Abstract
UPF1 functions in both Staufen 1 (STAU1)-mediated mRNA decay (SMD) and nonsense-mediated mRNA decay (NMD), which we show here are competitive pathways. STAU1- and UPF2-binding sites within UPF1 overlap so that STAU1 and UPF2 binding to UPF1 appear to be mutually exclusive. Furthermore, down-regulating the cellular abundance of STAU1, which inhibits SMD, increases the efficiency of NMD, whereas down-regulating the cellular abundance of UPF2, which inhibits NMD, increases the efficiency of SMD. Competition under physiological conditions is exemplified during the differentiation of C2C12 myoblasts to myotubes: The efficiency of SMD increases and the efficiency of NMD decreases, consistent with our finding that more STAU1 but less UPF2 bind UPF1 in myotubes compared with myoblasts. Moreover, an increase in the cellular level of UPF3X during myogenesis results in an increase in the efficiency of an alternative NMD pathway that, unlike classical NMD, is largely insensitive to UPF2 down-regulation. We discuss the remarkable balance between SMD and the two types of NMD in view of data indicating that PAX3 mRNA is an SMD target whose decay promotes myogenesis whereas myogenin mRNA is a classical NMD target encoding a protein required for myogenesis.
Keywords: Staufen1-mediated mRNA decay, nonsense-mediated mRNA decay, Staufen1, UPF proteins, premature termination codon, myogenesis
Nonsense-mediated mRNA decay (NMD) and Staufen 1 (STAU1)-mediated mRNA decay (SMD) generally provide mammalian cells with mechanisms to degrade mRNAs that harbor a translation termination codon upstream of a splicing-generated exon junction complex (EJC) of proteins (Behm-Ansmant et al. 2007; Chang et al. 2007; Isken and Maquat 2007; Mühlemann 2008; Shyu et al. 2008) or a STAU1-binding site (SBS), respectively (Kim et al. 2005, 2007). Each pathway requires UPF1, which is a member of the RNA helicase superfamily 1 that manifests RNA-dependent ATP hydrolytic and 5′–3′ ATP-dependent unwinding activities in vitro (Bhattacharya et al. 2000; Cheng et al. 2007; Chamieh et al. 2008). During classical NMD, when translation terminates sufficiently upstream of a post-splicing EJC, UPF1 binding to the EJC via UPF2 is thought to trigger mRNA decay (Lykke-Andersen et al. 2000; Kim et al. 2005; Kashima et al. 2006; Wittmann et al. 2006; Singh et al. 2007; Isken et al. 2008). Somewhat analogously, during SMD, when translation terminates sufficiently upstream of an SBS, UPF1 binding to the SBS via the dsRNA-binding protein STAU1 is thought to trigger mRNA decay (Kim et al. 2005, 2007). NMD often involves translation termination at a premature termination codon (PTC) and functions to down-regulate not only aberrant gene expression but also physiologic gene expression. In contrast, SMD usually involves translation termination at a normal termination codon and provides a means to conditionally down-regulate the expression of genes encoding mRNAs that contain an SBS in their 3′ untranslated region (UTR). Notably, NMD, unlike SMD, also involves UPF3X (also called UPF3b) and UPF2 in a process whereby UPF3X anchors UPF2 to EJCs (Lykke-Andersen et al. 2000; Serin et al. 2001; Singh et al. 2007; Chamieh et al. 2008).
Pathways that share a common component have the potential to compromise one another under conditions where one out-competes the other. To date, there is very little information on how the efficiency of either NMD or SMD is regulated. We show here that SMD and NMD are mechanistically related mRNA decay pathways and are indeed in competition. First, SMD, like NMD, requires translation since both pathways are inhibited by cycloheximide or a constitutively inactive form of eukaryotic initiation factor 2α (eIF2α). Second, STAU1- and UPF2-binding sites within UPF1 overlap so that immunoprecipitation (IP) of STAU1 precludes the detectable co-IP of UPF2 and vice versa. Third, down-regulating STAU1, which inhibits SMD (Kim et al. 2005), increases the efficiency of NMD, whereas down-regulating UPF2, which inhibits NMD (Mendell et al. 2002; Wang et al. 2002; Kim et al. 2005; Wittmann et al. 2006; Matsuda et al. 2007; Isken et al. 2008; Woeller et al. 2008), increases the efficiency of SMD. Our finding that the differentiation of C2C12 myoblasts (MBs) to myotubes (MTs) is accompanied by an increase in the efficiency of SMD and a decrease in the efficiency of classical (i.e., UPF2 siRNA-sensitive) NMD exemplifies the consequence of competition in light of the fact that the cellular abundance of STAU1 relative to UPF1 decreases less than the cellular abundance of UPF2 relative to UPF1. Consistent with ongoing competition during myogenesis that reflects the relative abilities of STAU1 and UPF2 to bind UPF1, the efficiency of an alternative NMD pathway that is relatively insensitive to UPF2 siRNA increases, as might be expected from the appreciable increase in the cellular abundance of UPF3X relative to UPF1 during C2C12 differentiation. We discuss the physiological consequences of competition between SMD and NMD in view of data demonstrating that PAX3 mRNA, whose decay promotes myogenesis (Epstein et al. 1995), is an SMD target, whereas myogenin mRNA, which encodes a protein required for myogenesis (Wright et al. 1989), is a classical NMD target.
Results
Amino acids 1–244 of UPF1 interact with STAU1
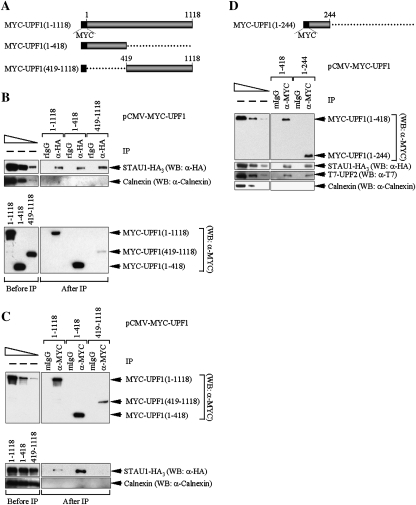
The region of UPF1 that interacts with STAU1 was mapped with the intent of understanding its position relative to the UPF1 amino acids that bind UPF2. Initially, two deletion variants of pCMV-MYC-UPF1(1–1118) (Isken et al. 2008) were generated and analyzed. pCMV-MYC-UPF1(1–1118) encodes full-length human UPF1 containing an N-terminal MYC tag (Fig. 1A). One deletion variant produces UPF1 consisting of amino acids 1–418, which is precisely the region that was previously shown to bind UPF2 (Mendell et al. 2000); the other deletion variant produces UPF1 containing amino acids 419–1118 (Fig. 1A). Cos cells were transiently transfected with pSTAU1-HA3, which encodes the 55-kDa isoform of human STAU1 containing three C-terminal HA tags (Luo et al. 2002, where it was called phSTAU1-HA3), and pCMV-MYC-UPF1(1–1118), pCMV-MYC-UPF1(1–418) or pCMV-MYC-UPF1(419–1118). Notably, none of the tagged proteins was expressed above the level of the corresponding endogenous protein (Supplemental Fig. S1; data not shown demonstrating using transient GFP expression and fluorescence microscopy that >90% of cells were transfected). Two days later, cell lysates were immunoprecipitated using anti-HA, which immunoprecipitates STAU1-HA3, or rat IgG (rIgG), to control for IP specificity.
Figure 1.
Amino acids 1–244 of UPF1 interact with STAU1 or UPF2. (A) Diagram of full-length MYC-UPF1 (1–1118) and two deletion variants. (B) Lysates of Cos cells (3 × 107 cells per 150-mm dish) that had been transiently transfected with 10 μg of pSTAU1-HA3 and 5 μg of pCMV-MYC-UPF1(1–1118), 4 μg of pCMV-MYC-UPF1(1–418), or 15 μg of pCMV-MYC-UPF1(419–1118) (plus a variable amount of pUC19 for a total of 25 μg of transfecting DNA) were immunoprecipitated using anti(α)-HA or, to control for nonspecific IP, rIgG. Equivalent amounts of immunoprecipitated proteins were then analyzed by Western blotting (WB) before (−) IP (refer to the left-most lane when dilutions were analyzed) or after IP using anti-HA, anti-MYC or, as a control for IP specificity, anti-Calnexin. The top three left-most lanes represent threefold dilutions of protein before (−) IP and demonstrate that the conditions used for Western blotting are semiquantitative. (C) As in B, except lysates were immunoprecipitated using anti-MYC or, to control for nonspecific IP, mIgG. (D, top) Diagram of MYC-UPF1(1–244). (D, bottom) As in C, except cells were transfected with 10 μg of pSTAU1-HA3, 10 μg of pCI-NEO-T7-UPF2, and 5 μg of either pCMV-MYC-UPF1(1–418) or pCMV-MYC-UPF1(1–244). Results are representative of five independently performed experiments.
Western blotting using anti-HA demonstrated that STAU1-HA3 was immunoprecipitated using anti-HA in each of the three IPs (Fig. 1B, top). The IP was specific since STAU1-HA3 was not immunoprecipitated using rIgG (Fig. 1B, top), and Calnexin was not immunoprecipitated using either anti-HA or rIgG (Fig. 1B, top). Western blotting using anti-MYC revealed that each MYC-UPF1 variant was expressed (Fig. 1B, bottom, Before IP), and a larger amount of STAU1-HA3 was coimmunoprecipitated with MYC-UPF1(1–418) than with either MYC-UPF1(1–1118) or MYC-UPF1(419–1118) (Fig. 1B, bottom, After IP). We conclude that STAU1 preferentially interacts with the N terminus of UPF1, although it appears to also bind additional residues. The co-IP of more STAU1-HA3 with MYC-UPF1(1–418) compared with MYC-UPF1(1–1118) may reflect greater accessibility of amino acids 1–418 in the absence of C-terminal residues present in the full-length protein (see also below).
As an additional means to define those amino acids of UPF1 that interact with STAU1, cell lysates were immunoprecipitated using anti-MYC or, as a control for IP specificity, mouse IgG (mIgG). Each MYC-UPF1 protein was immunoprecipitated using anti-MYC (Fig. 1C, top). The IP was specific since mIgG failed to immunoprecipitate any MYC-UPF1 protein (Fig. 1C, top), and neither anti-MYC nor mIgG immunoprecipitated Calnexin (Fig. 1C, bottom). Western blotting using anti-HA revealed that STAU1-HA3 coimmunoprecipitated more efficiently with MYC-UPF1(1–418) than with MYC-UPF1(1–1118) (Fig. 1C, bottom; it is not possible to detected STAU1-HA3 in an IP of MYC-UPF1(419–1118) at least in part because limited amounts of MYC-UPF1(419–1118) can be immunoprecipitated). These results are consistent with the finding that MYC-UPF1(1–418) preferentially coimmunoprecipitates with STAU1-HA3 (Fig. 1B).
To further delineate the amino acids of UPF1 that interact with STAU1, IPs were performed using either anti-MYC or mIgG and lysates from Cos cells that had been transfected with pSTAU1-HA3, pCI-NEO-T7-UPF2 (Serin et al. 2001), and either pCMV-MYC-UPF1(1–244) (Fig. 1D, top) or, for the sake of comparison, pCMV-MYC-UPF1(1–418). MYC-UPF1(1–244) coimmunoprecipitated with both STAU1-HA3 or T7-UPF2 with essentially the same efficiency as did MYC-UPF1(1–418) (Fig. 1D, bottom). Taken together, all data are consistent with the possibility that the STAU1- and UPF2-binding sites within UPF1 overlap and that STAU1 and UPF2 compete for binding to UPF1.
For the sake of completeness, the region of STAU1 that interacts with UPF1 was also determined. The results of two-hybrid analyses indicate that human UPF1 interacts with a region of human STAU1 containing the fourth RNA-binding domain (RBD4) and the tubulin-binding domain (TBD) (Supplemental Fig. S2A; Kim et al. 2005). Therefore, three deletion derivatives of pSTAU1-HA3 were generated and analyzed: pSTAU1Δ(RBD4)-HA3, pSTAU1Δ(TBD)-HA3, and pSTAU1Δ(RBD5)-HA3 (Supplemental Fig. S2A). Our finding that the interaction of MYC-UPF1(1–1118) with STAU1-HA3 was abolished with deletion of the TBD (Supplemental Fig. S2B) suggests that the TBD of STAU1 interacts with UPF1, although it is possible that deleting the TBD results in protein misfolding and occlusion of the UPF1-binding site.
Evidence that STAU1 and UPF2 compete for binding to UPF1
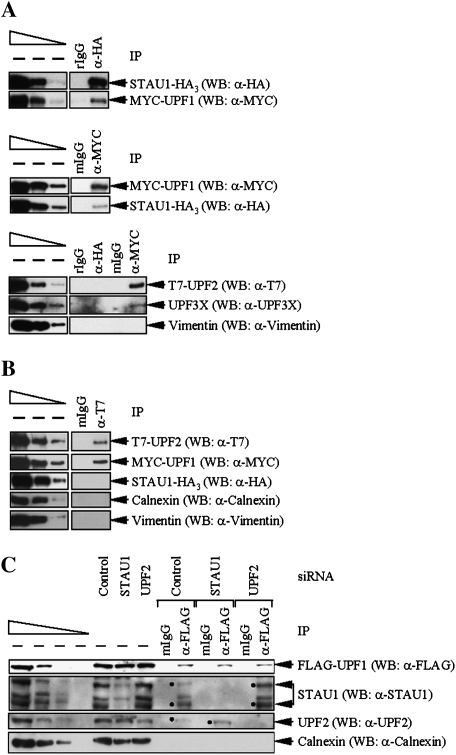
If the region of UPF1 that interacts with STAU1 overlaps with the region of UPF1 that interacts with UPF2, STAU1 and UPF2 would be expected to compete for binding to UPF1. To test for competition, Cos cells were transiently transfected with pSTAU1-HA3, pCMV-MYC-UPF1(1–1118), and pCI-NEO-T7-UPF2. Protein was analyzed by Western blotting before and after IP using anti-HA or, as a control, rIgG. Alternatively, Western blotting was performed before and after IP using anti-MYC or, as a control, mIgG. Using anti-HA, STAU1-HA3 was found to coimmunoprecipitate with MYC-UPF1(1–1118) (hereafter called MYC-UPF1; Fig. 2A, top) but not detectably with T7-UPF2 or, as a negative control, endogenous UPF3X or Vimentin (Fig. 2A, bottom). Using anti-MYC, MYC-UPF1 was found to coimmunoprecipitate with STAU1-HA3 (Fig. 2A, middle) as well as T7-UPF2 and endogenous UPF3X but not Vimentin (Fig. 2A, bottom). Therefore, UPF1 that coimmunoprecipitates with STAU1 is not detectably associated with UPF2.
Figure 2.
Evidence that STAU1 and UPF2 compete for binding to UPF1. (A) Cos cells (3 × 107) were transiently transfected with 10 μg of pSTAU1-HA3, 10 μg of pCMV-MYC-UPF1, and 10 μg of pCI-NEO-T7-UPF2. Protein was purified before (−) and after IP using anti(α)-HA or, to control for nonspecific IP, rIgG (top); anti-MYC or, to control for nonspecific IP, mgG (middle); or all four antibodies (bottom). Equivalent amounts of immunoprecipitated proteins were analyzed by Western blotting (WB) using antibody to the appropriate epitope tag or Vimentin. (B) Cos cells were transiently transfected as in A, and protein was purified before and after IP using anti-T7 or, to control for nonspecific IP, mIgG. Western blotting was performed using the antibody to the appropriate epitope, Calnexin or Vimentin. (C) HeLa cells (3 × 107) that stably express Flag-UPF1 were transiently transfected with 100 nM of STAU1 siRNA, UPF2 siRNA, or a nonspecific Control siRNA. Three days later, protein was purified before and after IP using anti-Flag or, as a control, mIgG that had been covalently conjugated to an agarose affinity gel. Protein was then analyzed by Western blotting using anti-Flag, anti-STAU1, anti-UPF2, or anti-Calnexin. Dots specify the two STAU1 isoforms or UPF2. For each panel, protein from the same number of cells was analyzed in the left-most lane of the dilution standards and in each IP.
In related experiments, Cos cells were similarly transfected with the three plasmids, and protein was analyzed before and after IP using anti-T7 or, as a control, mIgG. Western blotting demonstrated that T7-UPF2 coimmunoprecipitated with MYC-UPF1 but not with STAU1-HA3 or the Calnexin or Vimentin negative control (Fig. 2B). Therefore, UPF1 that coimmunoprecipitates with UPF2 is not detectably associated with STAU1. Taken together, all of these results indicate that UPF1 can bind to either STAU1 or UPF2, suggesting that STAU1 and UPF2 compete for binding to UPF1.
As an additional measure of competition between STAU1 and UPF2 for binding to UPF1, HeLa cells that stably express Flag-UPF1 at approximately twofold above the level of endogenous UPF1 (Pal et al. 2001) were transiently transfected with STAU1 siRNA, UPF2 siRNA, or a nonspecific Control siRNA (Kim et al. 2005). Two days later, protein was purified before and after IP using anti-Flag or, as a control, mIgG and analyzed by Western blotting using anti-Flag, anti-STAU1, anti-UPF2, or anti-Calnexin. Using the level of Calnexin to control for variations in protein loading, STAU1 siRNA down-regulated the cellular abundance of two (63- and 55-kDa) STAU1 isoforms to ∼13% of normal, and UPF2 siRNA down-regulated the cellular abundance of UPF2 to ∼21% of normal (Fig. 2C, where normal is defined as the level in the presence of Control siRNA). Down-regulating STAU1 increased the amount of UPF2 that coimmunoprecipitated with Flag-UPF1 3.1-fold (Fig. 2C; data not shown for STAU1(A) siRNA, which targets a different sequence than does STAU1 siRNA; Kim et al. 2005). Additionally, down-regulating UPF2 increased the amount of STAU1 that coimmunoprecipitated with Flag-UPF1 2.4-fold (Fig. 2C; data not shown for a UPF2(A) siRNA, which targets a different sequence than does UPF2 siRNA; Kim et al. 2005). Thus, all results indicate that STAU1 and UPF2 do indeed compete for binding to UPF1, suggesting that SMD and NMD may compete as well.
Down-regulating STAU1 increases the efficiency of NMD whereas down-regulating UPF2 increases the efficiency of SMD
SMD, like NMD, is inhibited by cycloheximide or constitutively inactive eIF2α (Supplemental Figs. S3, S4). Therefore, SMD, like NMD, depends on translation. To determine if SMD and NMD are competitive pathways, HeLa cells were transiently transfected with STAU1 siRNA, UPF2 siRNA, or Control siRNA. Notably, down-regulating STAU1 inhibits SMD but not NMD (Kim et al. 2005, 2007), whereas down-regulating UPF2 inhibits NMD but not SMD (Mendell et al. 2002; Wang et al. 2002; Kim et al. 2005; Matsuda et al. 2007; Isken et al. 2008; Woeller et al. 2008). Two days later, cells were transfected with (1) pmCMV-Gl Norm or pmCMV-Gl 39 Ter, which contains a β-globin gene that, respectively, does not or does harbor a PTC (referred to as Ter) at position 39 (Zhang et al. 1998), (2) pcFLUC-SBS, which consists of firefly (F) luciferase (LUC) cDNA harboring a C-terminal deletion followed by the 3′ UTR of ADP ribosylation factor 1 (ARF1) cDNA that harbors the SBS (Kim et al. 2005, 2007), and (3) phCMV-MUP, which encodes major urinary protein mRNA (Belgrader and Maquat 1994). The level of Gl 39Ter mRNA relative to the level of Gl Norm mRNA provides a measure of NMD, the level of FLUC-SBS mRNA serves to quantify SMD, and the level of MUP mRNA controls for variations in the efficiencies of cell transfection and RNA recovery. After an additional 24 h, lysates were generated for Western blotting and RT–PCR.
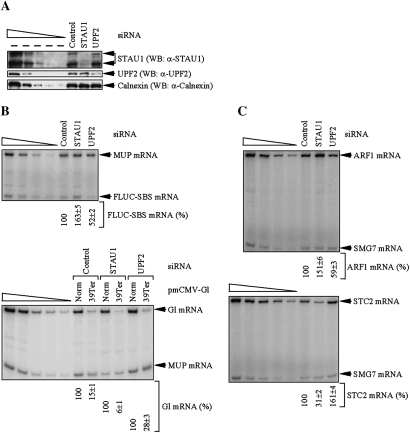
Using the level of Calnexin to control for variations in protein loading, Western blotting demonstrated that STAU1 or UPF2 siRNA, relative to Control siRNA, down-regulated the level of the two STAU1 isoforms or UPF2 to ∼15% or ∼18% of normal, respectively (Fig. 3A). RT–PCR demonstrated that down-regulating STAU1 inhibited the SMD of FLUC-SBS mRNA as evidenced by the ∼1.5-fold increase in the level of FLUC-SBS mRNA relative to the level of MUP mRNA (Fig. 3B, top, cf. 100% and 163 ± 5%; see Supplemental Table S1 for corroborating RT–quantitative PCR data). However, down-regulating UPF2 decreased the level of FLUC-SBS mRNA approximately twofold (Fig. 3B, top, cf. 100% and 52 ± 2%; Supplemental Table S1). Furthermore, down-regulating UPF2 also decreased the level of cellular ARF1 mRNA, which is a natural target of SMD (Kim et al. 2005), ∼1.5-fold relative to the level of cellular suppressor with morphological effect on genitalia (SMG)7 mRNA (Fig. 3C, top, cf. 100% and 59 ± 3%; Supplemental Table S1), which is not targeted for either SMD or NMD (Kim et al. 2005, 2007). These data indicate that down-regulating UPF2 increases the efficiencies with which FLUC-SBS and cellular ARF1 mRNAs are targeted for SMD.
Figure 3.
Decreasing the cellular abundance of STAU1 increases the efficiency of NMD, and decreasing the cellular abundance of UPF2 increases the efficiency of SMD. HeLa cells (2 × 106) were transiently transfected with the 100 nM of STAU1 siRNA, UPF2 siRNA, or Control siRNA. Two days later, cells were retransfected with 0.3 μg of the specified pmCMV-Gl test plasmid, 0.3 μg of the specified pcFLUC test plasmid, and 0.2 μg of the phCMV-MUP reference plasmid. After an additional 12 h, protein and RNA were analyzed. (A) Western blotting (WB) using anti(α)-STAU1, anti-UPF2, or anti-Calnexin. (B) RT–PCR analysis, where the level of FLUC-SBS or Gl mRNA was normalized to the level of MUP mRNA, and the normalized level of FLUC-SBS mRNA in the presence of Control siRNA or Gl Norm mRNA in the presence of each siRNA was defined as 100. (C) RT–PCR analysis, where the level of ARF1 or STC2 mRNA was normalized to the level of SMG7 mRNA, and the normalized value in the presence of Control siRNA was defined as 100. All results are representative of three independently performed experiments that did not vary by more than the amount shown.
As expected, UPF2 siRNA inhibited the NMD of Gl Ter mRNA as evidenced by the approximately twofold increase in the level of Gl Ter mRNA relative to MUP mRNA (Fig. 3B, bottom, cf. 28 ± 3% and 15 ± 1%; Supplemental Table S1). However, down-regulating STAU1 decreased the level of Gl Ter mRNA ∼2.5-fold (Fig. 3B, bottom, cf. 6 ± 1% and 15 ± 1%; Supplemental Table S1). Furthermore, down-regulating STAU1 decreased by approximately threefold the level of cellular stanniocalcin 2 (STC2) mRNA (Fig. 3C, bottom, cf. 100% and 31 ± 2%; Supplemental Table S1), which is a natural target of NMD because it contains an upstream ORF (uORF) (Fig. 3C, bottom, cf. 100% and 161 ± 4; Supplemental Table S1; Mendell et al. 2004). Consistent with STAU1 siRNA increasing the efficiency of STC2 NMD, the results of two independently performed microarray analyses in which the abundance of HeLa cell STAU1 had been down-regulated to ∼4% of normal revealed that the level of HeLa cell STC2 mRNA was decreased approximately fourfold (Kim et al. 2007). Thus, down-regulating STAU1 increases the efficiencies with which Gl Ter mRNA and cellular STC2 mRNA are targeted for NMD.
We conclude that SMD and NMD are competing pathways in HeLa cells. Based on our evidence that STAU1 and UPF2 compete for UPF1 binding in Cos cells (Fig. 2) together with the high degree to which all three proteins are conserved among different mammals, it is likely that SMD and NMD are also competitive pathways in Cos cells and probably generally in mammalian cells (see also below).
Transcripts down-regulated by UPF2 siRNA include SMD targets, and transcripts down-regulated by STAU1 siRNA include NMD targets
During the course of our studies, Wittmann et al. (2006) identified a number of HeLa cell transcripts that are decreased in abundance by UPF2 siRNA by mechanisms that were not examined. In view of our findings, it seemed likely that some of these transcripts would be SMD targets. In particular, amphoterin-induced gene 2 (AMIGO2) and FLJ21870 mRNAs not only decreased in abundance when UPF2 was down-regulated (Wittmann et al. 2006) but also increased in abundance when STAU1 was down-regulated (Kim et al. 2007). To determine if AMIGO2 and FLJ21870 mRNAs are SMD targets, the level of each mRNA in HeLa cells that had been transfected with Control, STAU1, UPF1, or UPF2 siRNA was assessed by RT–PCR. Additionally, the consequence of these siRNAs to the abundance of interleukin 7 receptor (IL7R) and c-JUN mRNAs, which are bona fide SMD targets (Kim et al. 2007), was also analyzed.
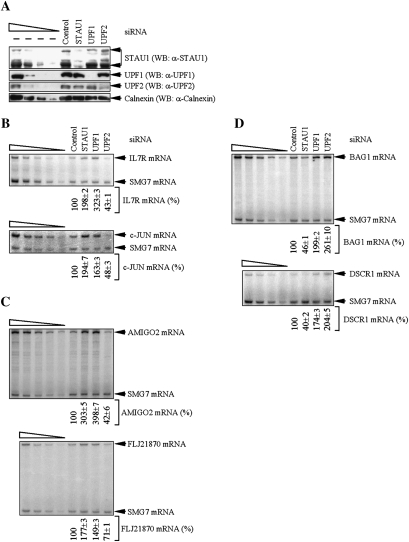
Using the level of Calnexin to control for variations in protein loading, Western blotting demonstrated STAU1, UPF1, or UPF2 siRNA, relative to Control siRNA, down-regulated the level of the corresponding protein to ∼20%, ∼10%, or ∼15% of normal, respectively (Fig. 4A). As expected, STAU1 or UPF1 siRNA increased the abundance of IL7R and c-JUN mRNAs relative to SMG7 mRNA (Fig. 4B; see Supplemental Table S2 for corroborating RT–quantitative PCR data). Consistent with competition between STAU1 and UPF2 binding to UPF1, UPF2 siRNA decreased the abundance of IL7R and c-JUN mRNAs relative to SMG7 mRNA (Fig. 4B; Supplemental Table S2). As predicted from Wittmann et al. (2006), UPF2 siRNA decreased the abundance of AMIGO2 and FLJ21870 mRNAs (Fig. 4C; Supplemental Table S2). Consistent with AMIGO2 and FLJ21870 mRNAs being SMD targets, STAU1 or UPF1 siRNA increased the abundance of each mRNA (Fig. 4C; Supplemental Table S2). We conclude that transcripts down-regulated upon UPF2 depletion include SMD targets.
Figure 4.
HeLa cell transcripts down-regulated upon UPF2 down-regulation include SMD targets. HeLa cells (2 × 107) were transiently transfected with 100 nM of the specified siRNA and harvested after an additional 48 h. (A) Western blotting (WB) demonstrated the degree of siRNA-mediated down-regulation of STAU1, UPF1, or UPF2. (B) RT–PCR analysis of IL7R or c-JUN mRNA after normalization to the level of SMG7 mRNA, where the level in the presence of Control siRNA was defined as 100. (C) As in B, except that AMIGO2 or FLJ21870 mRNA was analyzed. (D) As in B, except that BAG1 or DSCR1 mRNA was analyzed. All results are representative of three independently performed experiments that did not vary by more than the amount shown.
It appeared likely that the converse was also true; i.e., transcripts down-regulated by STAU1 siRNA include NMD targets. This idea was supported by our previous microarray data demonstrating that the BCL2-associated athanogene 1 (BAG1) transcript, which is an NMD target that is up-regulated upon UPF2 down-regulation because of an uORF (Gehring et al. 2005; Wittmann et al. 2006), was down-regulated upon STAU1 down-regulation (Kim et al. 2007). Results revealed that UPF2 or UPF1 siRNA increased the abundance of BAG1 mRNA relative to SMG7 mRNA (Fig. 4D; Supplemental Table S2) as would be expected of an NMD target. Consistent with competition between STAU1 and UPF2 binding to UPF1, STAU1 siRNA decreased the abundance of BAG1 mRNA relative to SMG7 mRNA (Fig. 4D; Supplemental Table S2). We also tested if a cellular PTC-containing alternatively spliced variant of the down syndrome critical region 1 (DSCR1) gene that is another NMD target (Mendell et al. 2004; Wittmann et al. 2006) is down-regulated upon STAU1 down-regulation (Kim et al. 2007). As expected, UPF1 or UPF2 siRNA increased the abundance of alternatively spliced DSCR1 mRNA relative to SMG7 mRNA (Fig. 4D; Supplemental Table S2). Furthermore, STAU1 siRNA decreased the abundance of this mRNA relative to SMG7 mRNA (Fig. 4D; Supplemental Table S2). We conclude that transcripts down-regulated upon STAU1 depletion include NMD targets. The modest effects seen on SMD after debilitating NMD by down-regulating UPF2 or on NMD after debilitating SMD by down-regulating STAU1 may reflect that neither UPF2 nor STAU1 function is completely dedicated to NMD or SMD, respectively: Each protein functions in additional pathways (Kim and Maquat 2006; Isken and Maquat 2008).
The differentiation of C2C12 MBs to MTs is accompanied by an increased efficiency of SMD and alternative NMD but a decreased efficiency of classical NMD
To test for competition between SMD and NMD under physiologic circumstances, we utilized the mouse skeletal muscle C2C12 cell line considering that the differentiation of C2C12 MBs to multinucleated MTs is accompanied by an increase in the efficiency of SMD (Kim et al. 2007).
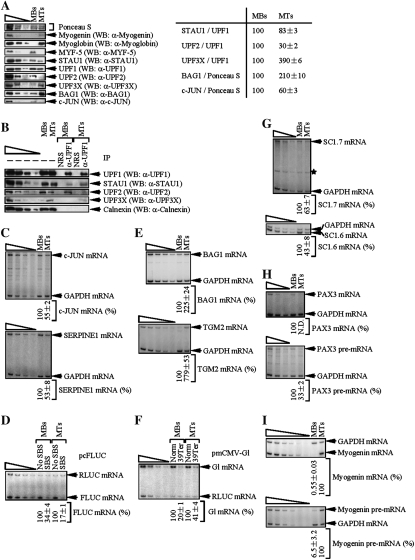
MBs were cultured in medium containing 15% fetal bovine serum, and differentiation was induced using medium containing 5% horse serum. MBs or MTs were transiently transfected with three plasmids: either (1) pmCMV-Gl Norm, pFLUC-No SBS (Kim et al. 2005), and pRLUC, which encodes Renilla LUC (RLUC), or (2) pmCMV-Gl Ter, pFLUC-SBS, and pRLUC. Differentiation was confirmed by increased levels of myogenin and myoglobin (Blais et al. 2005 and references therein) and a decreased level of MYF-5 (Deato and Tjian 2007) as evidenced using Western blotting (Fig. 5A, left) and Ponceau S staining to control for variations in protein loading (Deato and Tjian 2007). Differentiation was also accompanied by a slight decrease (from 100 to 83 ± 3) in the level of STAU1 relative to the level of UPF1 and a larger decrease (from 100 to 30 ± 3) in the level of UPF2 relative to the level of UPF1 (Fig. 5A, right), providing the first indication that competition between SMD and NMD could contribute to the increased efficiency of SMD and, possibly, a decreased efficiency of NMD during myogenesis. In further support of these ideas, IPs of equal amounts of UPF1 from lysates of MBs or MTs using anti-UPF1 revealed an increase in the amount of coimmunoprecipitated STAU1 and a decrease in the amount of coimmunoprecipitated UPF2 in MTs relative to MBs (Fig. 5B).
Figure 5.
Myogenic differentiation of C2C12 cells is accompanied by an increase in the efficiency of SMD, a decrease in the efficiency of classical NMD, and an increase in the efficiency of alternative NMD. MBs or MTs (2 × 107) were transiently transfected with three plasmids: 3 μg of pmCMV-Gl Norm or pmCMV-Gl Ter, 12 μg of pFLUC-No SBS or pFLUC-SBS, and 3 μg of pRLUC. Two days later, cells were harvested. (A, left) Western blotting (WB) of myogenin, myoglobin, MYF-5, STAU1, UPF1, UPF2, UPF3X, BAG1, or c-JUN. Ponceau S staining was used as a loading control. (A, right) Quantitation of the level of STAU1, UPF2, or UPF3X relative to UPF1, or BAG1 or c-JUN relative to Ponceau S staining, in MBs and MTs. Each level in MBs was defined as 100. (B) Western blotting of specified proteins before (−) or after IP using anti-UPF1 or, to control for nonspecific IP, normal rabbit serum (NRS). (C, top and bottom) RT–PCR analysis of c-JUN or SERPINE1 mRNA after normalization to the level of GAPDH mRNA, where each normalized level in MBs was defined as 100. (D) The level of FLUC-No SBS or FLUC-SBS mRNA was normalized to the level of RLUC mRNA, and the normalized level of FLUC-No SBS in MBs or MTs was defined as 100. (E, top and bottom) RT–PCR analysis of BAG1 or TGM2 mRNA after normalization to the level of GAPDH mRNA, where each normalized level in MBs was defined as 100. (F) The level of PTC-free (Norm) or PTC-containing (Ter) Gl mRNA was normalized to the level of RLUC mRNA, and the normalized level of G1 Norm mRNA in MBs or MTs was defined as 100. (G, top and bottom) RT–PCR analysis of SC1.7 or SC1.6 mRNA after normalization to the level of GAPDH mRNA, where each normalized level in MBs was defined as 100 (asterisk denotes a nonspecific RT–PCR product that does not interfere with the analysis). (H, top and bottom) As in G. However, PAX3 mRNA or pre-mRNA was analyzed. (I, top and bottom) As in G. However, myogenin mRNA or pre-mRNA was analyzed. All results are representative of at least three independently performed experiments that did not vary by more than the amount shown.
RT–PCR confirmed that the levels of cellular c-JUN and serpin peptidase inhibitor, clade E, member 1 (SERPINE1) mRNAs, which are SMD targets (Kim et al. 2007), were decreased upon differentiation (Fig. 5C, where the level of cellular GAPDH mRNA controlled for variations in RNA recovery; see Supplemental Table S3 for corroborating RT–quantitative PCR data). The decrease was attributable to an increased efficiency of SMD as evidenced by a concomitant decrease in the level of FLUC-SBS mRNA relative to the level of FLUC-No SBS mRNA, which provided a specific assay of SMD (Fig. 5D, where the level of RLUC mRNA controlled for variations in transfection efficiencies and RNA recovery; Supplemental Table S3). RT–PCR also demonstrated that the levels of cellular BAG1 and transglutaminase 2 (TGM2) mRNAs were increased upon differentiation (Fig. 5E; Supplemental Table S3). Notably, BAG1 and TGM2 mRNAs are not only up-regulated in mouse L cells by UPF1, UPF2, or UPF3X siRNA as would be expected of NMD targets (Supplemental Fig. S5) but also have HeLa cell counterparts that are NMD targets (Fig. 4D; Gehring et al. 2005; Wittmann et al. 2006). Consistent with a decreased efficiency of NMD upon differentiation, there was a concomitant increase in the level of Gl Ter mRNA relative to the level of Gl Norm mRNA, which provided a specific assay of NMD (Fig. 5F; Supplemental Table S3). In summary, based on changes during differentiation in the levels of STAU1 and UPF2 relative to UPF1 and differences in the abundance of natural or reporter SMD targets relative to natural or reporter NMD targets, we conclude that the efficiencies of SMD and NMD are altered in a reciprocal manner during myogenesis due to competition between the two pathways.
Unexpectedly, the ratio of the NMD factor UPF3X relative to UPF1 increased approximately fourfold during myogenesis (Fig. 5A). Since data indicate that UPF2 binding to EJCs is mediated by UPF3X (Lykke-Andersen et al. 2000; Hosoda et al. 2005; Kim et al. 2005; Singh et al. 2007), the increased ratio of UPF3X to UPF1 is not envisioned to augment the efficiency of classical NMD in MTs because of the very low and thus limiting level of UPF2 (Fig. 5A). However, the increased ratio may augment the efficiency of an alternative NMD pathway that is significantly less sensitive than classical NMD to UPF2 siRNA but, like classical NMD, depends on not only UPF1 but also UPF3X (Gehring et al. 2005; Saltzman et al. 2008; see below). RT–PCR revealed that the levels of cellular SC1.7 and SC1.6 mRNAs, which encode variants of the SC35 splicing factor generated by alternative splicing and are targets of this alternative NMD pathway (Gehring et al. 2005), were indeed decreased upon differentiation (Fig. 5G; Supplemental Table S3). Notably, the NMD of SC1.7 and SC1.6 mRNAs depends on UPF3X, as evidenced using UPF3X siRNA (Supplemental Fig. S5). Moreover, larger amounts of UPF3X immunoprecipitated with UPF1 in MTs compared with MBs despite MTs having less UPF2 than MBs (Fig. 5B), consistent with UPF3X functionally contributing to alternative NMD and reports that UPF3X and UPF1 can coimmunoprecipitate independently of UPF2 (Gehring et al. 2005; Kunz et al. 2006; Singh et al. 2007). Our findings support the idea that SMD and classical NMD are competitive pathways in a way that reflects that abilities of STAU1 and UPF2 to compete during myogenesis for binding to UPF1, which is of no measurable consequence to the alternative NMD pathway. Thus, SMD and the two modes of NMD together with their respective mRNA targets are likely to contribute to the myogenic process.
PAX3 mRNA is an SMD target whereas myogenin mRNA is a classical NMD target
We next aimed to understand how these mechanistically interconnected mRNA decay pathways contribute to myogenesis. We sought to define transcripts that function in either the maintenance of MBs or the differentiation of MBs to MTs and are affected by the competitive nature of SMD and NMD. Since globally down-regulating STAU1 or UPF factor function using siRNA would not accomplish this goal, we initially turned to microarray data to identify specific transcripts of interest.
Our previous microarray analyses of transcripts from HeLa cells in which STAU1 had been down-regulated demonstrated that the abundance of PAX3 mRNA, which encodes a transcription factor that inhibits myogenic differentiation (Epstein et al. 1995), was increased approximately fivefold whereas the abundance of myogenin mRNA was decreased approximately twofold (Kim et al. 2007). Other microarray analyses of transcripts from HeLa cells in which UPF1 had been down-regulated showed that the abundance of PAX3 mRNA was increased approximately twofold whereas the abundance of myogenin mRNA was increased ∼3.5-fold (Mendell et al. 2004). These findings, taken together, suggest that PAX3 mRNA could be an SMD target and myogenin mRNA could be a classical NMD target. Considering that the level of PAX3 mRNA is decreased in MTs relative to MBs (Boutet et al. 2007) in a way that promotes myogenesis (Epstein et al. 1995) but myogenin mRNA encodes a basic helix–loop–helix transcription factor that is required for and, thus, up-regulated during myogenesis (Wright et al. 1989), it seemed plausible that changes in the relative efficiencies of SMD and classical NMD during the differentiation of MBs to MTs would contribute to the differentiation process. RT–PCR revealed that, indeed, the decrease in the level of PAX3 mRNA and increase in the level of myogenin mRNA during myogenesis were at least partly attributable to changes in mRNA abundance as evidenced by using the level of the corresponding pre-mRNA to control for changes in mRNA abundance that were due to changes in gene transcription, pre-mRNA processing, or both (Fig. 5H,I; Supplemental Table S3).
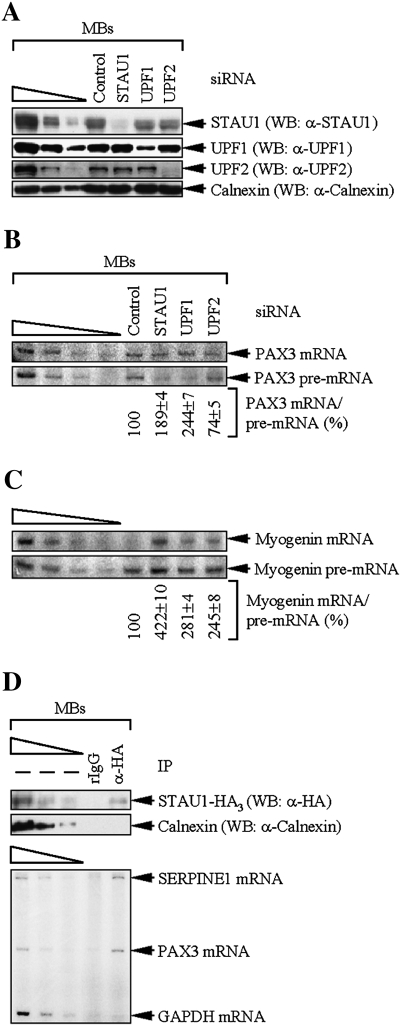
To test the ideas that PAX3 and myogenin mRNAs are SMD and classical NMD targets, respectively, STAU1, UPF1, or UPF2 was down-regulated in MBs using siRNA to 10%, 14%, or 5% of normal, respectively (Fig. 6A, where the abundance of each protein in the presence of Control siRNA was defined as 100%). Using the level of PAX3 pre-mRNA to control for changes in mRNA abundance that were due to changes in PAX3 gene transcription and pre-mRNA processing, the level of PAX3 mRNA was increased approximately twofold by STAU1 or UPF1 siRNA and decreased ∼1.4-fold by UPF2 siRNA (Fig. 6B; see Supplemental Table S4 for corroborating RT–quantitative PCR data), consistent with the prediction that PAX3 mRNA is an SMD target. Using the level of myogenin pre-mRNA to control for changes in mRNA abundance that were due to changes in myogenin gene transcription and pre-mRNA processing, the level of myogenin mRNA was increased approximately threefold by UPF1 siRNA and approximately twofold by UPF2 siRNA (Fig. 6C; Supplemental Table S4), consistent with the prediction that myogenin mRNA is a classical NMD target. The unexpected approximately fourfold increase in the level of myogenin mRNA abundance in the presence of STAU1 siRNA may reflect effects of STAU1 that counter the destabilizing effects of NMD (see the Discussion). Our finding that STAU1 siRNA increases the abundance of ∼1% of HeLa cell transcripts and decreases the abundance of a comparable percentage of other transcripts (Kim et al. 2007) suggests that the observed STAU1 siRNA-mediated increase in myogenin mRNA abundance could be attributable to STAU1 decreasing the concentration of a myogenin mRNA stabilizing factor or increasing the concentration of a myogenin mRNA destabilizing factor. Notably, the presence of an uORF in myogenin mRNA (NCBI ORF Finder) further substantiates that myogenin mRNA is an NMD target. All of these findings are compatible with data showing that myogenin mRNA half-life is extended upon myogenic differentiation (Figueroa et al. 2003) by multiple mechanisms that include a decreased efficiency of NMD.
Figure 6.
Evidence that PAX3 mRNA is an SMD target and myogenin mRNA is an NMD target. (A–C) MBs (1 × 106) were transiently transfected with 100 nM of the specified siRNA and harvested after an additional 48 h. (A) Western blotting (WB) demonstrated the degree of siRNA-mediated down-regulation of STAU1, UPF1, or UPF2. Notably, mouse STAU1, unlike human STAU1, is detected as a 55-kDa isoform but not a 63-kDa isoform. (B) RT–PCR analysis of PAX3 mRNA after normalization to the level of PAX3 pre-mRNA, where the level in the presence of Control siRNA was defined as 100. Notably, equal amounts of total cell RNA rather than equal amounts of PAX3 pre-mRNA were analyzed in each lane. (C) As in B, except that myogenin mRNA or pre-mRNA was analyzed. (D) MBs (2 × 107) were transiently transfected with 10 μg of pSTAU1-HA3. Two days later, cells were harvested, treated with formaldehyde to cross-link protein and RNA, and lysed using sonication. Lysates were immunoprecipitated using anti-HA or, to control for nonspecific IP, rIgG, and cross-links were reversed using heat. Protein was analyzed by Western blotting using anti-HA or anti-Calnexin. RNA was analyzed by RT–PCR for SERPINE1, PAX3, or GAPDH mRNA. All results are representative of at least three independently performed experiments that did not vary by more than the amount shown.
We sought additional support for data indicating that PAX3 mRNA is an SMD target by testing if PAX3 mRNA coimmunoprecipitates with STAU1. To this end, STAU1-HA3 was transiently expressed in MBs and immunoprecipitated using anti-HA (Fig. 6D, top). As shown earlier (Kim et al. 2007), SERPINE1 mRNA was immunoprecipitated using anti-HA but not rIgG, and GAPDH mRNA was not immunoprecipitated using either antibody (Fig. 6D, bottom). PAX3 mRNA was immunoprecipitated using anti-HA but not rIgG as is expected of an SMD target (Fig. 6D, bottom). Given the critical functions of PAX3 and myogenin to the maintenance of MBs and the differentiation of MBs to MTs, respectively, combined with our findings that PAX3 mRNA is an SMD target whereas myogenin mRNA is an NMD target, we conclude that the intricate balance between SMD and NMD factor abundance and the competitive nature of the UPF1 interaction with STAU1 and UPF2 contribute in important ways to the myogenic process.
Discussion
We show here that SMD and classical, UPF2-dependent NMD, both of which require UPF1 and translation (Supplemental Figs. S3, S4; Kim et al. 2005, 2007; Behm-Ansmant et al. 2007; Chang et al. 2007; Isken and Maquat 2007), compete with one another under conditions where an alternative NMD pathway that is relatively insensitive to UPF2 siRNA remains robust. First, not only STAU1, which functions in SMD but not NMD, but also UPF2, which functions in NMD but not SMD, interact with amino acids 1–244 of UPF1 (Fig. 1). The latter is in agreement with data published during the course of our studies demonstrating that UPF2 binds UPF1 residues 123–213 (Kadlec et al. 2006). It makes sense that STAU1 would bind the cysteine- and histidine-rich CH domain of UPF1 as does UPF2 in view of a recent report that the CH domain serves as an intramolecular regulator of UPF1 activities: Just as UPF2:UPF3X binding to the CH domain of UPF1 has been proposed to prevent the cis-inhibitory effect of the domain so as to stimulate UPF1 RNA-dependent ATPase and 5′-to-3′ helicase activities (Chamieh et al. 2008), so probably does STAU1 binding to the CH domain of UPF1. As a second indication that SMD and NMD are in competition, UPF1 that immunoprecipitates with STAU1 is not detectably associated with UPF2, and UPF1 that immunoprecipitates with UPF2 is not detectably associated with STAU1 (Fig. 2). These findings indicate that UPF1 can interact with either STAU1 or UPF2 but not both simultaneously. Third, down-regulating STAU1 increases the association of UPF1 and UPF2 while down-regulating UPF2 increases the association of UPF1 and STAU1 (Figs. 2, 5). Fourth, inhibiting SMD by down-regulating STAU1 increases the efficiency of NMD, and inhibiting NMD by down-regulating UPF2 increases the efficiency of SMD (Fig. 3; Supplemental Table S1). Additionally, HeLa cell transcripts that decrease in abundance upon UPF2 down-regulation include AMIGO2 and FLJ21870 mRNAs, which are SMD targets (Fig. 4; Supplemental Table S2). Furthermore, HeLa cell transcripts that decrease in abundance upon STAU1 down-regulation include BAG1 and DSCR1, which are NMD targets (Fig. 4; Supplemental Table S2).
Our finding that the differentiation of C2C12 MBs to MTs is accompanied by an increased efficiency of SMD and a concomitant decreased efficiency of classical (i.e., UPF2 siRNA-sensitive) NMD (Fig. 5; Supplemental Table S3) is consistent with competition between SMD and NMD. Competition indeed appears to reflect the relative abilities of STAU1 and UPF2 to bind to UPF1, as evidenced by the increased co-IP of STAU1 and UPF1 and the decreased co-IP of UPF2 and UPF1 in MTs relative to MBs (Fig. 5). Furthermore, the increased ratio of UPF3X to UPF1 during myogenesis increases not only the co-IP of UPF3X with UPF1 but also the efficiency of an alternative NMD pathway that is relatively insensitive to UPF2 siRNA (Fig. 5; Supplemental Fig. S5; Supplemental Table S3). The functional significance of competition between SMD and classical NMD is apparent with our finding that PAX3 mRNA, which encodes a protein required for the maintenance of MBs, is an SMD target, while myogenin mRNA, which encodes a protein required for the differentiation of MBs to MTs, is a classical NMD target (Fig. 6; Supplemental Table S4).
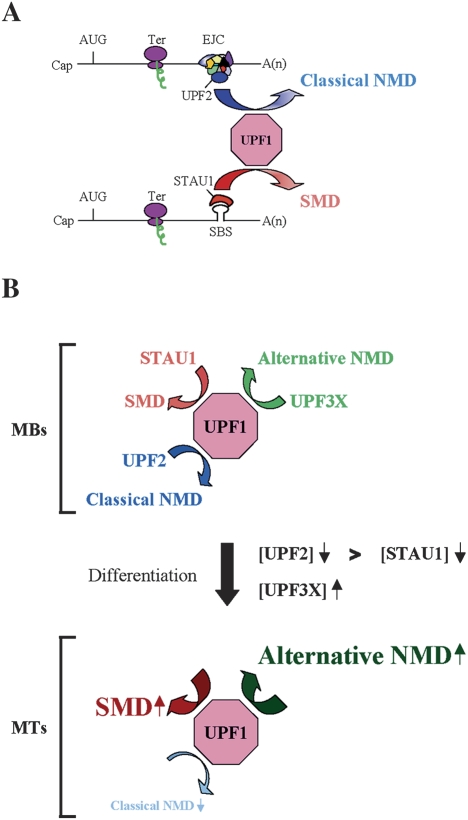
We propose a model in which SMD and the two types of NMD compete during myogenesis to a degree that at least in part reflects the relative abundance of STAU1, UPF2, UPF3X, and UPF1 as well as the number of mRNAs undergoing SMD or NMD (Fig. 7). UPF1, UPF2, UPF3X, and STAU1 are expressed in all cell types that have been examined. However, cell type-dependent differences in the abundance or nucleocytoplasmic distribution have been reported for STAU1 and UPF1, respectively, and differential splicing generates several STAU1 isoforms (Marion et al. 1999; Wickham et al. 1999; Monshausen et al. 2001; Chiu et al. 2003). Moreover, since STAU1, UPF1, UPF2, and UPF3X are phosphoproteins (Monshausen et al. 2001; Pal et al. 2001; Yamashita et al. 2001; Chiu et al. 2003; Isken et al. 2008; O. Isken and L.E. Maquat, unpubl.), phosphorylation and other post-translational modifications may influence the extent of competition between NMD and SMD. The physiological importance of competition undoubtedly manifests in the levels of proteins encoded by SMD and NMD targets. Significantly, data indicate that SMD and NMD targets are translated with efficiencies that are comparable with their counterparts that are not targeted for decay (Fig. 5; Supplemental Fig. S6; Supplemental Table S3; Chan et al. 2007).
Figure 7.
Model for competition between SMD and NMD. (A) UPF2, which is an EJC constituent and functions in the classical NMD pathway, and STAU1, which is an RNA-binding protein that functions in the SMD pathway, compete for binding to UPF1 and, thus, the recruitment of UPF1 to mRNA. UPF1 functions in both pathways to elicit mRNA decay when translation terminates sufficiently upstream of an EJC, in the case of NMD, or a SBS, in the case of SMD. Ter can be either a premature or normal termination codon. (B) As a consequence of C2C12 cell differentiation from MBs to MTs, the efficiency of SMD increases, the efficiency of classical (i.e., UPF2 siRNA-sensitive) NMD decreases, and the efficiency of an alternative NMD pathway that depends on UPF3X but not appreciably UPF2 increases. During myogenesis, a larger decrease in the abundance of UPF2, which drops to almost undetectable levels, relative to STAU1 permits STAU1 to out-compete UPF2 for binding to UPF1, and an approximately fourfold increase in the level of UPF3X supports an increase in the efficiency of the alternative NMD pathway.
Results reported here provide the first insight into how the efficiency of SMD is regulated, and they add to the one report currently available on NMD efficiency determinants: Variations in the efficiency of NMD among three different HeLa cell lines were shown recently to be attributable to differences in the cellular abundance of the EJC constituent RNPS1 (Viegas et al. 2007). Our findings illustrate that variations in factor abundance can affect not only the pathway in which the factor directly participates but also pathways that are indirectly connected to factor function.
While preparing this paper, it was reported that down-regulating STAU1 to <40% of the normal level in MBs, in contrast to down-regulating UPF1 in MBs, induced myogenin gene transcription and, as a result, differentiation to MTs (Yamaguchi et al. 2008). Considering that STAU1 is present in MTs, although at a reduced level than in MBs (Fig. 5), this new finding of a STAU1-dependent but UPF1-independent pathway that relies on the myogenin promoter (Yamaguchi et al. 2008) may further illustrate the delicate balance of STAU1-mediated pathways that must be achieved for proper MB and MT metabolism. Nevertheless, even though down-regulating STAU1 abundance in MBs to at least 30% of normal using either STAU1 or STAU1(A) siRNA (Kim et al. 2005) increased the level of myogenin pre-mRNA (Fig. 6; Supplemental Table S4), we neither detected myogenin in MBs nor induced the differentiation of MBs to MTs (data not shown). Reasons for discrepancies between our data and those of Yamaguchi et al. (2008) remain unclear. However, our data suggest that myogenin expression alone is insufficient to induce myogenesis, consistent with published reports (Melo et al. 1996; Osses and Brandan 2002). Alternatively or additionally, STAU1 siRNA may not only induce myogenin expression but may also result in events that counter the differentiation process.
Future studies of SMD and classical NMD in specific cell types and under particular growth conditions will be required to fully appreciate the pervasiveness and functional significance of competition between the two pathways. Roles for the alternative NMD pathway that is relatively insensitive to UPF2 siRNA and, thus, STAU1 siRNA so as to provide a means by which NMD can occur unimpeded by competition also merit investigation. As described here, our results portend that mRNA decay, like transcriptional and other post-transcriptional events, is undoubtedly regulated by complex and interdependent processes that, when appropriately balanced, contribute to the maintenance of or progression to distinct cell types.
Materials and methods
Plasmid constructions
See the Supplemental Material.
Cell culture and transfection, and protein and RNA purification
Human HeLa or monkey Cos cells (2 × 106 per 60-mm dish or 7.5 × 107 per 150-mm dish) were grown in DMEM (GIBCO-BRL) containing 10% fetal bovine serum (GIBCO-BRL). Cells were transiently transfected using Lipofectamine Plus Reagent (Invitrogen) as indicated in the figure legends. Mouse C2C12 cells (2 × 107) were propagated as MBs in DMEM containing 15% fetal bovine serum and induced to differentiate to MTs using DMEM containing 5% horse serum (GIBCO-BRL). Cells were transiently transfected using Lipofectamine 2000 (Invitrogen) as indicated in the figure legends. Importantly, MTs were harvested using 0.15% trypsin rather than by scrapping so as to leave behind undifferentiated reserve cells (Deato and Tjian 2007). Protein was isolated using passive lysis buffer (Promega), and RNA was purified using Trizol Reagent (Invitrogen).
siRNA-mediated down-regulation of human or mouse STAU1, UPF1, or UPF2
HeLa cells were transiently transfected with 100 nM of in vitro-synthesized UPF1, UPF2, UPF2(A), STAU1, or STAU1(A) siRNA or a nonspecific Control siRNA using Oligofectamine (Invitrogen) as described previously (Kim et al. 2005). When specified, cells were harvested after an additional 24 h or retransfected 48 h later with plasmids and harvested 24 h thereafter. C2C12 cells were transiently transfected with 100 nM of in vitro-synthesized UPF1 (Kim et al. 2005), UPF2 (Ambion), or STAU1 (Kim et al. 2005) siRNA or Negative Control #1 siRNA (Ambion) using Lipofectamine 2000.
RT–PCR
Reverse transcriptions were performed as described previously (Kim et al. 2005). cDNA from FLUC-SBS, FLUC-No SBS, ARF1, Gl, GPx1, MUP, or SMG7 mRNA was amplified as described previously (Chiu et al. 2004; Kim et al. 2005). cDNA from HeLa cell STC2, c-JUN, IL7R, AMIGO2, FLJ21870, BAG1, or DSCR1 mRNA was amplified using the primer pairs 5′-CCTGTCCCTGCAGAATACAG-3′ (sense) and 5′-GTTCACGAGGTCCACGTAGG-3′ (antisense), 5′-CTTGAAAGCTCAGAACTCGG-3′ (sense) and 5′-TCAGCCCCCGACGGTCTCTC-3′ (antisense), 5′-AAGTGGCTATGCTCAAAATG-3′ (sense) and 5′-TTCAGGCACTTTACCTCCAC-3′ (antisense), 5′-CTGCTAGAGCCGGATAAAGAG-3′ (sense) and 5′-TTCATCAGCGGAGGCATCAC-3′ (antisense), 5′-AACGACCTGTTAAAGGCCACTC-3′ (sense) and 5′-TCCTTACTTTGCAGGCATCCAG-3′ (antisense), 5′-AAGATGGTTGCCGGGTCATG-3′ (sense) and 5′-TGTTCTGCTCCACTGTGTCAC-3′ (antisense), or 5′-GCACCAGCTCAAGAAGGAACC-3′ (sense) and 5′-GGGACTCAAATTTGGCCCTGG-3′ (antisense), respectively. cDNA from C2C12 cell GAPDH, c-JUN, SERPINE1, BAG1, TGM2, SC1.7, SC1.6, PAX3, or myogenin mRNA was amplified using the primer pair 5′-CCAGGAGCGAGACCCCACTAAC-3′ (sense) and 5′-GGCGGAGATGATGACCCTTTTG-3′ (antisense), 5′- CTGCAAAGATGGAAACGACC-3′ (sense) and 5′-CGGAGGCTCACTGTGCAGGC-3′ (antisense), 5′-TTGCTTGCCTCATCCTGGGC-3′ (sense) and 5′-GTCATTGATCATACCTTTGG-3′ (antisense), 5′-CCCACAGCAATGAGAGGTATGAC-3′ (sense) and 5′-TTGCAGGTGGTTAGCTATCTTCTC-3′ (antisense), 5′- CCGAGTGGGGGACAGTATGAGC-3′ (sense) and 5′-GGCCCCGCACCTTGATGAG-3′ (antisense), 5′-TAAGAAAATGCTGCGGTCTCC-3′ (sense) and 5′-ACAACTGCGCCTTTTTAATAGC-3′ (antisense), 5′-TAAGAAAATGATGAATCGGC-3′ (sense) and 5′-ACAACTGCGCCTTTTTAATAGC-3′ (antisense), 5′-TTCCACAAGCTGTGTCAGATCC-3′ (sense) and 5′-AGTCCCATTACCTGAGGTGAGAG-3′ (antisense) or 5′-CCCATGGTGCCCAGTGAATG-3′ (sense) and 5′-GCGAGCAAATGATCTCCTGGG-3′ (antisense), respectively. cDNA from C2C12 cell PAX3 or myogenin pre-mRNA was amplified using the primer pair 5′-ACTGAACTTTCTCTGCTGGCC-3′ (sense) and 5′-TCGTCTGTCTAGAAACACGGG-3′ (antisense) or 5′-TTGGACCTTGGCCTTGTCTTG-3′ (sense) and 5′-GATTGGGACAGGGCTTATGG-3′ (antisense), respectively. cDNA from RLUC or RLUC-Gl mRNA was amplified using the primer pair 5′-CATGGTAACGCTGCCTCCAG-3′ (sense) and 5′-AGGGTAGGCCGTCTAACCTC-3′ (antisense). RT–PCR products were electrophoresed in 5% polyacrylamide and quantitated by PhosphorImaging (Molecular Dynamics).
IPs
IPs were as described (Kim et al. 2005), where anti-HA, anti-MYC, anti-T7, and anti-Flag derived from Roche, CalBiochem, Novagen, and Sigma, respectively. Anti-UPF1 was generated using protein that derived from pGEX-4T-1-UPF1(1–416) (Lykke-Andersen et al. 2000), which encodes GST-tagged UPF1 amino acids 1–416. Where specified, C2C12 MBs were exposed to 0.25% formaldehyde for 10 min at room temperature, lysed using sonication, and heated after IP for 45 min at 65°C to dissociate cross-linked molecules essentially as described previously (Niranjanakumari et al. 2002).
Western blotting
Protein was electrophoresed in SDS-polyacrylamide, transferred to Hybond ECL nitrocellulose (Amersham) and probed with antibodies that recognize Flag (Sigma), HA (Roche), T7 (Novagen), c-MYC (BD Bioscience), Calnexin (StressGen), Vimentin (Santa Cruz Biotechnologies), UPF1, UPF2 (Serin et al. 2001), UPF3X (Serin et al. 2001), STAU1 (Wickham et al. 1999), myogenin (Santa Cruz Biotechnologies), myoglobin (Abcam), MYF-5 (Santa Cruz Biotechnologies), MUP (Berger and Szoka 1981), BAG1 (Santa Cruz Biotechnologies), c-JUN (Abcam), and GAPDH (Santa Cruz Biotechnologies). Immunoreactivity was assessed using SuperSignal West Pico or Femto (Pierce Biotechnology). After autoradiography, films were quantitated using ImageQuant (Molecular Dynamics). C2C12 cell protein was quantitated using Ponceau S staining (Sigma).
Acknowledgments
We thank Daiki Matsuda, Xenia Schafer, and Joshua Munger for experimental advice; Olaf Isken and Eric Phizicky for comments on the manuscript; Luc DesGroseillers for anti-STAU1, pSTAU1-HA3, and pSTAU1-HA3 variants; Susana de Lucas and Juan Ortín for anti-STAU1; Jens Lykke-Andersen for pGEX-4T-1-UPF1(1–416); Nahum Sonenberg for pMT2-HA-eIF2α wild type and pMT2-HA-eIF2α MUT; and Heinz Baumann and Bill Held for anti-MUP. This work was funded by R01 GM074593 to L.E.M. from the National Institutes of Health. C.F.W. was supported by 1F32 GM085968 from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1717309.
Supplemental material is available at http://www.genesdev.org.
References
- Behm-Ansmant I., Kashima I., Rehwinkel J., Sauliere J., Wittkopp N., Izaurralde E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Belgrader P., Maquat L.E. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol. Cell. Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F.G., Szoka P. Biosynthesis of the major urinary proteins in mouse liver: A biochemical genetic study. Biochem. Genet. 1981;19:1261–1273. doi: 10.1007/BF00484578. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A., Czaplinski K., Trifillis P., He F., Jacobson A., Peltz S.W. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D. An initial blueprint for myogenic differentiation. Genes & Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S.C., Disatnik M.H., Chan L.S., Iori K., Rando T.A. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Chamieh H., Ballut L., Bonneau F., Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chan W.K., Huang L., Gudikote J.P., Chang Y.F., Imam J.S., MacLean J.A., II, Wilkinson M.F. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Muhlrad D., Lim M.K., Parker R., Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.Y., Serin G., Ohara O., Maquat L.E. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.Y., Lejeune F., Ranganathan A.C., Maquat L.E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes & Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato M.D., Tjian R. Switching of the core transcription machinery during myogenesis. Genes & Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J.A., Lam P., Jepeal L., Maas R.L., Shapiro D.N. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J. Biol. Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- Figueroa A., Cuadrado A., Fan J., Atasoy U., Muscat G.E., Muñoz-Canoves P., Gorospe M., Muñoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell. Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Kunz J.B., Neu-Yilik G., Breit S., Viegas M.H., Hentze M.W., Kulozik A.E. Exon–junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Hosoda N., Kim Y.K., Lejeune F., Maquat L.E. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O., Kim Y.K., Hosoda N., Mayeur G.L., Hershey J.W., Maquat L.E. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J., Guilligay D., Ravelli R.B., Cusack S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA. 2006;12:1817–1824. doi: 10.1261/rna.177606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Maquat L.E. Staufen1-mediated mRNA decay: A Upf1-dependent pathway. In: Maquat L.E., editor. Nonsense-mediated mRNA decay. Georgetown, TX: Landes Bioscience; 2006. pp. 229–236. [Google Scholar]
- Kim Y.K., Furic L., Desgroseillers L., Maquat L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L.E. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J.B., Neu-Yilik G., Hentze M.W., Kulozik A.E., Gehring N.H. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Duchaine T.F., DesGroseillers L. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem. J. 2002;365:817–824. doi: 10.1042/bj20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.D., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Marion R.M., Fortes P., Beloso A., Dotti C., Ortín J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D., Hosoda N., Kim Y.K., Maquat L.E. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat. Struct. Mol. Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- Melo F., Carey D.J., Brandan E. Extracellular matrix is required for skeletal muscle differentiation but not myogenin expression. J. Cell. Biochem. 1996;62:227–239. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C227::AID-JCB11%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi S.M., Lake R.G., Noensie E.N., Dietz H.C. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys C.M., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Monshausen M., Putz U., Rehbein M., Schweizer M., DesGroseillers L., Kuhl D., Richter D., Kindler S. Two rat brain staufen isoforms differentially bind RNA. J. Neurochem. 2001;76:155–165. doi: 10.1046/j.1471-4159.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- Mühlemann O. Recognition of nonsense mRNA: Towards a unified model. Biochem. Soc. Trans. 2008;36:497–501. doi: 10.1042/BST0360497. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Osses N., Brandan E. ECM is required for skeletal muscle differentiation independently of muscle regulatory factor expression. Am. J. Physiol. Cell Physiol. 2002;282:C383–C394. doi: 10.1152/ajpcell.00322.2001. [DOI] [PubMed] [Google Scholar]
- Pal M., Ishigaki Y., Nagy E., Maquat L.E. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A.L., Kim Y.K., Pan Q., Fagnani M.M., Maquat L.E., Blencowe B.J. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G., Gersappe A., Black J.D., Aronoff R., Maquat L.E. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol. Cell. Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B., Wilkinson M.F., van Hoof A. Messenger RNA regulation: To translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Jakob S., Kleedehn M.G., Lykke-Andersen J. Communication with the exon–junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol. Cell. 2007;27:780–792. doi: 10.1016/j.molcel.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Viegas M.H., Gehring N.H., Breit S., Hentze M.W., Kulozik A.E. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense-mediated decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Vock V.M., Li S., Olivas O.R., Wilkinson M.F. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J. Biol. Chem. 2002;277:18489–18493. doi: 10.1074/jbc.M111781200. [DOI] [PubMed] [Google Scholar]
- Wickham L., Duchaine T., Luo M., Nabi I.R., DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J., Hol E.M., Jack H.M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeller C.F., Gaspari M., Isken O., Maquat L.E. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 2008;9:446–451. doi: 10.1038/embor.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W.E., Sassoon D.A., Lin V.K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Oohinata R., Naiki T., Irie K. Stau1 negatively regulates myogenic differentiation in C2C12 cells. Genes Cells. 2008;13:583–592. doi: 10.1111/j.1365-2443.2008.01189.x. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun X., Qian Y., Maquat L.E. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]