Abstract
OBJECTIVE—The present study was conducted to confirm possible associations between candidate genes from genome-wide association studies and type 2 diabetes in Japanese diabetic patients and a community-based general population. A total of 11 previously reported single-nucleotide polymorphisms (SNPs) from the TCF7L2, CDKAL1, HHEX, IGF2BP2, CDKN2A/B, SLC30A8, and KCNJ11 genes were analyzed.
RESEARCH DESIGN AND METHODS—Candidate SNPs were genotyped in 506 type 2 diabetic patients and 402 control subjects and meta-analyzed with six previous association studies in Japanese patients. Associations with fasting plasma insulin levels were investigated in a general population sample (n = 1,963, 61 ± 13 years).
RESULTS—In our case-control subjects, susceptibility to type 2 diabetes was replicated in TCF7L2 (rs12255372), CDKAL1 (rs7756992, rs7754840), HHEX (rs7923837), IGF2BP2 (rs4402960 and rs1470579), CDKN2A/B (rs10811661), and SLC30A8 (rs13266634). In addition to these polymorphisms, meta-analysis confirmed the association of type 2 diabetes susceptibility with KCNJ11 rs5219, TCF7L2 rs7903146, and HHEX rs1111875. The TCF7L2 rs12255372 polymorphism showed the highest odds ratio (OR) for type 2 diabetes (OR 1.714 [1.298–2.263]). Odds ratio of other polymorphisms ranged from 1.13 to 1.41. The risk allele of CDKAL1 rs7756992 was significantly associated with lower insulin levels in type 2 diabetic patients after adjustment for other confounding factors.
CONCLUSIONS—Type 2 diabetes susceptibility of seven candidate genes was confirmed in Japanese. Conservation of susceptible loci for type 2 diabetes was independent of ethnic background.
Agreat number of studies in various populations have suggested an association between several single-nucleotide polymorphisms (SNPs) and type 2 diabetes. For example, transcription factor 7-like 2 (TCF7L2) is a highly reliable predisposing gene for type 2 diabetes (1–3). In addition, recent genome-wide association studies (GWASs) have provided new susceptible loci for type 2 diabetes (4–10). A GWAS in French subjects, for example, identified rs13266634, a nonsynonymous SNP (R325W) on the solute carrier family 30 member 8 (SLC30A8) gene, as a polymorphism involved in type 2 diabetes susceptibility (4). The study also reported an association between type 2 diabetes and rs1111875, as well as rs7923837, located in the hematopoietically expressed homeobox gene (HHEX). These associations were replicated in three independent GWASs in various populations (5–7).
Additional susceptible SNPs were independently identified in the insulin-like growth factor 2 mRNA-binding protein 2 gene (IGF2BP2, rs4402960, and rs1470569) (5,6). Involvement of SNPs rs10811661, located upstream of cyclin-dependent kinase inhibitor genes CDKN2A and CDKN2B, and “rs7754840/rs7756992,” located in the CDK5 regulatory subunit-associated protein 1-like 1 gene (CDKAL1), has also been suggested (5,6,8,9). A recent population-based study in Danish subjects replicated the susceptible association of HHEX rs111875, CDKN2A/B rs10811661, and IGF2BP2 rs4402960 with type 2 diabetes (10).
Findings from previous GWASs, however, cannot be extrapolated to other populations with different lifestyles and environmental backgrounds. In particular, the genetic background for type 2 diabetes development in East Asians, who show lower basal insulin secretion and a marked decrease in insulin release in response to development of glucose tolerance (11), appears to be different from that in Caucasians or individuals of European origin. Further, SNP frequency differences are suggested to be an additional factor influencing type 2 diabetes susceptibility.
Here, based on a recent GWAS (4–10), we conducted a replication study of candidate SNPs associated with type 2 diabetes in Japanese diabetic subjects, as well as in a general Japanese population sample.
RESEARCH DESIGN AND METHODS
Case and control subjects.
Basic clinical characteristics of subjects are summarized in Supplemental Table 1 (located in an online appendix at http://dx.doi.org/10.2337/db07-1785). All type 2 diabetic subjects (n = 506) were inpatients or outpatients evaluated by diabetes specialists at Ehime University Hospital and Ehime Prefectural Hospital in Japan. Diabetes was diagnosed based on the 1998 American Diabetes Association criteria (12). Nondiabetic control subjects (n = 402) were chosen based on the absence of a history of diabetes in the subject and among first-degree relatives, as well as either normal glucose tolerance, confirmed by a 75-g oral glucose tolerance test, or A1C levels <5.6 with fasting plasma glucose levels <110 mg/dl. All case and control subjects were native Japanese. Selection criteria details have been described in a previous study (13). A total of 139 type 2 diabetic patients and 136 control subjects were overlapped with the previous meta-analysis for TCF7L2 polymorphisms (14).
General population.
The general population subjects were selected from residents of a community of 11,000 inhabitants in Ehime Prefecture, a largely rural area located in western Japan (15). Subjects were recruited through a community-based annual medical checkup process for self-employees, including farmers and foresters, employees of small companies, and elderly without fixed employment. The sample population consisted of 1,963 middle-aged to elderly residents (Supplemental Table 2). Overnight fasting plasma samples for the measurement of plasma insulin concentrations were available for all sample subjects. Baseline clinical characteristics were obtained from personal health records evaluated during the medical checkup. All study procedures were approved by the ethics committee of the Ehime University Graduate School of Medicine, and informed consent was obtained from each participating subject.
Genotyping.
Genomic DNA was extracted from peripheral blood (QIAamp DNA blood kit; QIAGEN, Hilden, Germany). All SNPs were analyzed by TaqMan probe assay (Applied Biosystems, Foster City, CA) using commercially available primers and probes purchased from the Assay-on-Demand system (Supplemental Table 3).
Statistical analysis.
Data are expressed as means ± SD. Linkage disequilibrium was assessed using the Haploview software (Broad Institute, Cambridge, MA) (16). Frequency differences in each genotype were assessed by the χ2 test. The pooled odds ratios for allele frequency with those of six other association studies in Japanese (17–22) were estimated using the fixed-effects model (Mantel-Haenszel method). Differences in plasma insulin levels among genotypes (ANOVA and multiple regression analysis [additive model] adjusted for age, sex, and BMI) were assessed using a commercially available statistical software package (SPSS Version 14.0; SPSS, Chicago, IL). Current treatment of hyperglycemia was further adjusted in type 2 diabetic patients when appropriate. Null hypotheses were rejected at a level of significance of P < 0.05.
RESULTS
Table 1 summarizes the association between 11 candidate SNPs and type 2 diabetes in case-control subjects. The T allele of TCF7L2 (rs12255372) was significantly associated with type 2 diabetes. A tendency to association was also observed with SNP rs7903146, which was in linkage disequilibrium with rs12255372 (D′ = 0.854, r2 = 0.421). However, the risk allele frequency of these SNPs was considerably low, which is in agreement with previous reports in Japanese subjects (17,18). The post hoc calculated statistical power of these SNPs (allele frequency) was 36.1% and 25.4% for rs12255372 and rs7903146, respectively, with a 5% type 1 error rate.
TABLE 1.
Association of candidate SNPs with type 2 diabetes in case and control subjects
| Gene (rs number) | Type 2 diabetes frequency | Risk allele | Hardy-Weinberg equilibrium (P) (control) | Allele
|
Dominant
|
Recessive
|
Additive (P) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||||
| Control | ||||||||||
| TCF7L2 (rs12255372) | TT/TG/GG | T | 0.721 | |||||||
| 1/33/453 | 2.082 (1.112–3.898) | 0.022 | 2.059 (1.089–3.893) | 0.026 | – | – | 0.065 | |||
| 0/14/384 | ||||||||||
| TCF7L2 (rs7903146) | TT/TC/CC | T | 0.501 | |||||||
| 2/45/434 | 1.589 (0.978–2.582) | 0.061 | 1.549 (0.941–2.551) | 0.085 | – | – | 0.132 | |||
| 0/26/372 | ||||||||||
| CDKAL1 (rs7756992) | GG/GA/AA | G | 0.053 | |||||||
| 155/217/119 | 1.307 (1.084–1.577) | 0.005 | 1.081 (0.796–1.467) | 0.618 | 1.887 (1.381–2.578) | 6.8 × 10−5 | 2.0 × 10−4 | |||
| 78/217/102 | ||||||||||
| CDKAL1 (rs7754840) | CC/CG/GG | C | 0.189 | |||||||
| 117/225/149 | 1.321 (1.093–1.596) | 0.004 | 1.209 (0.912–1.604) | 0.187 | 1.866 (1.316–2.645) | 4.6 × 10−4 | 0.002 | |||
| 57/203/137 | ||||||||||
| HHEX (rs1111875) | CC/CT/TT | C | 0.593 | |||||||
| 44/211/235 | 1.086 (0.885–1.334) | 0.430 | 1.141 (0.876–1.488) | 0.328 | 1.018 (0.639–1.620) | 0.942 | 0.602 | |||
| 35/158/203 | ||||||||||
| HHEX (rs79203837) | GG/GA/AA | G | 0.381 | |||||||
| 17/178/295 | 1.286 (1.015–1.630) | 0.037 | 1.432 (1.085–1.891) | 0.011 | 0.920 (0.454–1.866) | 0.817 | 0.026 | |||
| 15/111/273 | ||||||||||
| IGF2BP2 (rs4402960) | TT/TG/GG | T | 0.972 | |||||||
| 66/196/231 | 1.239 (1.011–1.517) | 0.039 | 1.175 (0.902–1.530) | 0.232 | 1.714 (1.103–2.663) | 0.016 | 0.050 | |||
| 33/163/203 | ||||||||||
| IGF2BP2 (rs1470579) | CC/CA/AA | C | 0.940 | |||||||
| 77/198/216 | 1.334 (1.091–1.630) | 0.005 | 1.260 (0.967–1.643) | 0.087 | 1.929 (1.263–2.947) | 0.002 | 0.007 | |||
| 35/165/198 | ||||||||||
| CDKN2A/B (rs10811661) | TT/TC/CC | T | 0.394 | |||||||
| 189/222/85 | 1.227 (1.016–1.482) | 0.034 | 1.116 (0.792–1.572) | 0.531 | 1.454 (1.098–1.925) | 0.009 | 0.031 | |||
| 119/206/75 | ||||||||||
| SLC30A8 (rs13266634) | CC/CT/TT | C | 0.395 | |||||||
| 162/259/72 | 1.103 (0.913–1.332) | 0.311 | 1.439 (1.103–2.044) | 0.042 | 0.983 (0.742–1.300) | 0.902 | 0.090 | |||
| 133/188/79 | ||||||||||
| KCNJ11 (rs5219) | TT/TC/CC | T | 0.302 | |||||||
| 83/232/169 | 1.181 (0.974–1.432) | 0.090 | 1.156 (0.878–1.523) | 0.301 | 1.436 (0.983–2.099) | 0.061 | 0.154 | |||
| 50/195/152 | ||||||||||
n = 908. Type 2 diabetes is defined by fasting blood glucose ≥126 mg/dl, or occasional blood glucose ≥200 mg/dl and/or current use of antidiabetic agents. Differences in genotype frequency between diabetic patients and normal control subjects, as well as deviations from the Hardy-Weinberg equilibrium in control subjects, were assessed using the χ2 test.
In addition to TCF7L2 polymorphisms, a significant association was observed between type 2 diabetes and polymorphisms in CDKAL1 (rs7756992 [power: 51.0%], rs7754840 [52.9%]; D′ = 0.920; r2 = 0.648), HHEX (rs7923837 [30.4%]), IGF2BP2 (rs4402960 [31.3%], rs1470579 [51.1%]; D′ = 0.997; r2 = 0.918), CDKN2A/B (rs10811661 [32.6%]), and SLC30A8 (rs13266634 [10.7%]), but not HHEX (rs1111875 [8.5%]). Further, a marginally significant association was observed between type 2 diabetes and the KCNJ11 polymorphism (rs5219 [21.8%])). Compared with control subjects of European descent, risk allele frequencies in Japanese control subjects were higher in the CDKAL1 gene (rs7756992 G allele, 0.470 vs. 0.258) and lower in the HHEX (rs1111875 C allele, 0.288 vs. 0.598; rs7923837 G allele, 0.177 vs. 0.597), CDKN2A/B (rs10811661 T allele, 0.555 vs. 0.850), SLC30A8 (rs13266634 C allele, 0.568 vs. 0.699), and KCNJ11 (rs5219 T allele, 0.372 vs. 0.464) genes (4,6,8). In contrast, no significant frequency differences were observed in the CDKAL1 rs7754840 (C allele, 0.399 vs. 0.360) and IGF2BP2 rs4402960 (T allele, 0.287 vs. 0.304) polymorphisms.
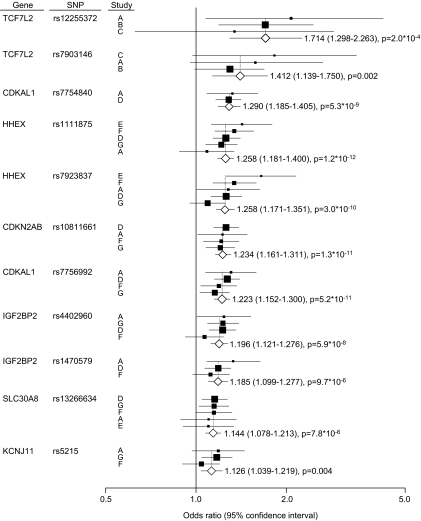
However, except for the CDKAL polymorphisms, statistical significance was not reached in the observed associations using Bonferroni's correction, possibly due to limited statistical power. To further clarify type 2 diabetes susceptibility, seven association studies in Japanese subjects (17–22), including our present data, were meta-analyzed (Fig. 1). Type 2 diabetes susceptibility was confirmed in all analyzed polymorphisms, both before and after Bonferroni's adjustment. Further, two SNPs (rs1111875 in HHEX and rs5219 in KCNJ11), which were not replicated in our data, were confirmed as susceptible polymorphisms for type 2 diabetes.
FIG. 1.
Meta-analysis of type 2 diabetes genetic association studies in Japanese. Estimation of odds ratios and 95% CIs in each study are displayed as a closed square and horizontal line, respectively. Square size represents the study weighting. Combined odds ratio is represented as the diamond. Study A, present study; Study B, Hayashi et al. (18); Study C, Horikoshi et al. (17); Study D, Horikawa et al. (19); Study E, Furukawa et al. (20); Study F, Horikoshi et al. (21); Study G, Omori et al. (22).
To further clarify the pathophysiological significance of the susceptibility of these seven genes for type 2 diabetes, associations with plasma insulin levels were evaluated in a community-derived population sample (Table 2). Although differences in plasma insulin levels among the CDKAL1 rs7756992 genotype did not reach statistical significance, probably due to the limited statistical power (post hoc calculated statistical power: 28.3% for type 2 diabetic patients, 31.8% for control subjects), multiple regression analysis involving the genotype as an additive model showed significantly lower insulin levels in type 2 diabetic subjects with risk genotypes after adjusting for age, sex, and BMI. The association of CDKAL1 rs7756992 remained significant after further adjustment for the current treatment of hyperglycemia (n = 67, P = 0.021). The risk allele of CDKAL1 rs7754840 also tended to be associated with lower insulin levels. However, no significant associations were observed in other SNPs.
TABLE 2.
Association of candidate SNPs with plasma insulin levels in the general population sample
| Gene (rs number) | Risk allele (T) | Plasma insulin (μU/ml)
|
P
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | ANOVA | Multivariate | |||||
| TCF7L2* (rs12255372) | |||||||||
| Control | 5.5 ± 2.7 | (2) | 6.2 ± 4.7 | (52) | 6.5 ± 4.8 | (1,770) | 0.661 | 0.749 | |
| Type 2 diabetes | — | (0) | 4.2 ± 1.0 | (3) | 9.0 ± 7.3 | (136) | 0.256 | 0.368 | |
| TCF7L2* (rs7903146) | T | TT | TC | CC | |||||
| Control | 6.8 | (1) | 6.5 ± 4.3 | (111) | 6.5 ± 4.8 | (1,712) | 0.967 | 0.336 | |
| Type 2 diabetes | — | (0) | 5.1 ± 2.3 | (8) | 9.1 ± 7.4 | (131) | 0.130 | 0.226 | |
| CDKAL1 (rs7756992) | G | GG | GA | AA | |||||
| Control | 6.3 ± 5.2 | (428) | 6.5 ± 4.6 | (895) | 6.6 ± 4.7 | (501) | 0.227 | 0.457 | |
| Type 2 diabetes | 7.9 ± 7.0 | (38) | 8.8 ± 6.3 | (75) | 10.7 ± 10.0 | (26) | 0.269 | 0.020 | |
| CDKAL1 (rs7754840) | C | CC | CG | GG | |||||
| Control | 6.1 ± 4.7 | (315) | 6.4 ± 4.8 | (868) | 6.7 ± 4.7 | (641) | 0.083 | 0.157 | |
| Type 2 diabetes | 7.1 ± 5.1 | (32) | 9.0 ± 6.5 | (67) | 10.3 ± 9.4 | (40) | 0.154 | 0.097 | |
| HHEX (rs1111875) | C | CC | CT | TT | |||||
| Control | 6.4 ± 4.6 | (185) | 6.4 ± 4.4 | (755) | 6.6 ± 5.1 | (884) | 0.867 | 0.883 | |
| Type 2 diabetes | 9.8 ± 11.6 | (15) | 7.7 ± 5.2 | (62) | 9.9 ± 7.7 | (62) | 0.323 | 0.470 | |
| HHEX (rs7923837) | G | GG | GA | AA | |||||
| Control | 6.1 ± 4.1 | (112) | 6.5 ± 4.9 | (692) | 6.5 ± 4.7 | (1020) | 0.818 | 0.749 | |
| Type 2 diabetes | 7.3 ± 4.6 | (9) | 8.8 ± 6.9 | (56) | 9.1 ± 7.8 | (74) | 0.811 | 0.789 | |
| IGF2BP2 (rs4402960) | T | TT | TG | GG | |||||
| Control | 6.2 ± 3.9 | (215) | 6.7 ± 5.2 | (767) | 6.3 ± 4.5 | (842) | 0.276 | 0.308 | |
| Type 2 diabetes | 12.2 ± 10.0 | (15) | 7.8 ± 5.4 | (72) | 9.5 ± 8.4 | (52) | 0.186 | 0.843 | |
| IGF2BP2 (rs1470579) | C | CC | CA | AA | |||||
| Control | 6.1 ± 3.8 | (239) | 6.8 ± 5.2 | (781) | 6.3 ± 4.5 | (804) | 0.118 | 0.314 | |
| Type 2 diabetes | 11.6 ± 9.6 | (17) | 7.8 ± 5.4 | (72) | 9.5 ± 8.5 | (50) | 0.276 | 0.869 | |
| CDKN2A/B (rs10811661) | T | TT | TC | CC | |||||
| Control | 6.4 ± 4.7 | (591) | 6.5 ± 4.6 | (886) | 6.6 ± 5.2 | (347) | 0.719 | 0.858 | |
| Type 2 diabetes | 8.1 ± 8.0 | (41) | 8.9 ± 6.5 | (72) | 10.2 ± 8.0 | (26) | 0.346 | 0.169 | |
| SLC30A8 (rs13266634) | C | CC | CT | TT | |||||
| Control | 6.5 ± 4.9 | (671) | 6.4 ± 4.4 | (847) | 6.6 ± 5.2 | (306) | 0.939 | 0.910 | |
| Type 2 diabetes | 9.8 ± 7.1 | (51) | 7.9 ± 5.6 | (68) | 9.8 ± 11.5 | (20) | 0.151 | 0.332 | |
| KCNJ11 (rs5219) | T | TT | TC | CC | |||||
| Control | 6.5 ± 4.8 | (222) | 6.3 ± 4.6 | (836) | 6.7 ± 4.9 | (766) | 0.088 | 0.156 | |
| Type 2 diabetes | 6.8 ± 3.8 | (28) | 8.8 ± 7.2 | (68) | 10.3 ± 8.7 | (43) | 0.144 | 0.205 | |
Data are means ± SD. n = 1,963. Number of subjects in each genotype is provided in parentheses. Statistical significance was assessed using log-transformed insulin value. Multiple regression analysis involving each genotype as an additive model adjusted for age, sex, and BMI.
T allele dominant model.
DISCUSSION
In the present study, we replicated the associations of several candidate genes derived from a recent GWAS (4–10). However, several conflicting results were observed with other replication studies in Japanese diabetic patients. Horikoshi et al. (21) and Furukawa et al. (20) observed a markedly strong association between type 2 diabetes and variants of HHEX rs1111875, whereas no association was observed in our study. However, results of our meta-analysis (Fig. 1) clearly indicate the type 2 diabetes susceptibility of all candidate genes, including HHEX rs1111875. Conservation of susceptible loci for type 2 diabetes was independent of ethnic background.
However, the attributable risk of these SNPs susceptible for type 2 diabetes was different from that of European ancestries. For example, the pooled odds ratios of the TCF7L2 gene polymorphisms were slightly higher than those in European ancestries (6,23), whereas the risk allele frequencies were considerably lower. Alternatively, for HHEX gene polymorphisms, odds ratios were slightly higher in Japanese (7). Very recently, genome-wide screening in a Japanese population identified the KCNQ1 gene polymorphism as a new susceptible loci for type 2 diabetes (24). These authors reported that the risk alleles of rs2237892 and other SNPs in linkage disequilibrium with rs2237892 were associated with an increased risk of type 2 diabetes. However, apparent associations of the KCNQ1 SNPs were not observed in previous GWAS in populations of European descent (4–10). This discrepancy may be due mainly to the differences in allele frequencies of the susceptible SNPs.
It has been suggested that several candidate SNPs identified from genome-wide screening contribute to diabetes susceptibility primarily through effects on insulin secretion. In our quantitative trait analysis in a general Japanese population, we observed lower basal plasma insulin levels in type 2 diabetic patients carrying the risk genotype of CDKAL1 rs7756992 SNP. Although the function of the CDKAL1 gene product is unknown, one study suggested that CDKAL1 has a role in the inhibition of cyclin-dependent kinase 5 (CDK5) activity in pancreatic β-cells (8), which prevents a decrease in insulin gene expression resulting from glucotoxicity. That study also observed reduced insulin secretion in response to glucose loading in homozygous carriers of the CDKAL1 rs7756992 polymorphism risk allele (8). Pascoe et al. (25) also reported lower insulin secretion after glucose loading in risk allele carriers of another SNP of the CDKAL1 gene. Our study is the first to show a possible association between this SNP and basal insulin levels in type 2 diabetic patients. This observation provides supporting evidence for the pathophysiological role of the CDKAL1 gene products in the progression of type 2 diabetes, as well as the disease susceptibility of this genetic variant.
Several limitations of this study warrant mention. First, differences in linkage disequilibrium between Japanese and European subjects means that tracking of the causal variants may not be possible in SNPs based on the association study in European ancestries. Although our meta-analysis showed an association between type 2 diabetes susceptibility and the analyzed candidate SNPs, causal variants may also be strongly represented by other SNPs in Japanese subjects. Studies with multiple tag-SNPs at loci chosen based on the linkage disequilibrium pattern in Japanese may provide further clarification of this issue. Second, we did not investigate the class of antihyperglycemic drugs, including insulin treatment, in the general population sample. Although each drug may have affected fasting plasma insulin differently, association of the CDKAL1 genotype with plasma insulin levels was statistically significant after further adjustment for current treatment for hyperglycemia.
In the present study, we replicated several genetic variants as risk markers for type 2 diabetes susceptibility in Japanese by performing a case-control analysis and meta-analysis. These findings may be useful in advanced clinical practice and public health genomics.
Supplementary Material
Acknowledgments
This study was supported by a Grant-in-Aids for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology of Japan; The Ministry of Health, Labor and Welfare of Japan; the Japan Arteriosclerosis Prevention Fund; and a Research Promotion Award from Ehime University.
No potential conflicts of interest relevant to this article were reported.
We greatly appreciate the support of Dr. Masaaki Ochi, Wataru Nishida, Yasunori Takata, and Yasuhisa Fujii and their help with sample collection.
Published ahead of print at http://diabetes.diabetesjournals.org on 25 November 2008.
Y.T. and H.O. contributed equally to this work.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K: Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320–323, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Reynisdottir I, Thorleifsson G, Benediktsson R, Sigurdsson G, Emilsson V, Einarsdottir AS, Hjorleifsdottir EE, Orlygsdottir GT, Bjornsdottir GT, Saemundsdottir J, Halldorsson S, Hrafnkelsdottir S, Sigurjonsdottir SB, Steinsdottir S, Martin M, Kochan JP, Rhees BK, Grant SF, Frigge ML, Kong A, Gudnason V, Stefansson K, Gulcher JR: Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34–q35.2. Am J Hum Genet 73: 323–335, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, Weitgasser R, Nejjari C, Patsch W, Chikri M, Meyre D, Froguel P: TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med 85: 777–782, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research, Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, Wellcome Trust Case Control Consortium (WTCCC), McCarthy MI, Hattersley AT: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MC, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RC, Andersen G, Borch-Johnsen K, Jorgensen T, van Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JC, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K: A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39: 770–775, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, Clausen JO, Rasmussen SS, Jorgensen T, Sandbaek A, Lauritzen T, Schmitz O, Hansen T, Pedersen O: Studies of association of variants near the HHEX, CDKN2A/B and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects validation and extension of genome-wide association studies. Diabetes 56: 3105–3111, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Fukushima M, Suzuki H, Seino Y: Insulin secretion capacity in the development from normal glucose tolerance to type 2 diabetes. Diabetes Res Clin Pract 66: S37–S43, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Expert Committee on the Diagnosis and Classification of Diabetes Mellitus: Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 26: S5–S20, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Osawa H, Yamada K, Onuma H, Murakami A, Ochi M, Kawata H, Nishimiya T, Niiya T, Shimizu I, Nishida W, Hashiramoto M, Kanatsuka A, Fujii Y, Ohashi J, Makino H: The G/G genotype of a resistin single-nucleotide polymorphism at −420 increases type 2 diabetes mellitus susceptibility by inducing promoter activity through specific binding of Sp1/3. Am J Hum Genet 75: 678–686, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyake K, Horikawa Y, Hara K, Yasuda K, Osawa H, Furuta H, Hirota Y, Yamagata K, Hinokio Y, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Yamamoto K, Tokunaga K, Takeda J, Makino H, Nanjo K, Kadowaki T, Kasuga M: Association of TCF7L2 polymorphisms with susceptibility to type 2 diabetes in 4,087 Japanese subjects. J Hum Genet 53: 174–180, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Osawa H, Tabara Y, Kawamoto R, Ohashi J, Ochi M, Onuma H, Nishida W, Yamada K, Nakura J, Kohara K, Miki T, Makino H: Plasma resistin, associated with single nucleotide polymorphism −420, is correlated with insulin resistance, lower HDL cholesterol, and high-sensitivity C-reactive protein in the Japanese general population. Diabetes Care 30: 1501–1506, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Horikoshi M, Hara K, Ito C, Nagai R, Froguel P, Kadowaki T: A genetic variation of the transcription factor 7-like 2 gene is associated with risk of type 2 diabetes in the Japanese population. Diabetologia 50: 747–751, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Hayashi T, Iwamoto Y, Kaku K, Hirose H, Maeda S: Replication study for the association of TCF7L2 with susceptibility to type 2 diabetes in a Japanese population. Diabetologia 50: 980–984, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Horikawa Y, Miyake K, Yasuda K, Enya M, Hirota Y, Yamagata K, Hinokio Y, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Yamamoto K, Tokunaga K, Takeda J, Kasuga M: Replication of genome-wide association studies of type 2 diabetes susceptibility in Japan. J Clin Endocrinol Metab 93: 3136–3141, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Furukawa Y, Shimada T, Furuta H, Matsuno S, Kusuyama A, Doi A, Nishi M, Sasaki H, Sanke T, Nanjo K: Polymorphisms in the IDE-KIF11-HHEX gene locus are reproducibly associated with type 2 diabetes in a Japanese population. J Clin Endocrinol Metab 93: 310–314, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, Froguel P, Kadowaki T: Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia 50: 2461–2466, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, Kawamori R, Nakamura Y, Maeda S: Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes 57: 791–795, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Groves CJ, Zeggini E, Minton J, Frayling TM, Weedon MN, Rayner NW, Hitman GA, Walker M, Wiltshire S, Hattersley AT, McCarthy MI: Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes 55: 2640–2644, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, Yamagata K, Hinokio Y, Wang HY, Tanahashi T, Nakamura N, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Takeda J, Maeda E, Shin HD, Cho YM, Park KS, Lee HK, Ng MC, Ma RC, So WY, Chan JC, Lyssenko V, Tuomi T, Nilsson P, Groop L, Kamatani N, Sekine A, Nakamura Y, Yamamoto K, Yoshida T, Tokunaga K, Itakura M, Makino H, Nanjo K, Kadowaki T, Kasuga M: Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 40: 1092–1097, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, Weedon MN, Mari A, Hattersley AT, McCarthy MI, Frayling TM, Walker M, RISC Consortium, U.K. Type 2 Diabetes Genetics Consortium: Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes 56: 3101–3104, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.