Abstract
An integrated microdevice is developed for the analysis of gene expression in single cells. The system captures a single cell, transcribes and amplifies the mRNA, and quantitatively analyzes the products of interest. The key components of the microdevice include integrated nanoliter metering pumps, a 200-nL RT-PCR reactor with a single-cell capture pad, and an affinity capture matrix for the purification and concentration of products that is coupled to a microfabricated capillary electrophoresis separation channel for product analysis. Efficient microchip integration of these processes enables the sensitive and quantitative examination of gene expression variation at the single-cell level. This microdevice is used to measure siRNA knockdown of the GAPDH gene in individual Jurkat cells. Single-cell measurements suggests the presence of 2 distinct populations of cells with moderate (≈50%) or complete (≈0%) silencing. This stochastic variation in gene expression and silencing within single cells is masked by conventional bulk measurements.
Keywords: lab-on-a-chip, microfabrication, RNAi, stochastic gene expression
Single-cell analysis is a powerful approach for understanding changes in gene expression within an isogenic cell population. Traditional gene expression analysis techniques such as microarrays (1) and serial analysis of gene expression (2) are not sensitive enough to analyze changes at the single-cell level and only report on the ensemble average behavior of a large numbers of cells. Recently, a variety of highly sensitive and specialized techniques have been developed for probing gene expression in single cells (3–11). Although many of these approaches offer the advantage of real-time monitoring, the protocols required are laborious, often require cellular engineering, and have limited multiplexing capabilities.
Newly developed microfluidic technologies and methods enable single-cell analysis in a format that can be scaled to large numbers of cells (11–13). Microfluidic devices present a powerful platform for probing single cells because the intrinsic length (1–100 μm) and volume scales (picoliters–nanoliters) are close to the size and volume of single cells (≈1 pL). The biggest advantage microfluidics offers is the ability to integrate all processing steps into a single device, eliminating sample contamination and product loss, which would preclude sensitive, reproducible, and quantitative single-cell analysis.
The 3 steps that must be integrated into a microdevice to perform single-cell gene expression analysis are cell selection and localization, enzymatic reaction, and quantitative detection of the analyte of interest. Although many microfluidic systems have demonstrated 1 or 2 of these elements, the successful integration of all 3 is extremely challenging. Early microfluidic systems successfully coupled PCR chambers to capillary electrophoresis (CE) separation channels (14, 15). Recent integrated microsystems have demonstrated a significant increase in detection sensitivity (16, 17), the handling of crude samples (18), and massive parallelism (12). Despite these advances, no integrated microfluidic device has successfully coupled all 3 steps into a single platform to measure changes in gene expression directly from single cells. The fundamental hurdle has been the efficient transfer of analyte between each nanoliter-processing step.
Results and Discussion
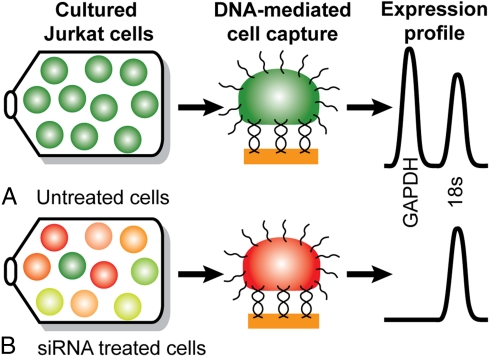
To address this challenge, we have developed an integrated microfluidic device with all of the necessary elements for single-cell gene expression profiling and used it to perform a study of single-cell gene silencing [Fig. 1 and supporting information (SI) Fig. S1]. Cells are functionalized with a 20-base oligonucleotide on their surface to enable capture on a gold pad by DNA hybridization (19, 20). Multiplex gene expression analysis from GAPDH mRNA and control 18S rRNA is performed on 2 cell populations. The first cell population consists of untreated Jurkat T lymphocyte cells grown under normal conditions exhibiting homogenous high expression of both target genes (Fig. 1A). The second cell population is treated with siRNA directed at GAPDH mRNA (Fig. 1B). The degree to which GAPDH mRNA is silenced in individual cells is probed relative to the 18S rRNA control.
Fig. 1.
Overview of single-cell gene silencing assay. Jurkat cells are cultured and surface-labeled, a single cell is captured on a target pad via DNA duplex formation, and an RT-PCR expression profile is generated. (A) Cells under normal growth conditions exhibit homogenous high expression of GAPDH (green cells) compared with a control 18S rRNA. (B) Cells treated with siRNA directed at GAPDH exhibit varying levels of mRNA knockdown.
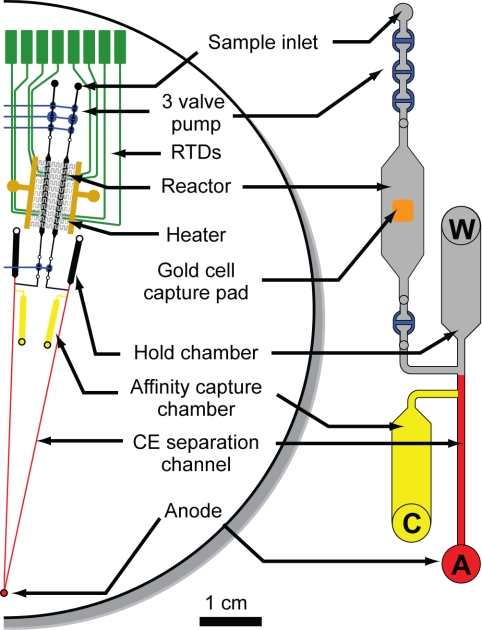
The gene expression microdevice contains 4 independently addressable arrayed analysis systems on a 100-mm-diameter glass wafer (Fig. 2). Each of the identical microsystems contains 4 distinct regions that are integrated to enable maximal transfer efficiency between processing steps. The first region is a 3-valve pump for moving material from the sample inlet through the reactor region (21). In the reactor region, single cells are captured, lysed, and the mRNA of interest is reverse transcribed and amplified by RT-PCR (22). The affinity capture region comprises a hold chamber that acts as a reservoir and a capture chamber where amplicons are immobilized, purified, and concentrated in an affinity capture gel matrix (23). Finally, the system contains a CE separation channel for the size-based separation and quantitation of products.
Fig. 2.
Microfluidic device layout. Schematic showing half of the device (2 of the 4 complete systems) for single-cell gene expression profiling. The 4-layer glass–PDMS–glass–glass microdevice contains 4 distinct regions. The first region at the top is a 3-valve pump. The reactor region consists of a photolithographically defined gold cell-capture pad in the center of a 200-nL reaction chamber along with RTDs and a microfabricated heater for thermal cycling. The affinity capture region comprises a hold chamber and an affinity capture chamber (yellow). Finally, the thermally released amplicons are analyzed on the CE separation channel (red). Each device contains 4 independently addressable systems enabling the analysis of 4 single cells in parallel. All channels are etched to a depth of 20 μm.
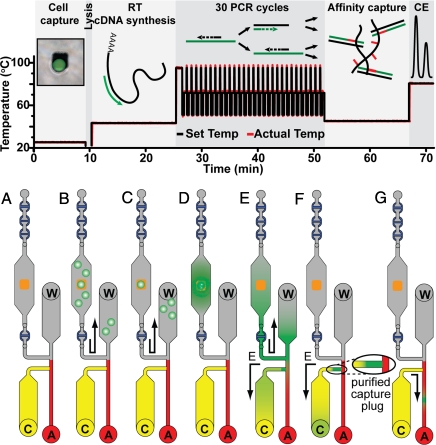
The complete analysis from single-cell capture to CE separation and detection is performed in <75 min as outlined in Fig. 3A. First, Jurkat cells are functionalized with a 20-base oligonucleotide (Fig. 3B). Jurkat cells are grown with a synthetic peracetylated N-azidoacetylmannosamine (Ac4ManNAz) sugar that is metabolized by the cell and results in the presentation of azido groups on the cells' surface (19, 20, 24, 25). Phosphine-modified ssDNA is reacted with the azido group via Staudinger ligation, generating a population of cells functionalized with ≈270,000 ssDNA molecules per cell. Inside the reactor, the complementary 20-base strand of thiol-modified capture DNA is immobilized on a photolithographically defined 25- × 25-μm2 size-limiting gold pad with a gold–thiol linkage. Cells are flowed into the reactor and immobilized on the gold pad via DNA hybridization. The size of the pad ensures that only 1 cell will bind. In previous reports we have shown that the combination of metabolic engineering and DNA-based attachment leads to significantly less cellular activation than antibody or lectin-based methods (19, 20, 24, 25). Thus, although any attachment method would be expected to have some effect on cell behavior, the DNA-based method minimizes these effects.
Fig. 3.
Schematic of the biochemical steps performed in the integrated gene expression microdevice. (Upper) The analysis is complete in <75 min. (Lower) (A) Depiction of the operation of the single-cell gene expression microsystem. (B) First, cells functionalized with a 20-base oligonucleotide on their cell membrane are flowed into the reactor. (C) A single cell is captured on a size-limiting 25- × 25-μm2 gold pad when the ssDNA on its exterior binds to the complementary capture strand immobilized on the gold pad. (D) The immobilized cell is freeze–thaw lysed, and mRNA is reverse-transcribed into a stable cDNA strand (15 min). PCR amplification (30 cycles) is completed in 25 min. (E) Amplified fragments and unreacted RT-PCR mixture are pumped from the reactor into the hold chamber and electrophoretically driven from the waste (W) to the cathode (C) reservoirs. (F) Fragments of interest with complementarity to the affinity capture probe are concentrated and immobilized at the entrance of the capture chamber creating a purified capture plug. (G) Finally, the products are thermally released at 80 °C from the affinity capture gel and electrophoretically separated as they migrate toward the anode (A). Fluorescently labeled amplicons are detected by confocal fluorescence to determine their amount and identity.
After washing the residual uncaptured cells out of the reactor, the captured cell is prepared for analysis (Fig. 3C). After a rapid 30-s freeze–thaw lysis, the target mRNA is reverse-transcribed into a stable cDNA strand during a 15-min incubation at 42 °C. Then, 30 cycles of PCR amplification are completed in the same 200-nL reactor in 25 min. All RT-PCR products are then quantitatively transferred from the reactor to the separation channel for size analysis (23). Amplified fragments and unreacted RT-PCR mixture are pumped from the reactor into the hold chamber and electrophoretically driven from the waste to the cathode reservoir. Fragments of interest with complementarity to the affinity capture probe are quantitatively concentrated and immobilized at the entrance of the capture chamber, creating a purified capture plug. The affinity capture matrix comprises a linear polyacrylamide (LPA) gel copolymerized with two 20-base oligonucleotide capture probes complementary to the fragments of interest (20 μM). This capture process uses sequence-specific helix invasion to immobilize the dsDNA amplicons (23). The capture probes are complementary to sequences 23 and 47 bases from the end of the GAPDH and 18S rRNA amplicons, respectively. By placing the probes internal to the priming sites, the capture gel also acts as a purification matrix to remove unreacted high-molarity FAM-labeled primers. Finally, the purified and concentrated products are thermally released at 80 °C from the affinity capture gel, electrophoretically separated, and quantitated by confocal fluorescence detection (Table S1 and Fig. S2).
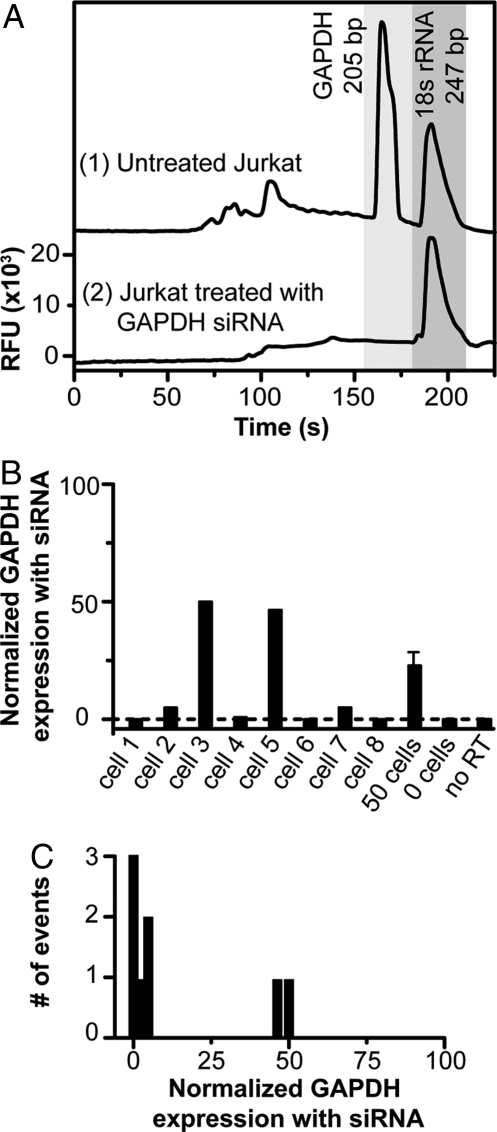
The integrated microfluidic gene expression analysis system yields quantitative data on the gene silencing of individual cells. As expected, the untreated Jurkat cells exhibit normal expression of GAPDH mRNA and 18S rRNA (Fig. 4A). The representative electropherogram shows 2 strong peaks migrating at 160 s and 185 s for the 200-bp GAPDH and the 247-bp 18S rRNA targets, respectively (Figs. S3 and S4). Moreover, the use of the capture matrix to immobilize the fragments of interest removes all unreacted primers from the separation, enabling us to look at both small and large amplicons without interference. A single Jurkat cell electroporated with siRNA directed at GAPDH mRNA produces only a single peak for 18S rRNA at 185 s. Single-cell experiments from 8 individual cells show expression of the GAPDH mRNA at 0, 5, 50, 1, 48, 0, 5, and 0% of untreated Jurkat cells (Fig. 4B and Table S2). This analysis indicates that single cells fall into populations with moderate (≈50%) or complete silencing (≈0%). These single-cell measurements differ fundamentally from a bulk measurement performed on 50 Jurkat cells under the same conditions where the expression of GAPDH is reduced to 21 ± 4% (n = 4) of its original value. Thus, the ensemble average measured for gene silencing masks the stochastic diversity of individual cellular response. A control assay where no cell is captured on the pad exhibits no products, verifying that there is no carryover contamination in the system. Similarly, a PCR control without reverse transcriptase shows no amplification, ensuring that the amplification template is RNA and not DNA.
Fig. 4.
Gene expression and silencing at the single-cell level. (A) Representative gene expression electropherograms from individual Jurkat cells (1). A single wild-type cell with primers targeting GAPDH (200 bp) and 18S rRNA (247 bp) generates 2 strong peaks migrating at 160 s and 185 s, respectively (2). A single cell electroporated with siRNA directed at GAPDH mRNA shows only a single peak for 18S rRNA. (B) Gene expression of GAPDH for Jurkat cells treated with GAPDH siRNA relative to normal untreated cells. GAPDH expression has been normalized to a control 18S rRNA for comparison. Experiments from 8 individual cells show GAPDH mRNA levels at 0, 5, 50, 1, 48, 0, 5, and 0% of normally untreated Jurkat cells. However, a representative bulk measurement from 50 cells shows GAPDH expression at 21 ± 4%. When no cell is captured on the pad there is no amplification. Similarly, a PCR control with no reverse transcriptase shows no amplification. (C) Histogram of the number of events for siRNA treated cells shows that there are 2 distinct populations of cells whose expression levels are very distinct from the population average.
To ensure that the variation in silencing behavior is not a simple function of the amount of siRNA introduced during the electroporation process, cells were treated with a fluorescently labeled siRNA. Cells grown under normal conditions showed an average uptake of the Cy3-labeled siRNA of 17 ± 2 relative fluorescence units (rfu) (Table S1) with 4 of 30 cells appearing at a slightly elevated level of 22 rfu. If electroporation variability were the cause of the 2 cellular populations, we would expect 13% of the cells to be characterized as completely silenced and 87% as moderately silenced. The single-cell gene expression analysis performed in the microfluidic device shows the opposite trend (Fig. 4B). In addition, the GAPDH gene was sequenced to examine whether the population of cells experiencing moderate silencing arises from a heterozygotic polymorphism in the siRNA-binding domain. The sequence was found to be without mutation (Fig. S5). Thus, the 2 populations of cells revealed here are not a trivial result of siRNA delivery, genetic variation, or cell viability.
The occurrence of 2 distinct cellular populations with different levels of gene silencing has been detected (26). Measurements of enhanced green fluorescent protein production in human embryonic stem cells (hESCs) have shown an all-or-nothing response to siRNA treatment, but the underlying mechanism was not characterized. Our system demonstrates the unique ability to perform quantitative transcript analysis, revealing that although 80% of the cells exhibited the expected complete inhibition, 20% exhibited 50% inhibition. This suggests the presence of a genetic or phenotypic bistability or switch that controls the degradation of the siRNA, blocks its target binding, or inhibits transcript degradation. Of these only the latter 2 mechanisms would provide a nontrivial explanation for the 50% inhibition level. A more exhaustive study is now called for to verify the biphasic expression trend observed here and to explore its mechanistic origin. Because the fabrication of highly parallel structures is one of the key advantages of our approach, scaling up to 96-analyzers would provide the throughput necessary for this type of study (27).
Our ability to perform 1-step RT-PCR amplification in our expression microdevice with only 30 cycles of PCR in a single reactor is a direct result of efficient integration. First, the use of glass rather than porous polymeric materials prevents product absorption. Second, the high thermal conductivity of glass enables rapid thermal cycling and increased reaction efficiency. Third, the use of pneumatic valves and pumps allows for the efficient transfer of nanoliter bolus material. Finally, the affinity capture, purification, and concentration process enables the quantitative analysis of all generated products, a dramatic improvement over the use of a traditional cross-injector (28) or hydrodynamic pressure injector (18), which only permits a small portion (<1%) of the products to be analyzed. These advantageous attributes of our integrated device point the way to a wide variety of bioanalytical studies on the properties and behavior of single cells.
Here, we have performed single-cell measurements on the variation of mRNA knockdown as a result of siRNA treatment. This assay suggests a unique biphasic gene knockdown efficiency in individual cells that was masked by bulk measurements. Because the analysis step utilizes a size-based separation, the multiplex capabilities are determined by the number of products that can be generated and analyzed, suggesting that the expression of 5–10 targets could be studied in parallel. By coupling this microdevice with laser capture microdissection, the heterogeneous nature of tumors could be investigated at the single-cell level (29). It should also be possible to perform a quantitative single-cell analysis of the effects of siRNA treatment on expression in hESCs when Oct4 mRNA targeting is used to trigger differentiation into trophoblast-like cells (30, 31). Moreover, based on our previous detection of <11 mRNA molecules per reactor (22), our microfluidic device may ultimately enable studies of expression from individual cells at the single-transcript level, once improved product capture, purification, and injection processes are fully enabled and integrated. Overall, our approach offers many exciting prospects for revealing the stochastic variation in gene expression that underlies the ensemble average.
Materials and Methods
Additional procedures are detailed in SI Text.
Bioprocessor Fabrication.
The fabrication protocol is similar to that used in previous nucleic acid amplification microdevices (22). Briefly, to form the pneumatic manifold wafer, valve seats and actuation channels were photolithographically defined and etched to a depth of 38 μm on a 0.5-mm-thick borofloat 100-mm glass wafer. Valve actuation access holes were drilled, and the manifold was diced into reusable 9-mm × 6-cm strips. Removable polydimethylsiloxane (PDMS) elastomer valves were formed by activating both sides of the 254-μm PDMS membrane with a UV ozone cleaner for 1.5 min to improve PDMS-glass bonding and then sandwiching the membrane between the manifold and the bonded channel wafers (21).
The reactor/channel wafer was fabricated on a 0.5-mm-thick borofloat glass wafer. Fluidic channels for pumping were photolithographically defined on the front side and etched to a depth of 38 μm. Reaction, hold, and capture chambers along with separation channels were photolithographically defined on the back side and etched to a depth of 20 μm. Electrophoresis reservoirs, resistance temperature detection (RTD) access holes, and valve via holes were diamond drilled. To form the RTD wafer, a 0.5-mm-thick borofloat glass wafer sputter deposited with 200 Å of Ti and 2,000 Å of Pt (Ti/Pt; UHV Sputtering) was photolithographically patterned and etched with 90 °C aqua regia to form the 30-μm-wide RTD elements and 300-μm-wide leads. The drilled reactor/channel wafer was aligned and thermally bonded to the RTD wafer by using a programmable vacuum furnace at 655 °C for 6 h.
To form the removable modular heater, a 0.5-mm-thick borofloat glass wafer was sputter-deposited with 2,200 Å of Ti/Pt. Heater leads were formed by electroplating 6 μm of gold onto photolithographically defined areas. Ti/Pt serpentine resistive heater elements connecting the gold leads were formed by anisotropically etching photolithographically exposed Ti/Pt in an ion mill.
Jurkat Cell Preparation.
T lymphocyte Jurkat cells were cultured in 50-mL flasks (Nalge–Nunc International) for 48 h in 10 mL of medium (RPMI medium 1640; Invitrogen) containing 1% penicillin/streptomycin (1% P/S; Invitrogen) and 25 μM Ac4ManNAz resulting in the display of N-azidoacetylsialic acids on the cell surface glycans (24). For the first 24 h of growth, the cells were cultured with 10% FBS (JR Scientific). The Jurkat cells were washed and incubated in serum-depleted medium containing 25 μM Ac4ManNAz for an additional 24 h for synchronization. Fresh DNA-functionalized Jurkat cells were prepared 1 h before the analysis. Cells were washed twice with 5 mL of PBS (Ambion) containing 1% FBS and reacted with 125 μM phosphine-modified ssDNA (5′-phos-GTA ACG ATC CAG CTG TCA CT-3′) in 1% FBS/PBS for 1 h at 37 °C (20). The cells were then rinsed 3 times with 5 mL of 1% FBS/PBS solution before introduction into the microfluidic device.
siRNA Treatment.
For gene-silencing studies, 150,000 Jurkat cells were electroporated with 2.5 μg of double-stranded GAPDH siRNA (sense, 5′-GGU CAU CCA UGA CAA CUU UdTdT-3′; Ambion). Cells were suspended in 75 μL of siPORT electroporation buffer (AM1629; Ambion) and a single pulse is performed in a 1-mm cuvette (Bio-Rad) for 250 μs at 250 V. Cells were then grown and prepared in the same manner as described in the Jurkat cell preparation section above. For negative control studies, cells were electroporated with 150 pmol of Cy3-labeled siRNA that does not bind to mRNA.
RT-PCR Mixture.
Multiplex RNA RT-PCR was performed on GAPDH and 18S rRNA transcripts directly from Jurkat cells. A 25-μL RT reaction mixture comprises a Cell-to-cDNA II kit [4 units of Moloney murine leukemia virus (Mo-MLV) reverse transcriptase, 0.4 unit of RNase inhibitor, 0.1 μM dNTPs, 1× RT buffer (Ambion)], 0.08 unit of platinum Taq polymerase (Invitrogen), along with 800 nM forward and reverse primers for the GAPDH gene and 20 nM forward and reverse primer for the 18S rRNA target. The GAPDH forward (5′-AGG GCT GCT TTT AAC TCT GG-3′) and reverse (5′-FAM-TTG ATT TTG GAG GGA TCT CG-3′) primers generate a 200-bp amplicon. The 18S rRNA forward (5′-CGG CTA CCA CAT CCA AGG AAG-3) and reverse (5′-FAM-CGC TCC CAA GAT CCA ACT AC-3′) primers generate a 247-bp amplicon. Controls without RT and without template were performed on the microdevice by removing the Mo-MLV RT and Jurkat cells from the reaction mixture, respectively.
Matrix Synthesis.
A DNA affinity capture gel is synthesized by copolymerizing LPA with 2 5′-acrydite-modified capture oligonucleotides. The affinity capture matrix is synthesized at 4 °C by sparging a 2-mL solution containing 6% wt/vol acrylamide, 1× TTE, and 40 nmol of the 2 acrydite-modified oligonucleotides (IDT) for 2 h with argon followed by the addition of 0.015% wt/vol ammonium persulfate (APS; Fisher Scientific) and tetramethylethylenediamine (TEMED; Fisher Scientific). The affinity capture matrix contains capture probes for GAPDH (5′-Acry-ATC CCA TCA CCA TCT TCC AG-3′, TM = 54.2; 50 mM monovalent salt, 20 μM) and 18S rRNA (5′-Acry-GCA GCC GCG GTA ATT CCA GC-3′, TM = 61.9; 50 mM monovalent salt, 20 μM). The GAPDH capture oligonucleotide is complementary to a 20-base sequence in the 200-bp amplicon, 23 bases from the 5′ FAM-labeled terminus. The 18S rRNA capture oligonucleotide is complementary to a 20-base sequence in the 247-bp amplicon, 60 bases from the 5′ FAM-labeled terminus.
Reactor Preparation.
The glass surface is derivatized with polydimethylacrylamide (PDMA) by using a modified Hjerten coating protocol to prevent nonspecific cell adhesion (32). First, the reactors glass surface is deprotonated by incubating with 1 M NaOH for 1 h. The NaOH solution is replaced with a 0.6% (vol/vol) (γ-methacryloxypropyl)trimethoxysilane solution (γ, Sigma) in 3.5 pH H2O. The bifunctional γ-solution prepares the glass surface for acrylamide polymer nucleation. During γ-solution incubation, 250 μL of dimethylacrylamide is dissolved in 4.75 mL of H2O and sparged with Ar for 1 h. After Ar sparging, 100 μL of isopropyl alcohol (IPA), 20 μL of TEMED, and 25 μL of 10% (vol/vol) APS were sequentially added to the acrylamide solution to form linear PDMA. The γ-solution is removed from the channel, and PDMA solution incubates in the channel for 1 h. The channel is then rinsed and dried with acetonitrile.
Next, the photolithographically defined 25-μm × 25-μm gold pad in the center of the reaction chamber is functionalized with ssDNA by incubating for 1 h with Tris(2-carboxyethyl)phosphine (TCEP, 200 μM; Invitrogen) deprotected thiol-DNA (5′-thiol-AGT GAC AGC TGG ATC GTT AC-3′, 20 μM). The chamber is then rinsed and dried to remove unbound DNA.
Gel Loading Sequence.
The device is prepared for affinity capture and separation by treating the separation channels, hold chambers, and capture chambers with a dynamic coating diluted in methanol for 1 min (1:1; DEH-100; The Gel Company) to suppress electroosmotic flow. Multiplex affinity capture matrix (20 μM, 6%, yellow) is loaded from each cathode (C) reservoir up to the separation channel cross at room temperature. Separation matrix (red) is then loaded from the central anode (A) past the capture chamber to the sample load cross. With all of the valves opened, the rest of the system is hydrated by adding 3 μL of RNase-free water at the sample ports and applying a vacuum at the waste (W) reservoirs. The microdevice is then placed on a 44 °C temperature-controlled stage. An additional 2 μL of water is flushed through the system to remove thermally expanded gel from the sample load cross.
Bioprocessor Operation.
As depicted in Fig. 3, the microdevice operation begins by preparing the reaction chamber for cell capture. Cell modified to contain ssDNA on their surface were suspended in the reaction mixture and drawn into the reaction by vacuum. When a cell flows over the gold cell capture pad, DNA hybridization occurs between the ssDNA on the surface of the cell and the complementary ssDNA immobilized on the gold pad. To maximize capture efficiency, DNA-mediated cell capture was performed for 15 min. The uncaptured cells were washed out of the system and removed at the waste port, and the valves were closed for thermal cycling. During this study, all 4 reactors were used in parallel. This enabled simultaneous analysis from 4 individual cells. After capture, the morphology of each cell is recorded to ensure that it has remained viable. The total data collection time for the single-cell studies was 1 month.
Freeze–thaw lysis and 1-step RT-PCR thermal cycling were performed in a single 200-nL reactor. A piece of dry ice was placed over the reaction chambers of all 4 reactors for 30 s to freeze–thaw lyse the captured Jurkat cell. Freeze–thaw lysis was used in this 1-step RT-PCR to prevent early unwanted activation of the Hot Start Taq polymerase, to prevent denaturation of the reverse transcriptase enzyme, and to minimize RNA degradation by RNases (33, 34). Next, a linear 15-min cDNA synthesis from the cells' RNA was performed at 42 °C by using primers complementary to the RNA transcripts of interest (GAPDH and 18S rRNA). After cDNA synthesis, the Mo-MLV RT was denatured, and the platinum Taq polymerase was activated at 95 °C for 60 s followed by 30 cycles of PCR at 95 °C for 5 s, 47 °C for 20 s, and 72 °C for 25 s. Because of the rapid heating and cooling rates (>15 °C s−1), each cycle of PCR is completed in 50 s, and the total reaction time is 46 min.
After thermal cycling, affinity capture, purification and concentration of the products of interest were performed. The reactor contents were pumped into the hold chamber by using a 5-step pump cycle. A 350-ms actuation was used with each step, resulting in a 30-nL stroke volume. A 23-s delay was used between each pump cycle to allow sufficient time for the analyte to migrate into the capture region and to prevent analyte accumulation in the hold chamber. A constant 100-V/cm field between the waste (W) and cathode (C) reservoirs electrophoretically drives the analyte toward the capture chamber. Analytes complementary to the capture probe were hybridized at the entrance of the capture chamber, creating a sample plug. The electric field between the waste and cathode was maintained until residual PCR reactants (excess primer, salts, and buffer) were washed into the cathode reservoir thus resulting in a purified amplicon sample plug. Thirty pump cycles were used resulting in a total capture and wash time of 12.2 min. After the capture process was completed, the temperature of the entire device was raised to 80 °C to thermally release the captured DNA fragments thermally from the affinity capture gel, and the sample was separated with a field of 150 V/cm between the cathode and the anode. The electrophoretically separated FAM-labeled products from all 4 lanes were detected by using laser-induced fluorescence with the Berkeley rotary confocal scanner (27). The entire capture and release process was performed on a temperature-controlled stage on the scanner to prevent thermal gradients.
Supplementary Material
Acknowledgments.
We thank Eric Chu for assistance with microfabrication and James Yang, Palani Kumaresean, Terry Johnson, Teris Liu, and Robert Blazej for valuable discussions. This work was supported by National Institutes of Health Grant HG003329 and by the Physical Biosciences, Materials, and Chemical Sciences Divisions of the U.S. Department of Energy under Contract DE-AC02-05CH11231. N.M.T. was supported by National Institutes of Health Molecular Biophysics Training Grant T32GM08295. E.S.D. was supported by a National Science Foundation fellowship.
Footnotes
Conflict of interest statement: R.A.M. has a financial interest in Microchip Biotechnologies, Inc., which is commercially developing aspects of the technologies presented here.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806355106/DCSupplemental.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 3.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 4.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 5.Shav-Tal Y, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capodieci P, et al. Gene expression profiling in single cells within tissue. Nat Methods. 2005;2:663–665. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 8.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Xiao J, Ren XJ, Lao KQ, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 11.Huang B, et al. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
- 12.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 13.Kumaresan P, Yang CJ, Cronier SA, Blazej RG, Mathies RA. High-throughput single-copy DNA amplification and cell analysis in engineered nanoliter emulsions. Anal Chem. 2008;80:3522–3529. doi: 10.1021/ac800327d. [DOI] [PubMed] [Google Scholar]
- 14.Burns MA, et al. An integrated nanoliter DNA analysis device. Science. 1998;282:484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 15.Lagally ET, Simpson PC, Mathies RA. Monolithic integrated microfluidic DNA amplification and capillary electrophoresis analysis system. Sens Actuator B-Chem. 2000;63:138–146. [Google Scholar]
- 16.Blazej RG, Kumaresan P, Mathies RA. Microfabricated bioprocessor for integrated nanoliter-scale Sanger DNA sequencing. Proc Natl Acad Sci USA. 2006;103:7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blazej RG, Kumaresan P, Cronier SA, Mathies RA. Inline-injection microdevice for attomole-scale Sanger DNA sequencing. Anal Chem. 2007;79:4499–4506. doi: 10.1021/ac070126f. [DOI] [PubMed] [Google Scholar]
- 18.Easley CJ, et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci USA. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB. Programmable cell adhesion encoded by DNA hybridization. Angew Chem Int Edit. 2006;45:896–901. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 20.Douglas ES, Chandra RA, Bertozzi CR, Mathies RA, Francis MB. Patterning mammalian cells using DNA barcodes. Lab Chip. 2007;7:1442–1448. doi: 10.1039/b708666k. [DOI] [PubMed] [Google Scholar]
- 21.Grover WH, Skelley AM, Liu CN, Lagally ET, Mathies RA. Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices. Sens Actuator B-Chem. 2003;89:315–323. [Google Scholar]
- 22.Toriello NM, Liu CN, Mathies RA. Multichannel reverse transcription–polymerase chain reaction microdevice for rapid gene expression and biomarker analysis. Anal Chem. 2006;78:7997–8003. doi: 10.1021/ac061058k. [DOI] [PubMed] [Google Scholar]
- 23.Toriello NM, Liu CN, Thaitrong N, Blazej RG, Mathies RA. Integrated affinity capture, purification and capillary electrophoresis microdevice for quantitative double-stranded DNA analysis. Anal Chem. 2007;79:8549–8556. doi: 10.1021/ac0712547. [DOI] [PubMed] [Google Scholar]
- 24.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao SC, et al. DNA-coated AFM cantilevers for the investigation of cell adhesion and the patterning of live cells. Angew Chem Int Edit. 2008;47:8473–8477. doi: 10.1002/anie.200802525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu YP, Dambaeva SV, Dovzhenko OV, Garthwaite MA, Golos TG. Stable plasmid-based siRNA silencing of gene expression in human embryonic stem cells. Stem Cells Dev. 2005;14:487–492. doi: 10.1089/scd.2005.14.487. [DOI] [PubMed] [Google Scholar]
- 27.Shi YN, et al. Radial capillary array electrophoresis microplate and scanner for high-performance nucleic acid analysis. Anal Chem. 1999;71:5354–5361. doi: 10.1021/ac990518p. [DOI] [PubMed] [Google Scholar]
- 28.Harrison DJ, et al. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science. 1993;261:895–897. doi: 10.1126/science.261.5123.895. [DOI] [PubMed] [Google Scholar]
- 29.Emmert-Buck MR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 30.Martin MM, et al. Specific knockdown of Oc4 and β2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–668. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- 31.Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- 32.Hjerten S. High-performance electrophoresis: Elimination of electroendosmosis and solute adsorption. J Chromatogr. 1985;347:191–198. [Google Scholar]
- 33.Zabzdyr JL, Lillard SJ. Measurement of single-cell gene expression using capillary electrophoresis. Anal Chem. 2001;73:5771–5775. doi: 10.1021/ac0155714. [DOI] [PubMed] [Google Scholar]
- 34.Zabzdyr JL, Lillard SJ. A qualitative look at multiplex gene expression of single cells using capillary electrophoresis. Electrophoresis. 2005;26:137–145. doi: 10.1002/elps.200406126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.