can be grouped broadly into lymphoid, myeloid, erythroid, megakaryocytic, and other types. Stem cells become particular cell types by passing through sequences of partially restricted, but still pluripotent intermediates. The outcome of competing regulatory forces at work on the cells during the partially restricted intermediate stages ultimately forms the basis for the commitment decision and controls differentiation. In this review, we will discuss how this works as revealed in cells making the choice between the T-cell fate and its developmental alternatives.
Particular transcription factors can drive specific hematopoietic lineage choices. An example is the way erythro-megakaryocytic lineages separate from myeloid lineages (rev. by [1]). Here, the zinc finger factor GATA-1 apparently drives erythro-megakaryocytic fates in opposition to the E26 transformation-specific (Ets) family factor PU.1, whereas PU.1 plays a dominant role to direct myeloid fates in opposition to GATA family factors [2–6]. Downregulation of PU.1 in hematopoietic precursors provides one of the earliest indices that cells have undertaken an erythro-megakaryocytic lineage pathway [7,8]. In another dichotomy, Notch signals that activate the RBPJ transcription factor (also known as CSL, for CBF-1, Suppressor of Hairless, Lag-1) are pitted against the activity of the Paired-homeodomain transcription factor Pax5. Notch activation triggers and sustains the T-lymphocyte program while blocking the B-lymphocyte program, whereas Pax5 directs the B-cell program while blocking the T-cell program, in part by inhibiting Notch1 expression [9–11]. As these oppositions suggest, the dominance of each of these factors in a particular pathway is ultimately based not only on its ability to activate cell-type-specific genes, but also on its ability to repress or inhibit any factors important for alternative pathways (rev. in [12]).
The discovery of partially restricted progenitors, stable enough to identify and isolate prospectively, marked a profound advance toward understanding how hematopoietic lineage choice occurs [13–15]. The existence of these restricted progenitors revealed that when cells depart from the stem cell state they are not yet committed to adopt one particular developmental fate. Rather, they begin differentiating while equipped with a regulatory apparatus that could support any of several alternative fates. This suggests that fates may be grouped based on relatedness of the gene expression programs they activate, a grouping that ought to be reflected in the modes of action of their dominant transcription factors.
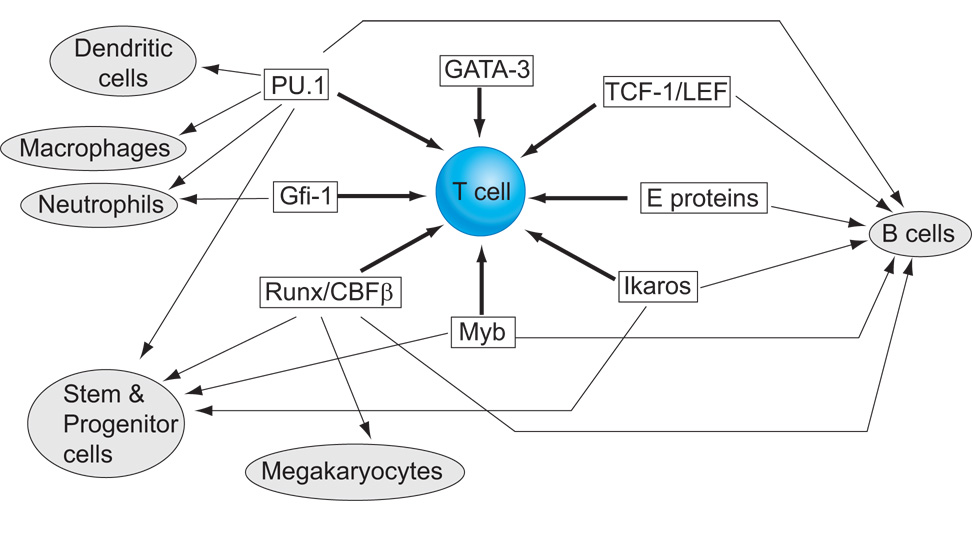
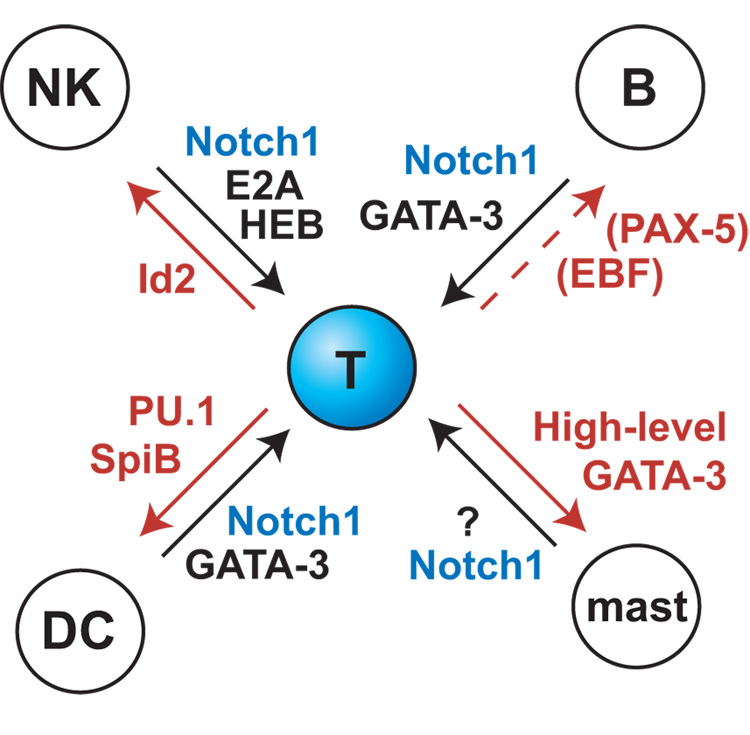
The exact relationships between these powerful regulators, in action, depend on whether lineage decisions are strictly hierarchical or not, with potential for more similar fates being combined longer than potential for more distinct fates. The notion that lymphocytes derive from a common lymphoid precursor and myeloid cells from a common myeloid precursor has been highly appealing. However, in a challenge to the hierarchical models, there is now abundant evidence that most T lymphoid precursors lose B lymphoid developmental potential earlier than they lose myeloid potential [16–19]. Most radically, T-cell development – like dendritic cell development – may not even be required to pass through a unique sequence of intermediates [20–25]. This can be explained by the fact that the essential T-lineage developmental program depends on a substantial overlapping of activities between transcription factors that are “T-cell specific” and factors capable of driving other fates (Figure 1). The specification of particular hematopoietic cell types by shifting, overlapping combinations of lineage-nonspecific factors has been noted for other lineages [26], and in all such cases it can only work if there is precise quantitative and stage-specific regulation of the factors involved to enable differentiation to progress. This overlap and mutual constraint is vividly illustrated in the relationships between three essential factors, Notch, PU.1, and GATA-3, during early T-cell development.
Figure 1.
T cell regulatory requirements in relation to other hematopoietic pathways. Schematic summary of the transcription factor requirements for T-lineage specification from hematopoietic stem cells (thick arrows), indicating alternative hematopoietic pathways that are naturally promoted by the same factors (thin arrows). For reviews and updated references, see [1,39,154–162]. Note that although GATA-3 is T-lineage specific under normal conditions shown here, it is also capable of driving a non-T differentiation program even in early T-lineage cells if expressed at an elevated level.
2. LANDMARKS FOR EARLY T-CELL DEVELOPMENT
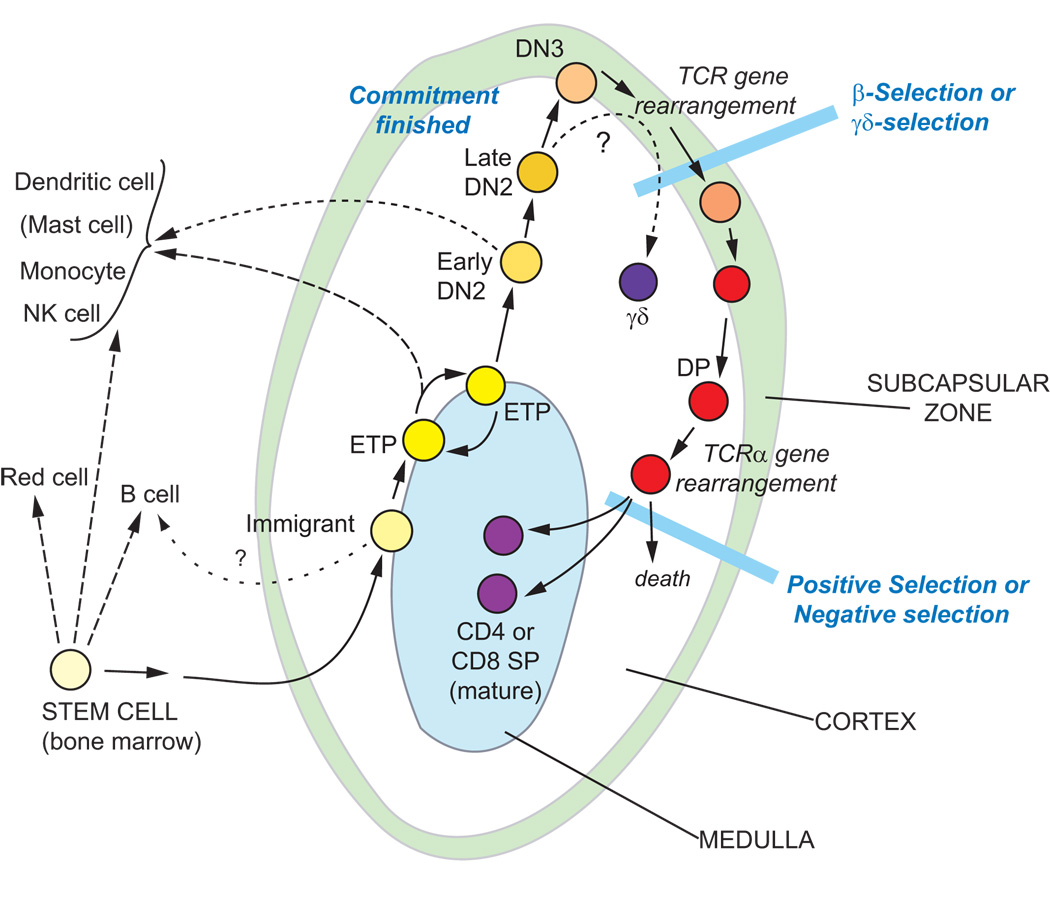
Early-stage T-cell precursors are partially restricted cells that migrate to the thymus either just after or immediately before losing the ability to give rise to B lymphocytes, but before they shut off molecularly distinct developmental alternatives including macrophage or dendritic-cell fates. Differentiation of these cells occurs in the thymic cortex over a period of many days and is accompanied by a flexible degree of proliferation. Fig. 2 summarizes the cell-surface and molecular phenotypic criteria that distinguish mouse thymocytes in successive stages toward T-lineage commitment [27–34].
Figure 2.
T cell developmental stages. Stages of early T-cell development in the young postnatal mouse are shown in the context of their ordered migration through different compartments of the thymus (rev. in [28,153]). Cells in the ETP and DN2 stages undergo extensive proliferation (>10 cell cycles in all), and then the only other major phase of proliferation is immediately following β-selection. To the left are shown the developmental fates that cells at each stage can display if they are removed from the thymic microenvironment at the indicated stages.
The earliest thymic subset, Early T-cell precursors (ETP) or “DN1” cells (CD3−, CD4−, CD8−, c-Kithi, CD44+, and CD25−)1, can undergo extensive intrathymic proliferation, especially in adult animals. But throughout this proliferation ETPs remain uncommitted to the T lineage, as they retain efficient competence to develop into natural killer, macrophage, and dendritic cells [16–19,35]. Transcription of Rag1, Rag2, and many T-lineage-specific genes is low or undetectable at the ETP stage. Then, a broad increase in T-lineage-associated gene expression begins as the cells pass to the next stage, called DN2 (CD3−, CD4−, CD8−, c-Kit+, CD44+, CD25+). Still, many uncommitted cells remain in the DN2 population, exhibiting dendritic-cell, macrophage, and NK potential, and some cells in the DN2 population can even develop into mast cells [36]. However, these alternatives disappear completely as the cells progress further to the DN3 stage (CD3−, CD4−, CD8−, c-Kitlow, CD44low, CD25+). DN3 cells express a full panoply of T-cell specific genes, indicating the maximal activation of positive regulators of T-lineage identity. At this stage, expression of Rag1 and Rag2 is fully upregulated, most cells slow or stop their proliferation, and the first complete rearrangements of TCRβ, TCRγ, and TCRδ genes occur. If successful, these rearrangements enable the cells to assemble a pre-TCR (rearrranged TCRβ + invariant pre-TCRα) or TCRγδ complex at the cell surface, and these complexes trigger signals that enable the cells to undergo β-selection or γδ-selection, respectively.
The stages of intrathymic development that lead up to β-selection (ETP to DN3) can be described as pro-T cell stages. T-cell development through these stages depends on a set of regulators that include both T-cell specific and shared transcription factors (Fig. 1)[30,32,37–42]. The T-cell specific transcription factors GATA-3 and TCF-1 are transcriptionally induced from a low level in prethymic precursors to a substantial level as early as the ETP stage, with further increase to the DN3 stage [43–47]. Another crucial transcription factor, the basic helix-loop-helix transcription factor E2A, is strongly upregulated through stabilization at the protein level, starting at the DN2 stage [48,49].
At the same time, a rich legacy of transcription factor gene expression inherited from stem cell precursors continues throughout early stages of T-cell development [43,46,50]. Some prethymically expressed factors are maintained all the way through lineage commitment (E.-S. David-Fung et al., unpublished results), and several regulators crucial for T-cell differentiation have stem cell provenance; e.g. Myb, Runx1, Ikaros, and Gfi-1 [51]. Nevertheless, there are also parts of the stem cell legacy that must be downregulated in a precisely staged way in order to allow T-cell development to proceed. The ETP population retains strong expression of a number of stem cell associated factors, among them PU.1 and SCL, and at least in some ETP cells, GATA-2 and/or C/EBPα as well [52–55]. All of these genes are shut off by the DN3 stage. PU.1, C/EBPα, and GATA-2 can all promote non-T fates when overexpressed [36,53,54,56–59], and their precisely regulated repression is important for lineage commitment, as discussed below.
3. CONTROL OF EARLY T-CELL DEVELOPMENT BY EXTERNAL SIGNALS
T-lineage specification is driven and maintained by signals from the environment. The transition from each DN stage to the next depends on the existing internal transcription factor complement in the cell together with changes in transcription factor function induced by external environmental signals. The most important of these signals, on current evidence, are cytokine signals, signals through the Wnt-β-catenin-TCF pathway, and repeated signals from the Notch pathway. As for other hematopoietic lineages, receptors for trophic signals are highly regulated in T-lineage cells and only select receptors are expressed.
For survival and proliferation, the earliest T cell precursors may rely on signaling through the cytokine receptor c-Kit [60], but also from ETP stage through the end of DN2 stage they are dependent on IL-7/IL-7R signals [61]. Among the transcription factors required at these stages, recent evidence suggests that the Runx family factors may help establish appropriate levels of proliferation through their impact on IL-7R expression and function [62,63]. Proliferation may compete with differentiation, especially for adult-derived immature thymocytes, as high-level IL-7/IL-7R or c-Kit signaling is seen to retard progression from DN2 to DN3 stage even while it promotes expansion [23,64,65]. However, these cytokine signals do not appear to promote differentiation, and are not sufficient to maintain viability without additional survival signals.
Wnts are suspected to be another part of the survival signal. This inference is based on the importance of the TCF-1 transcription factor and its relative LEF-1, which are the transcriptional effectors of the Wnt canonical signaling pathway. Both of these transcription factors are converted from repressors to activators by β-catenin, which is mobilized by signals from the canonical Wnt pathway. Activity of TCF-1 or LEF-1 is essential for passage through β-selection, and TCF-1 also plays a critical, nonredundant role in adult thymocytes in progression from ETP to DN2 [47,66,67]. TCF-1 target genes in thymocytes include many survival and proliferation genes [68]. It is supposed that most TCF/LEF functions are Wnt dependent, based on the severe developmental blocks caused by transgenic expression of Wnt inhibitory proteins, Dickkopf and ICAT [69,70]. However, the connections between the initial Wnt input, the effector function of TCF-1, and the β- or γ-catenin proteins that normally act as intermediates have remained somewhat difficult to parse. Forced activation of β-catenin in progenitor cells apparently causes them to arrest development in a multipotent stage and reverse commitment, rather than enhancing their commitment to the T-cell fate [71–73]. New data suggest that hematopoiesis and apparently normal T-cell development can occur even in the absence of both β- and γ-catenin [74], suggesting that some roles for TCF/LEF family transcription factors could depend on other mediators. This ambiguity raises the question of whether TCF-1 in DN2-stage cells could actually be playing an important role as a repressor rather than as an activator. If so, one potential repression target implicated at this stage is the PU.1 gene [75].
The most dominant environmental factor in T-cell specification is the availability of ligands for the transmembrane receptor, Notch. As reviewed elsewhere in detail, interaction between Notch1 and its ligands of the Delta-like class is a distinctive requirement for precursors entering and pursuing the T-cell pathway, from the generation and maintenance of ETPs all the way to the beginning of β-selection (see reviews [9,10,28,31,76–78]). Notch-Delta signaling contributes not only to T-lineage survival and growth but also to lineage choice. Notch-Delta signaling favors T-cell development and blocks B-cell development from multilineage prethymic precursors; then, even after B-lineage potential is permanently suppressed, it blocks ETP and DN2 cells from access to all other physiological alternatives to T-lineage choice [16,35,79,80].
4. CONTEXT-DEPENDENT MODULATION OF NOTCH SIGNALS
Notch signaling is not a Boolean operator that determines T-lineage fate at a stroke, but a quantitatively modulated participant in protracted lineage decisions. A sustained succession of interactions with Delta-class target ligands, through all the cell cycles from ETP throughout DN2 stages, is needed to elicit full commitment to the T lineage [35,36,45,81]. Notch/Delta interaction can drive many different starting populations of hematopoietic precursors into T-cell development [21,23,36], but the dosages required are subject to distinct thresholds at different stages [82–84], and different Notch-triggered effects require different signal intensities. Prethymic precursors from bone marrow require substantially higher levels of Notch signaling than even the earliest intrathymic precursors to initiate T-cell development in vitro [84]. However, only a low level of Notch signaling is needed to suppress and eliminate the B-cell option [35,81], while a stronger Notch signal intensity is needed to block the dendritic-cell and natural killer cell fate options [35,83,85]. ETP and the earliest fetal thymic immigrants begin T cell development in a Notch “primed” state, already showing upregulation of Notch target genes very early after entering the thymus [44,82,84,86,87]. Yet known Notch target gene expression peaks at the DN3 stage [43,88,89]. It is not clear yet which this apparently amplified response reflects: stronger Notch-Delta signals, or interaction of constant-level Notch signals with a developmentally altered regulatory context.
The biochemical mechanisms through which cells detect and respond differentially to different levels of Notch pathway signals are still being investigated. Notch engagement triggers proteolytic release of the intracellular domain of Notch, which moves to the nucleus and provides an activation domain for the pre-existing transcription factor RBPJ (CSL). Recent data provide a possible mechanism to make certain target genes more sensitive than others to the strength of Notch signaling. Head to head CSL binding sites, called SPS (sequence paired sites), preferentially direct cooperative assembly of transcriptionally active complexes of RBPJ, intracellular Notch, and the coactivator MAML1 as compared to single sites for RBPJ [90]. This suggests a mechanism for translating strength of Notch signaling through to the target gene. Furthermore, such SPS sites may favor cooperation with basic helix-loop-helix E proteins bound at neighboring elements [91]. Such cis-regulatory elements could structurally underlie the responses that depend on Notch-E protein cooperation [92] at many early stages of T-cell development[93]. At the same time, Notch signaling is highly modulated by antagonistic transcription factors such as LRF, or Zbtb7a [94]; by antagonists of MAML1 assembly like Msx2-interacting protein (MINT)[95]; and by feedback from negative regulators of the cytoplasmic part of the cascade such as Deltex1 and Nrarp [96,97]. One further way in which the Notch signal could be regulated to achieve gradations of signal strength and duration is by diversity in the sites of S3 cleavage of Notch [98]. This generates two types of Notch intracellular domain, one more stable than the other, with a resulting effect on intensity of transcriptional response.
In the past two years, Notch signaling has been suggested to affect T-lineage gene expression through yet additional pathways, and this is through gene-specific modulation of the effects of other transcription factors. Although the biochemical mechanisms remain to be determined, Notch pathway mediators can also modify the activities of GATA-3 and PU.1 as discussed below, effectively restraining their abilities to promote non-T fates.
5. ESSENTIAL ROLES FOR GATA-3 AND PU.1 IN EARLY T CELL DEVELOPMENT
Both GATA-3 and PU.1 are essential for T-cell development. GATA-3, a double zinc finger (C4-type) transcription factor, is expressed in an almost T lineage-specific pattern among hematopoietic cells. It is turned on among the earliest genes activated by Notch-Delta signaling in hematopoietic precursors, and is used throughout T cell development from the ETP stage all the way to post-thymic responses to antigen. The ETS-family transcription factor PU.1 is almost the reverse, with virtually no expression in most thymocytes or mature T cells. PU.1 is most prominent in B cells, multipotent hematopoietic progenitors, and myeloid cells of various lineages, where it has well-documented positive gene regulatory activities. The competitive antagonism between PU.1 and GATA family factors is thought to control myeloid vs. erythro-megakaryocytic fate decisions as already noted, and so a simple expectation would be that GATA-3 might dictate T-lineage fate while PU.1 should oppose it. Nevertheless, substantial expression of GATA-3 and PU.1 RNA overlaps in ETP and DN2 stage thymocytes.
The loss of GATA-3 at the germline or stem-cell stage eliminates all recognizable T-cell precursors at least as early as the DN2 stage [99–101]. If GATA-3 is conditionally deleted later, at different times after thymic entry, powerful effects are seen at multiple later stages of T-cell development (rev. by [38]). Similarly, germline disruption or early conditional deletion of PU.1 eliminates most or all T cell development [102–105]; a severe hypomorphic allele also causes profound inhibition of fetal T-cell development [106]. Because PU.1 expression in T-lineage cells is confined to the earliest DN stages, the knockout phenotype is also abnormal in early stages. PU.1 is needed for generation of precursors with the ability to migrate to the thymus; it is not yet clear for how long after their arrival intrathymic intermediates require PU.1 as well. However, throughout the extensive intrathymic proliferation by ETPs, these cells maintain a level of PU.1 expression similar to those found in multilineage prethymic precursors and stem cells [7,8,43,107], arguing for a continuing intrathymic role.
The target genes that GATA-3 and PU.1 are needed to regulate at these early stages are not clearly identified, in part because so little is known about the requirements of cells so close to the stem-cell stages. The blocks caused by losses of these factors long precede the blocks that are seen at β-selection due to lack of TCR rearrangements or any of the signaling components that transduce TCR signals. Thus, most T-lineage genes that have been suggested as targets for GATA-3, including the Rag recombinases, TCR genes, CD5, and the Th2 cytokine complex, are unlikely to account for its early essential role (see [38] for review). Both GATA factors and PU.1, however, can positively or negatively affect expression of the two most important growth factor receptors used in ETP and DN2 cells, namely c-Kit and IL-7Rα [36,108–112]. These results raise the possibility that balanced activity of GATA-3 and PU.1 is primarily needed in early thymocytes to support survival and growth.
The paradigm of competition between PU.1 and GATA factors in erythromyeloid development might make this cooperative action seem surprising, but it is not exceptional. First, the mutual interference that occurs between PU.1 and GATA factors is at the level of protein/protein interaction. This kind of mechanism should depend on diffusion, and should not benefit from specific co-recruitment to neighboring DNA sites to accelerate interaction. Therefore, interference should be highly concentration-dependent, and as long as both PU.1 and GATA protein concentrations are below a certain threshold, they may not interfere in fact. Second, there may be steric constraints to the antagonism between PU.1 and GATA factors at target genes where both factors engage DNA simultaneously. For example, the inhibition of PU.1 transactivation by GATA-1 depends on an interaction between the DNA binding domains of the factors, the Ets β3-β4 region of PU.1 with the C-terminal zinc finger of GATA-1 [4,113], whereas the inhibition of GATA-1 DNA binding by PU.1 requires polarized binding of the PU.1 N-terminus to the GATA-1 C-terminal finger [114]. There is clearly no antagonism at some mast-cell lineage target genes that depend on simultaneous GATA and PU.1 binding [115]. Thus, balanced activity of GATA-3 and PU.1 could selectively promote expression of a particular subset of target genes that respond to both factors, while inhibiting expression of targets that respond to one or the other exclusively.
6. GATA-3-DRIVEN LINEAGE INFIDELITY: DOSAGE AND NOTCH SENSITIVITY
With its T-lineage specific expression and recurrent T-lineage roles, GATA-3 has been an appealing candidate for a factor conferring T-lineage identity in early thymocytes (Fig. 1B). However, gain of function of GATA-3, in the presence or absence of Notch signals, fails to enhance either appearance or developmental progression of the earliest T-lineage cells [36,111,116,117]. Only around the time of lineage commitment, from the DN3 stage or its human equivalent onward, does high-level GATA-3 become compatible with modulation of fates within the T lineage [38,116,118–120]. Thus, high-level GATA-3 is inhibitory for some of the same stages of T-cell development that require normal levels of GATA-3. GATA family factors can activate a cell-cycle arrest mechanism [121,122], and so one possibility would be that excessive GATA-3 simply causes loss of cellularity. Indeed, high-level GATA-3 can inhibit survival of later-stage thymocytes, and a conserved KRR motif within GATA-3 specifically mediates this cytostatic activity [123]. However, this is not the only cause of interference with early T-cell precursors. High-level GATA-3 actually promotes an alternative, non-T lineage choice. The effects of high-level GATA-3 are context-dependent: whereas it can promote a megakaryocytic fate from bone marrow stem cells [117], it specifically drives a mast-cell development program when expressed in early T-lineage cells [36] (Figure 3).
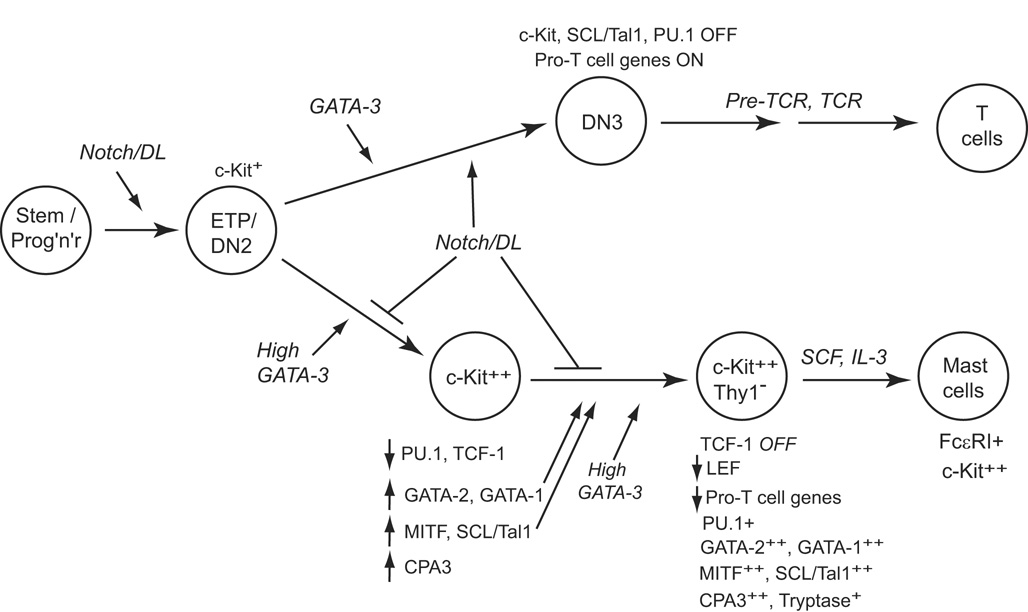
Figure 3.
Stepwise respecification of ETP and DN2 cells by GATA-3 overexpression. Early, intermediate, and late stages in the reprogramming of ETP or DN2 cells by forced expression of GATA-3. The stages where Notch signaling exerts inhibition are shown. Adapted from Supplementary Figure 6, ref. [36]. Note that susceptibility of cells to these interactions is limited to the stages before DN3, when an unidentified commitment function blocks access to the mast-cell pathway.
This lineage diversion is interesting for three reasons. First, the mast-cell program has not previously been shown to be related to any lymphoid program [124–126], and so its activation within T-lineage precursors by a “T-lineage-specific” transcription factor is remarkable. Second, the ability of thymocytes to respond to GATA-3 in this way is developmentally regulated, observing the same commitment boundary as other non-T fates. While ETP and DN2 stage cells can respond to GATA-3 in this way, DN3 cells cannot. This suggests that the diversion depends on additional intrinsic features of the earliest-stage cells which may be lost with commitment. Finally, the mast-cell pathway is effectively blocked in thymocytes by Notch signaling. When Notch-Delta interaction is also occurring, high-level GATA-3 simply blocks survival, and the mast-cell program cannot be completed [36].
Respecification is a slow, multistep process [36], as diagrammed in Fig. 3. Overexpression of GATA-3 in fetal thymocytes causes a number of early gene regulatory effects within the first 40 hr, but it does not materially alter the expression of most T-cell differentiation markers or regulatory genes. Among the most pronounced early changes are a strong, temporary downregulation of PU.1 (in the ETP and DN2 cells) and upregulation of the mast-cell transcription factor MITF (microphthalmia associated transcription factor) and the two other hematopoietic GATA factors, GATA-1 and GATA-2. These effects occur concurrently with a strong upregulation of c-Kit expression and can be detected, more or less, whether Notch signaling is present or not. Over the next days, further changes occur to activate an increasing set of mast-cell genes, including those encoding transcription factors SCL (Tal1) and Gfi-1b, while T-lineage-specific gene expression becomes increasingly broadly inhibited. The emergence of viable converted cells now depends on removal from Notch-Delta signals. It is likely that the additional steps depend on the newly induced (or reinduced) transcription factors as well as on GATA-3 itself (Fig. 3). The distinctness of this stage is reflected in restored expression of PU.1, to levels appropriate for the mast-cell fate [127]. However, there is still a substantial “hangover” of expression of a few T-cell genes, such as CD3ε and CD3γ, even in the cells that have undergone complete phenotypic conversion to c-Kithigh Thy-1low CD27− mast-cell precursors. Full maturation to a surface FcεRI+ state depends on further culture in mast-cell cytokines for at least another week (Fig. 3).
Under normal circumstances in the bone marrow, the crucial transcription factor for initiating mast-cell development is GATA-2 [125,128]. GATA-3 overexpression has similar or identical effects to forced expression of GATA-2 in early thymocytes, consistent with the extensive structural similarities between these related factors [36]. Thus GATA-3 probably opens up access to the mast-cell pathway by activating genes that are normally regulated in bone-marrow precursors by GATA-2. But the interesting point is that GATA-3 is normally a participant in the T-cell program, not the mast-cell program, and a collaborator with Notch signaling, not an antagonist. In the unstable developmental context of early T-lineage cells, its entire developmental impact on the cells depends on its level of expression. Thus avoiding excessive induction of GATA-3 in the ETP and DN2 stages is as important for T-cell development as guaranteeing that some GATA-3 will be present.
It remains to be determined how the committed state of DN3 cells blocks access to the mast-cell program. But how does Notch signaling prevent this lineage choice in the ETP and DN2 stages? All of the short-term effects of GATA-3 are slightly stronger when Notch signals are removed [36]. High-level GATA-3 actually appears to blunt Notch1 expression itself as well as the expression of certain Notch target genes, suggesting a competitive mechanism, but most T-cell transcription factors are minimally affected by GATA-3 overexpression, with or without Notch-Delta signaling. The largest Notch-dependent difference in GATA-3 effects that can be noted at this early point is that TCF-1 is repressed only when Notch signals are absent. Thus, Notch signaling may indirectly be acting either to maintain the transcriptional effector of the Wnt-β-catenin pathway, or else to preserve a repressive function of TCF-1 which may be important for T-lineage fidelity in this case. TCF-1 and increasingly LEF are then severely repressed in later stages of the conversion to a mast-cell precursor state [36]. If these factors are serving in thymocytes as repressors of PU.1 expression [75](see above), then their own downregulation as the cells become fully converted mast-cell precursors may help to explain why PU.1 expression resumes [36](Fig. 3, dashed lines). These results suggest that GATA-3 is linked with Notch signaling, TCF/LEF factors, and PU.1 in a network of dose-dependent, competitive interactions that determine access of early T-cell precursors to the mast-cell developmental pathway.
7. PU.1 AND MYELOID ALTERNATIVES TO THE T-LINEAGE PROGRAM
Although needed to start T-cell development, PU.1 also helps to maintain a progenitor-like developmental plasticity in the ETP and DN2 cells. During commitment, expression of PU.1 and SCL (Tal1) precipitously shuts off [129,130]. Forced gain of PU.1 function, beyond the stage when it is normally shut off [53,54,56–58,111], provides evidence that endogenous PU.1 expression in earlier T-cell precursors is probably rate-limiting for their developmentally regulated access to macrophage and dendritic-cell fates.
Forced PU.1 expression can undo the commitment of DN3 thymocytes and convert them to myeloid dendritic cell phenotype, passing through dual-lineage phenotypic intermediates that clearly show the transdifferentiation in progress [54,56–58](Figure 4). High-level, dysregulated PU.1 expression can inhibit growth under “lymphoid” cytokine conditions, but the converted cells thrive if myeloid growth factors are supplied [54] or if the cells express a Bcl2 transgene [57,58]. The mechanism PU.1 uses to promote these alternative fates appears to be intrinsically Notch-sensitive. Notch-Delta signaling can restrain exogenous PU.1 from causing respecification of DN2 or DN3 cells [54,58](Fig. 4), even when forced expression of PU.1 reaches levels more than ten times higher than normal ETP or DN2 levels. The important feature of this antagonism is that Notch signaling does not directly inhibit expression of PU.1 itself, while PU.1 also does not appear to inhibit Notch1 or Notch3 expression directly, at least not when overexpressed in DN2 or DN3 stage cells [58]. Thus PU.1 as expressed in DN2 cells should be a conditional mediator of lineage plasticity, which ideally positions the cells in a regulatory state where responses to microenvironmental Notch signals can dictate their fates.
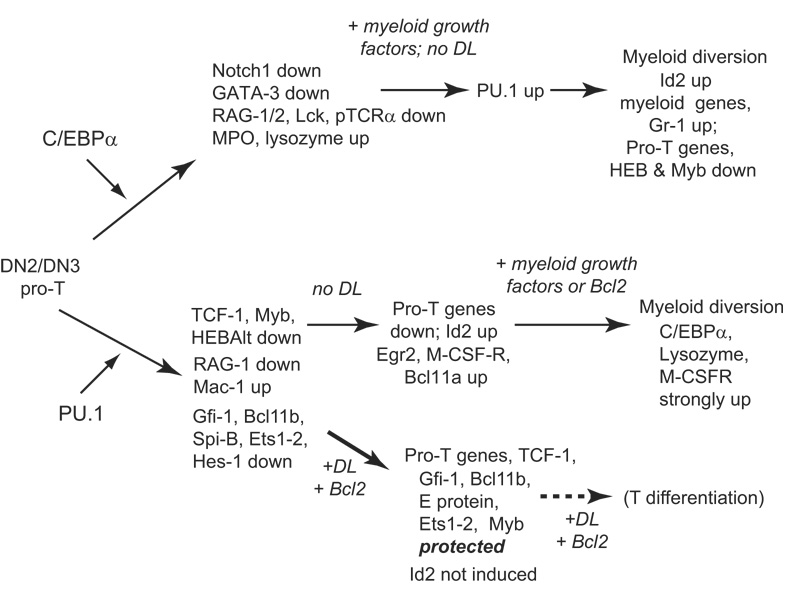
Figure 4.
Myeloid diversion pathways for advanced pro-T cells by re-introduction of C/EBPα or PU.1. Comparison of the sequences of gene expression changes induced by transduction of pro-T cells with PU.1 or with C/EBPα, from first effects (day 1–2) through lineage conversion (4–9 days). Effects summarized in the figure were measured on purified DN3 pro-T cells from adult mouse thymus, transduced with C/EBP factors [54] or on DN2/3 stage fetal thymocytes, transduced with PU.1 [58]. “Pro-T genes” include Rag1, Lck, LAT, and ZAP70, the products of which are needed for TCR function. Continued progress along the myeloid pathway depends on the lack of Notch-Delta signals in PU.1 overexpressing cells. The pathway branch indicated by “+DL” shows how reintroduction of Notch-Delta signaling can stall or reverse the reprogramming of fetal pro-T cells by PU.1. Although high-level PU.1 is inhibitory for proliferation and progression of T-lineage cells beyond the DN3 stage, Notch signaling and neutral viability support (a Bcl2 transgene) can restore substantial T-lineage differentiation even beyond β-selection. Major gene targets that are differentially affected by PU.1 in the presence or absence of Notch-Delta signaling are shown (data from ref. [58]).
Even in the DN3 stage, the committed state remains intrinsically rather flimsy with respect to myeloid and dendritic-cell alternatives. Other exogenous genes can also undo the commitment of DN3 cells. Forced expression of the myeloid factor C/EBPα or its relative C/EBPβ can also respecify DN3 cells to myeloid lineage cells very efficiently, to an inflammatory macrophage fate rather than a myeloid dendritic cell fate [54](Fig. 4). Furthermore, surprisingly, DN3 cells that have downregulated PU.1 express peak levels of the PU.1 relative Spi-B, and yet if artificially expressed at sufficiently high levels, Spi-B itself can also cause conversion of the cells to a dendritic-cell phenotype [56]. Thus the commitment of normal DN3 cells may be a composite of the recent repression of PU.1, absence of C/EBP factors, quantitative limits on the expression of Spi-B, and the relatively weaker myeloid-promoting potency per molecule of the Spi-B which they do express. Spi-B expression here may provide the cells with continuation of any T-lineage promoting functions that PU.1 formerly may have served; for example, PU.1 (and probably Spi-B) may actually upregulate expression of the T-lineage markers Cd25 and Ptcra [57,111,131].
Somewhat like PU.1, C/EBPα effects on DN3 cells are blunted by Notch signaling [54]. However, C/EBPα has a much more direct antagonistic effect on expression of Notch1 itself. Preliminary evidence suggests also that Notch-Delta signaling may actively inhibit C/EBPα expression in immature fetal thymocytes, implying a more direct binary opposition than that of Notch and PU.1 [36,58]. Thus, sustained C/EBPα expression would be incompatible with passage through the Notch-dependent early stages of T-cell development. Indeed, endogenous C/EBPα may very well contribute to the robust myeloid potential of many ETPs, but its expression is already very low by the DN2 stage [54,132](E.-S. David-Fung & E. V. R., unpublished results). However, even in DN2 cells reinduction of C/EBPα can be triggered by ectopic growth factor receptor stimulation [59], especially in the absence of Notch signaling. It is likely to be the combination of this upregulated C/EBPα with endogenous PU.1 that explains why ectopic growth factor receptor signaling is sufficient to drive ETP and DN2 cells into myeloid lineage conversion [79,133].
Both C/EBPα and PU.1 drive pro-T cells to myeloid identities through paths of several distinct steps over a period of days (Fig. 4). Each of these steps represents a potential opportunity for regulatory intervention, either by Notch signals or by other signaling pathways. Strikingly, and in contrast to GATA-3 promoted lineage diversion, both myeloid factors start by undermining T-lineage identity, through downregulation not only of differentiation genes but also of multiple DN3-expressed regulatory genes. C/EBPα represses Notch1, GATA-3, and E2A, while PU.1 represses Myb, TCF-1, Gfi-1, E proteins, certain other Ets family transcription factors, and Ikaros [54,57,58]. Some myeloid gene expression also begins. Then, to complete the transformation, each of these factors can reactivate the other’s expression after several days in the absence of Notch signals [54,58](Fig. 4). These cross-activations of myeloid regulators do not precede, but rather follow, the downregulation of T-cell regulatory factors. When Notch signaling neutralizes the effects of high-level PU.1, its effects are mostly to protect the expression of these T-lineage transcription factors rather than to block the earliest increases in myeloid-associated gene expression [58](Fig. 4, “+DL”). Later, C/EBPα reactivation may be inhibited in thymocytes receiving Notch signals [36,58], but it is not clear whether the effect is direct. This raises the possibility that among the T-cell factors that are initially targeted, there are Notch-dependent “gatekeeper” functions that would otherwise restrain the cells from the myeloid program, and that these functions need to be disabled before trans-differentiation can proceed.
PU.1 and C/EBPα do not reach the point of positive cross-regulation by a common route. Because these two factors have divergent initial effects on Notch, GATA-3, and Myb [54,58], the early intermediates in lineage conversion should be quite different (Fig. 3A). This is strikingly reminiscent of a recent report that forced sequential expression of GATA-2 and C/EBPα in common lymphoid progenitors, a “tabula rasa” cell type, leads to highly biased generation of either eosinophils or basophils, with the whole developmental outcome depending on the order in which these factors act [125]. The C/EBPα-driven program is therefore a different lineage alternative for DN3 cells than the pathway activated by PU.1. When artificially overexpressed, C/EBP factors can even force DN3 thymocytes to adopt a form of myeloid fate after their endogenous PU.1 genes have been excised [54]. The fact that these different myeloid pathways are just one gene away for DN3 cells shows that these newly-committed cells differ in relatively few respects from the regulatory state and gene accessibility pattern of multipotent precursors.
8. GATA-3 AS ANTAGONIST OF MYELOID LINEAGE OPTIONS
The functions mobilized by Notch to preserve T-lineage identity before lineage commitment need to be identified. GATA-3 is an obvious candidate for one of the T-lineage gatekeeper functions that antagonizes myeloid fates under normal conditions. Myeloid lineage conversion of thymocytes by either C/EBPα or PU.1 is inhibited if GATA-3 is co-expressed [54,134]. Furthermore, GATA-3 may be suspected of a role in lineage commitment itself, as overexpression of GATA-3 in early fetal thymocytes efficiently downregulates PU.1 RNA expression as already noted [36,111,118]. GATA-3 itself is encoded by one of the genes that is turned on earliest in progenitors as they begin the T-cell developmental program in response to Notch-Delta signaling. GATA-3 is also a direct target of Notch induction in the context of T helper 2 cells [135,136]. Therefore GATA-3 might be implicated in aspects of the Notch-dependent protection of developing T cells from myeloid diversion by PU.1 or C/EBPα.
In reality, the interplay between GATA-3 and myeloid determination is probably more subtle. Whether or not GATA-3 is able to protect cells from myeloid differentiation is very much dependent on cellular context and probably also dependent on its exact level of expression. Indeed, in non-T lineage cells GATA-3 can even trigger myeloid development, as observed in Pax5-deficient pro-B cells [137]. The simplest case of GATA-3 blocking the myeloid fate may be when C/EBPα expression is imposed on DN3 cells [54]. Here, one of the earliest effects of C/EBPα or C/EBPβ seems to be downregulation of GATA-3. Adding back GATA-3 can therefore be seen as an epistasis test, in which exogenous GATA-3 can restore the missing function. Coexpressed GATA-3 can block Mac-1 (CD11b) upregulation by C/EBPα, an aspect of the C/EBPα response that appears to reflect cross-induction of PU.1, and this inhibition is consistent with the ability of high GATA-3 to block PU.1 expression [36,111,118]. Even here, however, there is not clear evidence how much of the T-cell program GATA-3 can restore. For example, it is not clear whether GATA-3 can prevent C/EBPα from downregulating Notch and E protein expression [54], which would independently be required for full T-cell development.
As an antagonist for PU.1, the possible role of GATA-3 is more complex. High-level PU.1, unlike C/EBPα, has little immediate effect on expression of GATA-3 itself or of Notch1. When PU.1 effects are neutralized by Notch signaling, again there is little if any impact on levels of GATA-3 RNA; instead, the strongest protective effects are seen on RNAs encoding a completely different set of T-lineage regulators: Myb, E proteins, Gfi-1, and Bcl11b [58]. TCF-1, also downregulated by PU.1, is also partially protected by Notch signaling. Thus, when GATA-3 is co-expressed with PU.1, it is probably antagonizing PU.1 in a supraphysiological, gain-of-function mode. Thus, it is possible that GATA-3 coexpression with PU.1 does not really restore the T-lineage program but simply provides antagonism for PU.1 by titrating it at the protein level, preventing PU.1 from transactivating the myeloid genes. However, the biochemical basis for these effects remains unresolved. Many of the most immediate impacts of PU.1 on pro-T cells are the repression of critical T-cell genes. Yet GATA factors are not thought to block PU.1 binding to DNA, but simply PU.1 transactivation; it is completely unclear what effect they would have on any target genes where PU.1 acts as a direct repressor.
Ultimately, GATA-3 may only be one part of the mechanism antagonizing myeloid fates in developing T cells. Under normal circumstances, GATA-3 could also play a role in the programmed downregulation of PU.1 itself (D. D. S.-A., unpublished results), although current published evidence suggests that the normal control of PU.1 levels in developing thymocytes may be more affected by inputs from TCF/LEF family and Runx family transcription factors [75,138]. On the other hand, the abilities of the myeloid transcription factors to repress Gata3 expression or block GATA-3 DNA binding could play a substantial role in the timing of T-cell developmental progression, as discussed in the final section.
9. TRANSDIFFERENTIATION, NOT DEDIFFERENTIATION, IN RESPONSE TO REGULATORY PERTURBATION
Neither myeloid nor mast-cell programs are thought to be closely related to lymphocyte developmental programs, and indeed, conversion of thymocytes to dendritic cells by PU.1, inflammatory macrophages by C/EBPα, and mast cells by excess GATA-3 all involve a repression of T-lineage specific gene expression. Their diversionary activity is even more remarkable in view of the positive roles that two of these diversion-inducing factors normally play in T-cell development, when expressed at different times and levels. This raises the question of whether the effects of these factors are simply to cause T-cell precursors to revert to a more primitive, multipotent progenitor state, from which all these diverse pathways naturally emerge.
The question is more interesting because recent data indicate that in vivo, PU.1 downregulation coincides with the main onset of T-lineage specific gene expression from the DN2 to the DN3 stage, linking a major positive regulatory event in the T-cell program with the loss of alternative choices. T-lineage genes appear to be upregulated first in that subset of DN2 cells that have already begun to downregulate PU.1 [129,139]. PU.1 acts as a transcriptional repressor of genes encoding T-lineage transcription factors including TCF-1 and E proteins [58], and through protein-protein interactions it could antagonize GATA-3 functions needed for progression to the DN3 stage (D. D. S.-A. and E. V. R., unpublished results) as long as it is expressed. Conceivably, some of these effects could be strong enough even in the presence of Notch signaling to enable endogenous PU.1 to play a timing role for the triggering of particular aspects of normal T cell development that depend on levels of E proteins, TCF-1, and GATA-3.
If excess GATA-3, PU.1, and C/EBPα each caused reversion to a multipotent progenitor state before promoting redifferentiation, then the intermediate stages of the reconversion process should go through a common stage. Furthermore, this common stage should be similar to the phenotype of a natural multipotent progenitor. The transcription factor profile of natural multipotent progenitors is marked by high-level coexpression of at least four informative transcription factors: GATA-2, PU.1, SCL (Tal1), and Myb [43,140]. Indeed, forced high-level GATA-3 turns on GATA-2 and SCL in thymocytes that no longer express these factors, and it preserves the high-level Myb expression that thymocytes naturally share with progenitors. However, it sharply downregulates PU.1, in contrast to normal progenitors. On the other side, high-level expression of PU.1 in thymocytes fails to turn on GATA-2 (C. Franco & E. V. R., unpublished data) and actively represses Myb expression. Although the two perturbations have in common an ability to downregulate TCF-1 (encoded by Tcf7), this too may go through different pathways, as TCF-1 is almost completely protected from GATA-3 effects by Notch signaling, while it is still inhibited by PU.1 in the presence of Notch-Delta signals. Thus the intermediate regulatory states of the conversion process are completely different, and in both cases they differ markedly from natural multipotent progenitors, and even from ETP thymocytes. This shows that the paths to lineage diversion do not require backtracking to a common multipotent state, but instead involve alternative, direct reprogrammings of thymocytes from the midst of their T-lineage differentiation process.
10. CONCLUSIONS
The results reviewed here show that T-cell precursor specification is not only a protracted process but a remarkably precarious one at the regulatory level. This makes sense of much biology that could otherwise seem counterintuitive. An increasingly rich literature confirms that most T-cell precursors naturally retain access to non-T and non-lymphoid programs well beyond the stage when they have lost B-lineage potential [16–19,81,141–143]. The myeloid developmental choices can be triggered even more readily in immature thymocytes before the DN3 stage by manipulations that might have been imagined to leave the basic regulatory state intact, such as stimulation through ectopic cytokine receptors [79,133]. Although the divergence in gene expression program is less dramatic, until the DN3 stage these cells also have access to the distinct natural killer pathway, which can be enhanced through antagonism of E protein activity and/or removal of Notch-Delta signals [35,83,85,144–147]. This simultaneous access to multiple distinct developmental programs can now be seen as the consequence of a tug of war among regulatory forces acting normally within the cells between the ETP and the DN3 stage (Figure 5). Among the T, B, NK, mast, and dendritic-cell or myeloid fates, the only one prohibited to T-cell precursors at an early stage is the B-cell fate, due to the extreme incompatibility of initial B-cell specification with Notch signals. The other fates are actually promoted by factors which also continue to have important roles in the T-lineage pathway itself.
Figure 5.
Schematic of counterbalancing and dose-dependent regulatory forces that act on T-cell precursors through the ETP and DN2 stages. Regulators named in red denote forces diverting cells from the T-cell pathway. Of these, only EBF and Pax5 are not normally active in the thymus during these stages. Notch signaling acts as a constraint on all of these alternatives. This dynamic equilibrium is normally resolved at the DN3 stage, in part by the downregulation of PU.1, but almost certainly through other regulatory changes as well [163].
In this review, we have looked in detail at the interaction among three of these factors, PU.1, GATA-3, and Notch, and to a lesser extent at roles of C/EBP factors and TCF-1/LEF family members. The alternatives made available to early T-cell precursors by selective dysregulation of these factors require several steps of reprogramming, but not a return to a stem-cell condition. This is important because it shows that the dynamic balancing of these regulatory forces maintains a degree of true multipotentiality that is inherent in T-cell precursor states throughout the onset of T-lineage gene expression. Even in DN3 cells, where ectopic cytokine receptor signaling, withdrawal of Notch signals, and GATA-3 overexpression are no longer capable of causing respecification, reintroduction of C/EBP factors or PU.1 are sufficient to overcome the fragile new state of commitment. Yet later, the lineage identity of a mature T cell in the periphery is remarkably robust over arbitrarily large numbers of cell generations and through many storms of activation-induced regulatory change, including high-level GATA-3 expression [38,148], intense MAP Kinase activation [149–151], frequent deprivation of Notch signals, and even in some cases reinduction of PU.1 [152]. The contrast reveals that another level of commitment must exist for T-cell precursors, possibly through an epigenetic mechanism, which must occur after the DN3 stage and remains to be discovered and dissected.
ACKNOWLEDGMENTS
We are most grateful to the primary authors of the work cited in this review, including members and former members of this laboratory whose results were discussed in detail. We especially thank Thomas Graf for patient encouragement and valuable insights, and Howard Petrie, Cornelis Murre, Hiroshi Kawamoto and Avinash Bhandoola for exciting discussions of work before publication. The authors’ work was supported by NIH grants CA90233, CA98925, and HL089123, the Albert Billings Ruddock Professorship, and the Louis A. Garfinkle Memorial Laboratory Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
“ETP” is used here instead of “DN1” because the classically defined DN1 thymocyte population (CD44+ CD25−) includes a majority of cells that are not T lineage precursors.
REFERENCES
- 1.Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Graf T. Transcription factors that induce commitment of multipotent hematopoietic progenitors: lessons from the MEP system. In: Zon LI, editor. Hematopoiesis: a Developmental Approach. New York: Oxford University Press; 2001. pp. 355–362. [Google Scholar]
- 4.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 5.Rekhtman N, Radparvar F, Evans T, Skoultchi A. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P, Behre G, Pan J, Iwama A, Wara-aswapati N, Radomska HS, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 10.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 11.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 12.Rothenberg EV. Cell lineage regulators in B and T cell development. Nat Immunol. 2007;8:441–444. doi: 10.1038/ni1461. [DOI] [PubMed] [Google Scholar]
- 13.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 14.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 15.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 17.Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 18.Shen HQ, Lu M, Ikawa T, Masuda K, Ohmura K, Minato N, et al. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J Immunol. 2003;171:3401–3406. doi: 10.4049/jimmunol.171.7.3401. [DOI] [PubMed] [Google Scholar]
- 19.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 20.Wu L, Liu YJ. Development of dendritic-cell lineages. Immunity. 2007;26:741–750. doi: 10.1016/j.immuni.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Krueger A, Garbe AI, von Boehmer H. Phenotypic plasticity of T cell progenitors upon exposure to Notch ligands. J Exp Med. 2006;203:1977–1984. doi: 10.1084/jem.20060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zenke M, Hieronymus T. Towards an understanding of the transcription factor network of dendritic cell development. Trends Immunol. 2006;27:140–145. doi: 10.1016/j.it.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Garrett KP, Pelayo R, Zúñiga-Pflücker JC, Petrie HT, Kincade PW. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. J Immunol. 2005;175:4858–4865. doi: 10.4049/jimmunol.175.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, Traver D, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Sieweke MH, Graf T. A transcription factor party during blood cell differentiation. Curr Opin Genet Devel. 1998;8:545–551. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 27.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nat Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 28.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 29.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- 31.Ciofani M, Zúñiga-Pflücker JC. A survival guide to early T cell development. Immunol Res. 2006;34:117–132. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 32.Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 33.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J Exp Med. 2004;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho IC, Pai SY. GATA-3 - not just for Th2 cells anymore. Cell Mol Immunol. 2007;4:15–29. [PubMed] [Google Scholar]
- 39.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 40.Rothenberg EV, Taghon T. Molecular Genetics of T Cell Development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 41.Greenbaum S, Zhuang Y. Regulation of early lymphocyte development by E2A family proteins. Semin Immunol. 2002;14:405–414. doi: 10.1016/s1044532302000751. [DOI] [PubMed] [Google Scholar]
- 42.Staal FJT, Weerkamp F, Langerak AW, Hendriks RW, Clevers HC. Transcriptional control of T lymphocyte differentiation. Stem Cells. 2001;19:165–179. doi: 10.1634/stemcells.19-3-165. [DOI] [PubMed] [Google Scholar]
- 43.Tydell CC, David-Fung E-S, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 44.David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, et al. Progression of regulatory gene expression states in fetal and adult pro-T cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taghon TN, David E-S, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, Petrie HT. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- 47.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, Van de Wetering M, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 48.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–746. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- 50.Kawazu M, Yamamoto G, Yoshimi M, Yamamoto K, Asai T, Ichikawa M, et al. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J Immunol. 2007;179:5335–5345. doi: 10.4049/jimmunol.179.8.5335. [DOI] [PubMed] [Google Scholar]
- 51.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-Tα chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 53.Anderson MK, Weiss A, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T-cell development at the pro-T stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 54.Laiosa CV, Stadtfeld M, Xie H, Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 55.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, et al. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol. 2005;174:6184–6194. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- 57.Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, et al. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 58.Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, et al. Notch/Delta signaling constrains re-engineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu CL, King-Fleischman AG, Lai AY, Matsumoto Y, Weissman IL, Kondo M. Antagonistic effect of CCAAT enhancer-binding protein-α and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc Natl Acad Sci U S A. 2006;103:672–677. doi: 10.1073/pnas.0510304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur J Immunol. 2006;36:526–532. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 61.Kang J, Der SD. Cytokine functions in the formative stages of a lymphocyte's life. Curr Opin Immunol. 2004;16:180–190. doi: 10.1016/j.coi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008 doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Pierce LJ, Spangrude GJ. Distinct roles of IL-7 and stem cell factor in the OP9-DL1 T-cell differentiation culture system. Exp Hematol. 2006;34:1730–1740. doi: 10.1016/j.exphem.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balciunaite G, Ceredig R, Fehling HJ, Zúñiga-Pflücker JC, Rolink AG. The role of Notch and IL-7 signaling in early thymocyte proliferation and differentiation. Eur J Immunol. 2005;35:1292–1300. doi: 10.1002/eji.200425822. [DOI] [PubMed] [Google Scholar]
- 66.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 67.Schilham MW, Wilson A, Moerer P, Benaissa-Trouw BJ, Cumano A, Clevers HC. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 68.Staal FJT, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, et al. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J Immunol. 2004;172:1099–1108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
- 69.Weerkamp F, Baert MR, Naber BA, Koster EE, de Haas EF, Atkuri KR, et al. Wnt signaling in the thymus is regulated by differential expression of intracellular signaling molecules. Proc Natl Acad Sci U S A. 2006;103:3322–3326. doi: 10.1073/pnas.0511299103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pongracz JE, Parnell SM, Jones T, Anderson G, Jenkinson EJ. Overexpression of ICAT highlights a role for catenin-mediated canonical Wnt signalling in early T cell development. Eur J Immunol. 2006;36:2376–2383. doi: 10.1002/eji.200535721. [DOI] [PubMed] [Google Scholar]
- 71.Baba Y, Garrett KP, Kincade PW. Constitutively active β-catenin confers multilineage differentiation potential on lymphoid and myeloid progenitors. Immunity. 2005;23:599–609. doi: 10.1016/j.immuni.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 73.Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, et al. Hematopoietic stem cell and multilineage defects generated by constitutive β-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 74.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of β-catenin and γ-catenin. Blood. 2007 doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 75.Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 76.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garbe AI, von Boehmer H. TCR and Notch synergize in αβ versus γδ lineage choice. Trends Immunol. 2007;28:124–131. doi: 10.1016/j.it.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 78.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 79.King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci U S A. 2002;99:4508–4513. doi: 10.1073/pnas.072087899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lucas K, Vremec D, Wu L, Shortman K. A linkage between dendritic cell and T-cell development in the mouse thymus: the capacity of sequential T-cell precursors to form dendritic cells in culture. Dev Comp Immunol. 1998;22:339–349. doi: 10.1016/s0145-305x(98)00012-3. [DOI] [PubMed] [Google Scholar]
- 81.Garcia-Peydro M, de Yebenes VG, Toribio ML. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J Immunol. 2006;177:3711–3720. doi: 10.4049/jimmunol.177.6.3711. [DOI] [PubMed] [Google Scholar]
- 82.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 83.De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 2005;106:3498–3506. doi: 10.1182/blood-2005-02-0496. [DOI] [PubMed] [Google Scholar]
- 84.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–868. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 85.Lehar SM, Dooley J, Farr AG, Bevan MJ. Notch ligands Delta1 and Jagged1 transmit distinct signals to T cell precursors. Blood. 2005;105:1440–1447. doi: 10.1182/blood-2004-08-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 87.Harman BC, Jenkinson WE, Parnell SM, Rossi SW, Jenkinson EJ, Anderson G. T/B lineage choice occurs prior to intrathymic Notch signalling. Blood. 2005;106:886–892. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- 88.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 89.Huang EY, Gallegos AM, Richards SM, Lehar SM, Bevan MJ. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J Immunol. 2003;171:2296–2304. doi: 10.4049/jimmunol.171.5.2296. [DOI] [PubMed] [Google Scholar]
- 90.Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cave JW, Loh F, Surpris JW, Xia L, Caudy MA. A DNA transcription code for cell-specific gene activation by Notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- 92.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci U S A. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 97.Yun TJ, Bevan MJ. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- 98.Tagami S, Okochi M, Yanagida K, Ikuta A, Fukumori A, Matsumoto N, et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol Cell Biol. 2008;28:165–176. doi: 10.1128/MCB.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CA, Colucci F, et al. GATA-3 promotes maturation, IFN-γ production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 100.Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 101.Ting C-N, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 102.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dahl R, Ramirez-Bergeron DL, Rao S, Simon MC. Spi-B can functionally replace PU.1 in myeloid but not lymphoid development. EMBO J. 2002;21:2220–2230. doi: 10.1093/emboj/21.9.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Spain LM, Guerriero A, Kunjibettu S, Scott EW. T cell development in PU.1-deficient mice. J Immunol. 1999;163:2681–2687. [PubMed] [Google Scholar]
- 105.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 106.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 107.Back J, Allman D, Chan S, Kastner P. Visualizing PU.1 activity during hematopoiesis. Exp Hematol. 2005;33:395–402. doi: 10.1016/j.exphem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 108.Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Goff OR, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 109.Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, et al. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.DeKoter RP, Lee H-J, Singh H. PU.1 regulates expression of the Interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 111.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias A, Chen D, Rothenberg EV. Definition of regulatory network elements for T-cell development by perturbation analysis with PU.1 and GATA-3. Devel Biol. 2002;246:103–121. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- 112.Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 113.Liew CW, Rand KD, Simpson RJ, Yung WW, Mansfield RE, Crossley M, et al. Molecular analysis of the interaction between the hematopoietic master transcription factors GATA-1 and PU.1. J Biol Chem. 2006;281:28296–28306. doi: 10.1074/jbc.M602830200. [DOI] [PubMed] [Google Scholar]
- 114.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- 115.Henkel G, Brown MA. PU.1 and GATA: components of a mast cell-specific interleukin 4 intronic enhancer. Proc Natl Acad Sci USA. 1994;91:7737–7741. doi: 10.1073/pnas.91.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Taghon T, De Smedt M, Stolz F, Cnockaert M, Plum J, Leclercq G. Enforced expression of GATA-3 severely reduces human thymic cellularity. J Immunol. 2001;167:4468–4475. doi: 10.4049/jimmunol.167.8.4468. [DOI] [PubMed] [Google Scholar]
- 117.Chen D, Zhang G. Enforced expression of the GATA-3 transcription factor affects cell fate decisions in hematopoiesis. Exp Hematol. 2001;29:971–980. doi: 10.1016/s0301-472x(01)00670-1. [DOI] [PubMed] [Google Scholar]
- 118.Hernández-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 119.Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol. 2001;167:715–723. doi: 10.4049/jimmunol.167.2.715. [DOI] [PubMed] [Google Scholar]
- 120.Nawijn MC, Dingjan GM, Ferreira R, Lambrecht BN, Karis A, Grosveld F, et al. Enforced expression of GATA-3 in transgenic mice inhibits Th1 differentiation and induces the formation of a T1/ST2-expressing Th2- committed T cell compartment in vivo. J Immunol. 2001;167:724–732. doi: 10.4049/jimmunol.167.2.724. [DOI] [PubMed] [Google Scholar]
- 121.Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, et al. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ezoe S, Matsumura I, Nakata S, Gale K, Ishihara K, Minegishi N, et al. GATA-2/estrogen receptor chimera regulates cytokine-dependent growth of hematopoietic cells through accumulation of p21(WAF1) and p27(Kip1) proteins. Blood. 2002;100:3512–3520. doi: 10.1182/blood-2002-04-1177. [DOI] [PubMed] [Google Scholar]
- 123.Hernandez-Hoyos G, Anderson MK, Rothenberg EV. Distinct actions of GATA-3 in three phases of thymocyte development: specification, β-selection, and CD4/CD8 differentiation. 2001 Submitted for publication. [Google Scholar]
- 124.Kitamura Y, Oboki K, Ito A. Molecular mechanisms of mast cell development. Immunol Allergy Clin North Am. 2006;26:387–405. doi: 10.1016/j.iac.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 125.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. 2005;102:11408–11413. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 128.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 129.Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- 130.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Petersson K, Ivars F, Sigvardsson M. The pTα promoter and enhancer are direct targets for transactivation by E box-binding proteins. Eur J Immunol. 2002;32:911–920. doi: 10.1002/1521-4141(200203)32:3<911::AID-IMMU911>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 132.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K. Enforced Granulocyte/Macrophage Colony-Stimulating Factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J Exp Med. 2003;197:1311–1322. doi: 10.1084/jem.20021843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Laiosa CV. Lineage reprogramming of T lymphoid cells by myeloid transcription factors. Albert Einstein College of Medicine, Yeshiva University; 2006. Ref Type: Thesis/Dissertation. [Google Scholar]
- 135.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heavey B, Charalambous C, Cobaleda C, Busslinger M. Myeloid lineage switch of Pax5 mutant but not wild-type B cell progenitors by C/EBPα and GATA factors. EMBO J. 2003;22:3887–3897. doi: 10.1093/emboj/cdg380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 139.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 141.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 143.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 144.Kee BL. Helix-loop-helix proteins in lymphocyte lineage determination. Curr Top Microbiol Immunol. 2005;290:15–27. doi: 10.1007/3-540-26363-2_2. [DOI] [PubMed] [Google Scholar]
- 145.van den Brandt J, Voss K, Schott M, Hunig T, Wolfe MS, Reichardt HM. Inhibition of Notch signaling biases rat thymocyte development towards the NK cell lineage. Eur J Immunol. 2004;34:1405–1413. doi: 10.1002/eji.200324735. [DOI] [PubMed] [Google Scholar]
- 146.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci U S A. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heemskerk MHM, Blom B, Nolan G, Stegmann APA, Bakker AQ, Weijer K, et al. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- 149.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–433. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 150.Zamoyska R, Lovatt M. Signalling in T-lymphocyte development: integration of signalling pathways is the key. Curr Opin Immunol. 2004;16:191–196. doi: 10.1016/j.coi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 151.Alberola-Ila J, Hernández-Hoyos G. The Ras/MAPK cascade and the control of positive selection. Immunol Rev. 2003;191:79–96. doi: 10.1034/j.1600-065x.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 152.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 153.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ng SY, Yoshida T, Georgopoulos K. Ikaros and chromatin regulation in early hematopoiesis. Curr Opin Immunol. 2007;19:116–122. doi: 10.1016/j.coi.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 155.Talebian L, Li Z, Guo Y, Gaudet J, Speck ME, Sugiyama D, et al. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rothenberg EV. Regulatory factors for initial T lymphocyte lineage specification. Curr Opin Hematol. 2007;14:322–329. doi: 10.1097/MOH.0b013e3281de72a8. [DOI] [PubMed] [Google Scholar]
- 157.Hock H, Orkin SH. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr Opin Hematol. 2006;13:1–6. doi: 10.1097/01.moh.0000190111.85284.8f. [DOI] [PubMed] [Google Scholar]
- 158.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 160.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rothenberg EV, Anderson MK. Elements of transcription factor network design for T-lineage specification. Devel Biol. 2002;246:29–44. doi: 10.1006/dbio.2002.0667. [DOI] [PubMed] [Google Scholar]
- 162.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 163.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]