Abstract
A key process in the tsetse reproductive cycle is the transfer of essential nutrients and bacterial symbionts from mother to intrauterine offspring. The tissue mediating this transfer is the milk gland. This work focuses upon the localization and function of two milk proteins (milk gland protein (GmmMGP) and transferrin (GmmTsf)) and the tsetse endosymbionts (Sodalis and Wigglesworthia), in the context of milk gland physiology. Fluorescent in situ hybridization (FISH) and immunohistochemical analysis confirm that the milk gland secretory cells synthesize and secrete milk gland protein and transferrin. Knockdown of gmmmgp by double stranded RNA (dsRNA) mediated RNA interference results in reduction of tsetse fecundity, demonstrating its functional importance in larval nutrition and development. Bacterial species-specific in situ hybridizations of milk gland sections reveal large numbers of Sodalis and Wigglesworthia within the lumen of the milk gland. Sodalis is also localized within the cytoplasm of the secretory cells. Within the lumen, Wigglesworthia localize close to the channels leading to the milk storage reservoir of the milk gland secretory cells. We discuss the significance of the milk gland in larval nutrition and in transmission of symbiotic bacteria to developing offspring.
Keywords: Tsetse, reproduction, milk gland, Sodalis, Wigglesworthia
Introduction
Tsetse (Diptera: Glossinidae) are important vectors of African trypanosomes which cause disease of both medical and agricultural importance. Trypanosomes are the causative agents of sleeping sickness (Human African Trypanosomiasis – HAT) and nagana in sub-Saharan Africa. There are limited tools for control of these diseases in the mammalian host but control has been realized via tsetse population reduction methods in previous efforts. Given the low reproductive capacity of tsetse, additional knowledge on its reproductive physiology can provide new avenues for control.
Flies have developed various forms of viviparous reproduction. However, most of these undergo facultative viviparity, resulting in the deposition of developing embryos or larvae. Tsetse reproductive physiology is unique as the female carries and nourishes their offspring for their entire larval developmental cycle. Females develop a single oocyte at a time. The oocyte is ovulated, fertilized and undergoes embryonic development in the uterus. The resulting larva hatches and is carried and nourished in the intrauterine environment for the duration of its development. Within an hour of parturition, the larva burrows into the earth and pupates. This viviparous strategy is termed pseudo-placental unilarviparity and has been observed in three other families of flies, Hippoboscidae, Nycteribiidae and Streblidae (Meier et al. 1999). All of these families are close relatives of tsetse and are haematophagous (blood feeding). Viviparity and blood feeding may be associated due to the nutritional demands of the intrauterine larva. Blood is one of the few sources of nutrition rich enough support this reproductive strategy.
Tsetse reproductive physiology differs from other Diptera in significant ways. These differences accommodate the requirements of the viviparous reproductive strategy. The uterus is a modified vaginal canal that is covered with highly tracheated muscle tissue. The uterus has the capacity to hold a mature third instar larva equivalent in weight to the mother. Larval nutrition is provided via a modified accessory gland (milk gland) that empties into the uterus. The milk gland is connected to the dorsal side of the uterus and expands throughout the abdominal cavity of the fly as bifurcating tubules intertwining with fat body tissue (Tobe and Langley 1978). The lumen of the milk gland is surrounded by secretory and epithelial cells. The epithelial cells secrete and maintain the chitinous lining of the lumen. The secretory cells contain large nuclei and surround an extracellular secretory reservoir which changes size dynamically as it fills with and empties milk secretions over the course of pregnancy. The opening of the reservoir into the lumen is covered by a fibrous plug which is confluent with the lining of the lumen (Ma et al. 1975; Tobe et al. 1973).
Tsetse has a biological association with three bacterial species, Sodalis glossinidius, Wigglesworthia glossinidia and Wolbachia pipientis (Aksoy 2000). Wolbachia is predominantly localized in the ovaries and is passed from generation to generation by transovarial transmission. Sodalis and Wigglesworthia are both hypothesized to be transmitted to the larva via milk gland secretions (Denlinger and Ma 1975). Sodalis is detectable in multiple tissues within the fly, including the milk gland, as evidenced by a symbiont specific PCR amplification assay (Cheng and Aksoy 1999). Wigglesworthia resides in specialized cells (bacteriocytes) that form the bacteriome in the midgut. The transmission route of Wigglesworthia from mother to the developing progeny was also assumed to be via the milk. Electron microscopy analysis showed bacteria in the lumen of the milk gland and based upon their large size, these bacteria were thought to be Wigglesworthia (Denlinger and Ma 1975). However, the identity of these bacteria remains unconfirmed.
The milk secreted from the milk gland into the uterus consists primarily of protein and lipids (Cmelik et al. 1969). Two milk proteins were characterized, the milk gland protein (GmmMGP) and transferrin (GmmTsf) (Attardo et al. 2006; Guz et al. 2007). Other milk proteins have been detected and remain uncharacterized (Riddiford and Dhadialla 1990). GmmMGP and GmmTsf are synthesized by the adult female and are transferred to the developing larva. During the first gonotrophic cycle, gmmmgp expression correlates with larval development. The first oocyte is ovulated into the uterus between days 6–8 post-eclosion and begins embryonic development. At days 10–13 the embryo hatches and larval development begins. Development continues to between days 19–21 when the mother undergoes parturition. Over the course of larval development, gmmmgp transcript levels make a dramatic increase in abundance beginning around day 8 post eclosion through partuition. After the first gonotrophic cycle expression of gmmmgp remains constitutive. However, GmmMGP protein is almost undetectable in the mother upon larval deposition illustrating its transfer from mother to larva (Attardo et al. 2006).
Expression of gmmtsf differs somewhat from that of gmmmgp. In females transcript abundance of gmmtsf is cyclic and correlates with oogenesis and larvigenesis. Expression of gmmtsf also differs from gmmmgp as it is expressed in both males and females. GmmTsf protein levels are constitutive and can be found in the milk gland, hemolymph, reproductive tract and developing larva. An increase in GmmTsf is observed during larval development, however, it is not equivalent to observed levels of GmmMGP (Guz et al. 2007).
The primary goals of this research are: 1. to investigate the physiological and functional characteristics of the major milk proteins, GmmMGP and GmmTsf, in the specific context of the milk gland and 2. to identify and localize the symbionts residing in the milk gland. The role of milk proteins in fecundity and vertical transmission biology of tsetse’s symbionts are discussed.
Materials and Methods
Fly rearing
The Glossina morsitans morsitans colony maintained in the insectary at Yale University was originally established from puparia from fly populations in Zimbabwe. Newly emerged flies are separated by sex and mated at three to four days post-eclosion. Flies are maintained at 24±1°C with 50–55% relative humidity, and receive defibrinated bovine blood every 48h using an artificial membrane system (Moloo 1971).
Tissue dissection and sample preparation
Fat body and milk gland were collected from pregnant females. Fat body and milk gland were detached from the abdomen and reproductive tract and placed directly into 4% paraformaldehyde for fixation for one week.
Paraformaldehyde was replaced with several changes of phosphate buffered saline (PBS) over 2–3 hours. The tissue was then dehydrated through a graded ethanol series: 70% ethanol 2 times for 15 minutes, 95% ethanol 2 times for 30 minutes, 100% ethanol 2 times for 30 minutes. Samples were then moved to 100% butanol 2 times for 10 minutes and stored in butanol at 4°C for one week. Tissues were embedded in paraffin and cut on a rotary microtome at a thickness of 5 µm. The sections were placed on poly-L-Lysine glass slides, left at room temperature until dried and kept at 4°C until used.
Immunohistochemistry / Fluorescent in situ hybridization (FISH) and microscopy
For the following applications prepared slides were dewaxed by incubation in methylcyclohexane 2 times for 15 minutes, followed by incubation in 100% ethanol 2 times for 10 minutes. Samples were air dried.
The FISH protocol used for symbiont staining was published in (Anselme et al. 2006) and includes steps of membrane permeabilization by coating the sections with a drop of 70% acetic acid at 60°C for 2–3 minutes. Slides were then rinsed 2 times with phosphate buffered saline for 5 minutes, followed by a series of dehydration washes in 70% EtOH for 2 minutes, 95% EtOH for 2 minutes and a final wash in 100% EtOH 2 times for 2 minutes. The slides were then air dried. Deproteinization of slides was performed as follows. Slides were incubated in 0,01N HCl with 0,1mg/ml pepsin for 10 minutes at 37°C, followed by a repetition of the washing and dehydration steps above. Sections were then prehybridized at 45°C for 30 minutes and each slide was hybridized for 3 hours at 45°C with 200ng of 5’ end rhodamine labeled oligonucleotide probes targeting Wigglesworthia specific (5'-ACGATACTCTAGTTTATTAG-3') and Sodalis specific (5'-ACGAGACTCTAGCCT GCCAG-3') 16S ribosomal RNA, respectively. After hybdridization sections were washed in wash buffer (hybridization buffer + 0,1% SDS) , followed by a quick rinse in water and subsequently mounted with GelMount™ (Electron Microscopy Science, Hatfield, PA) with 4′,6-diamidino-2-phenylindole (DAPI). Co- staining of either gmmmgp or gmmtsf transcripts and protein was done by FISH and immunohistochemistry. Slides were dewaxed and air dried as described above. They were then prehybridized in probe less hybridization buffer (200 mM NaCl, 8.9 mM TrisHCl, 1.1 mM Tris Base, 6.5 mM NaH2PO4, 5 mM Na2HPO4, 5 mM EDTA, 50% high quality deionized formamide, 10% dextrane sulphate, 1 mg/ml tRNA, 1X Denhardts solution (.02% bovine serum albumin, .02% ficoll, .02% PVP)) for 30 minutes at 65°C. Digoxigenin labeled antisense RNA probes were generated using the MAXIscript In vitro transcription kit following the manufacturer’s protocol (Ambion, Austin, TX). Template for probe synthesis was the cloned open reading frame for either gmmmgp or gmmtsf within linearized T-vector plasmid (Invitrogen, Carlsbad, CA). Probes were diluted in hybridization buffer to a final concentration of 1 ng/µl. Hybridization solution including probe was added to the slides which were then incubated overnight in a humidified chamber at 65°C.
Following hybridization, slides were washed in wash buffer (1× SCC, 50% formamide and 0.1% Tween 20) at 65°C, 1 time for 15 minutes and 2 more times for 30 minutes. The slides were then washed 2 times for 10 minutes in PBST (1× PBS and 0.1% Tween 20). Slides were blocked in 1× blocking solution (Roche, Indianapolis, IN). Antibody solutions were made using 1× blocking solution, anti- DIG-rhodamine Fab fragments for FISH probe detection (1:200 dilution)(Roche) and either rabbit anti-GmmMGP (1:5000) or rabbit anti-GmmTsf (1:5000) antibodies. Antibody creation is described in (Attardo et al. 2006; Guz et al. 2007). Antibody solutions were added to the slides and incubated overnight in the dark at 4°C.
Slides were washed 3 times for 10 minutes in PBST in the dark. Secondary antibody solution was made using 1× blocking solution and Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) at a dilution of 1:500. Secondary antibody solution was added to the slides and incubated at room temperature in the dark for one hour. Slides were washed 3 times for 10 minutes in PBST and 1 time for 2 minutes in dH2O. Slides were mounted using GelMount™ medium with DAPI (Biomedia, Foster City, CA). Samples were observed using a Zeiss Axioskop2 microscope (Zeiss, Thornwood, NY) equipped with a fluorescent filter set with fluorescein, rhodamine and DAPI specificity. Samples were viewed and imaged at either 200 or 400 times magnification using the following lenses (Zeiss Achroplan 20×/0,45 Ph2, Zeiss Achroplan 40×/0,65 Ph2) and a 10× occular. Images were captured using an Infinity1 USB 2.0 camera and software (Lumenera Corporation, Ottawa, Ontario, Canda).
Milk Gland Protein RNAi
Flies for the knockdown experiment were collected within a 24 hour eclosion period to synchronize their reproductive status. Two cages were set up for the experiment each containing 30 female flies. The flies were reared according to standard protocols and were mated at day 3 post eclosion. One cage was given dsGFP injections while the other received dsMGP, respectively. dsRNA for gmmmgp and gfp were produced by in vitro transcription of PCR-generated DNA templates containing the T7 promoter sequence at both ends. The cDNA clone for gmmmgp and plasmid eGFP (BD Bioscience, San Jose, CA) served as templates for PCR amplification with gene-specific primer sets: milk gland protein T7 forward 5’-TAATACGACTCACTATAGGGGTTGGTCAATGGGGAACAAG-3’, milk gland protein T7 reverse 5’-TAATACGACTCACTATAGGGTAAGCAGTGCCTGGGGTATT-3’, GFP T7 forward 5’-TAATACGACTCACTATAGGGTCAGTGGAGAGGGTGAAG-3’, GFP T7 reverse 5’-TAATACGACTCACTATAGGCTAGTTGAACGGATCCATC-3’.
The PCR amplification conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 50°C for 30 s and 72°C for 45 s and by 1 cycle at 72°C for 10 min in a MJ Research (PTC-200) thermal cycler. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) prior to use for in vitro transcription. Sense and anti-sense RNA were synthesized using the MEGAscript® RNAi Kit (Ambion, Austin, TX) following the manufacturer’s protocol. The transcripts were precipitated using isopropanol, resuspended in RNase-free water and annealed by incubation at 65°C for 30 min followed by gradual cooling to room temperature. The concentration of dsRNA was determined by means of spectrophotometry. Injections of 5 µg of dsRNA were given on day 5 post-eclosion and day 13 post-eclosion. Cages were monitored daily for fly mortality and larval deposition. At the end of the experiment at day 40, all flies were dissected to confirm mating status by visual examination of spermathica for the presence of sperm and for the accumulation of unovulated oocytes (virgin flies do not ovulate).
Results
Milk gland protein and mRNA localization within the milk gland
Florescent light microscopy of sectioned milk gland and fat body tissue was performed to expand upon the previous whole mount immunohistochemical analysis of the milk gland (Attardo et al. 2006). Fat body and milk gland tubules were analyzed by in situ hybridization and immunohistochemical analysis to identify cellular and subcellular expression and localization of milk gland protein transcript and protein.
DAPI staining reveals the secretory cell nuclei (Fig 1a+e). In situ hybridization with digoxigenin labeled gmmmgp antisense probes shows uniform distribution of transcript throughout the cytoplasm of the milk gland secretory cells (Fig. 1b+f). Staining for gmmmgp is absent within the storage reservoir, lumen, nuclei or surrounding fat body tissue.
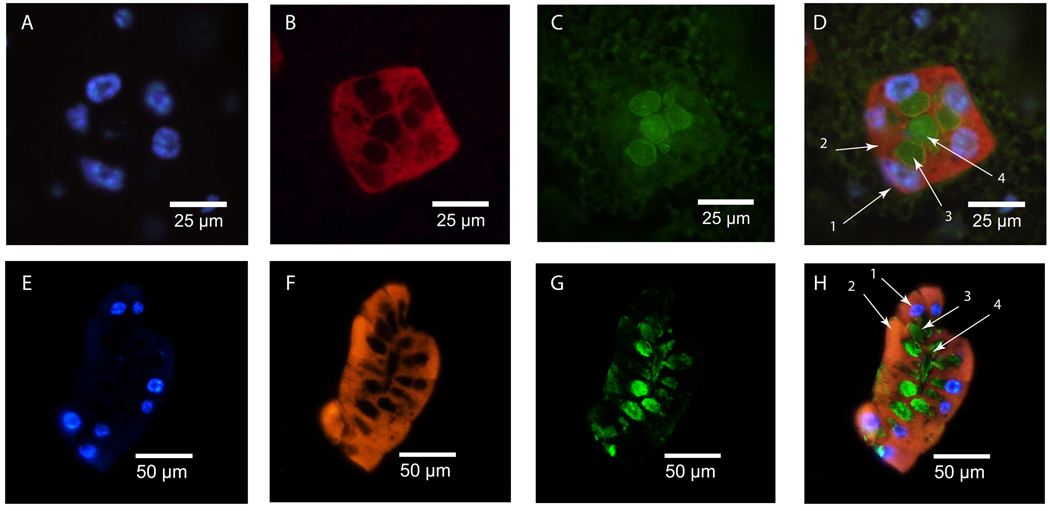
Figure 1. GmmMgp in situ and immunohistochemical staining in pregnant female milk gland.
Tissue sections were stained with DAPI, DIG-labeled antisense gmmmgp RNA probe, and Anti-GmmMGP antisera. (A–D: Transverse milk gland section; E–H: Longitudinal milk gland section) - A+E: DAPI staining of milk gland cell nuclei, B+F: In situ staining of gmmmgp transcript. Staining is visible throughout the cytoplasm of the secretory cells with no staining observed in the surrounding fat body. C+G: Immunohistochemical staining of GmmMGP. GmmMGP is concentrated within the extracellular secretory reservoir and the lumen of the milk gland. D+H merged images: D1- nucleus, D2- cytoplasm (gmmmgp transcript), D3- secretory reservoir (GmmMGP), D4- lumen (GmmMGP), H1- nucleus, H2- cytoplasm (gmmmgp transcript), H3- secretory reservoir (GmmMGP), H4- lumen (GmmMGP).
Immunohistochemical analysis with GmmMGP specific antibodies reveals the most intense staining localized to the storage reservoirs and the lumen (Fig. 1c+g). This staining pattern suggests that the milk gland is the specific site of GmmMGP synthesis.
Transferrin protein and mRNA localization within the milk gland
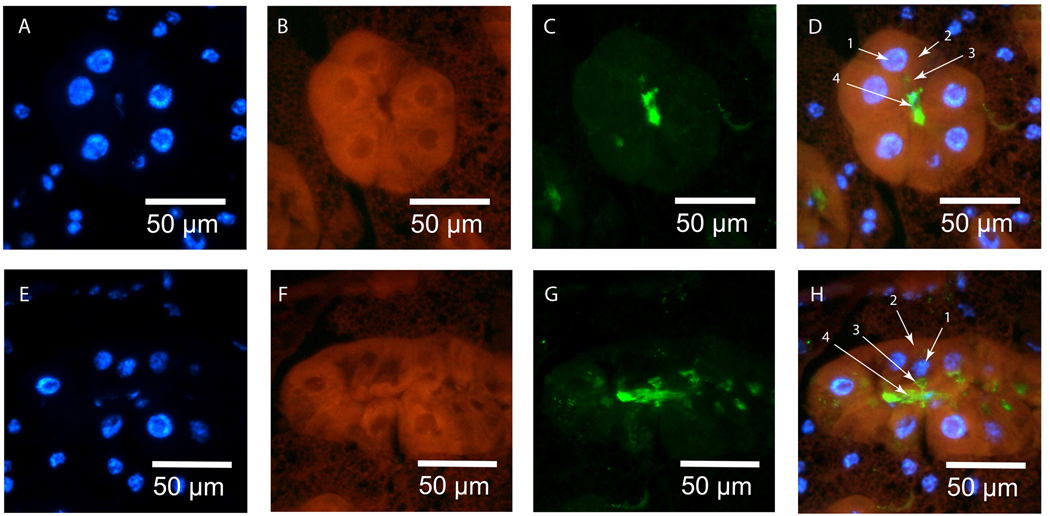
Visualization of transferrin (gmmtsf) transcripts and protein (GmmTsf) within the milk gland were performed in the same manner as for gmmmgp. Transcripts for gmmtsf are localized to the cytoplasm of the secretory cells with little staining detected in the surrounding fat body cells (Fig. 2b+f). Localization of GmmTsf protein is similar to that of GmmMGP as it is visualized within the secretory reservoir and the lumen of the milk gland (Fig. 2c+g).
Figure 2. GmmTsf in situ and immunohistochemical staining in pregnant female milk gland.
Tissue sections were stained with DAPI, DIG-labeled antisense gmmmgp RNA probe, and Anti-GmmMGP antisera. (A–D: Transverse milk gland section; E–H: Longitudinal milk gland section) - A+E: DAPI staining of milk gland cell nuclei. B+F: In situ staining of gmmtsf transcript. Transcript is concentrated within the cytoplasm of the secretory cells with light staining of the surrounding fat body. C+G: Immunohistochemical staining of GmmTsf. GmmTsf staining is specific to the secretory reservoir and the gland lumen. (Reservoirs appear small in this image as this female was approaching the end of the pregnancy cycle. At this point the larva takes milk secretions faster than they can accumulate). D+H: Merged images: D1- nucleus, D2- cytoplasm (gmmtsf transcript), D3- secretory reservoir (GmmTsf), D4- lumen (GmmTsf), H1- nucleus, H2, cytoplasm (gmmtsf transcript), H3- secretory reservoir (GmmTsf), H4- lumen (GmmTsf).
Milk gland protein RNAi effects on fecundity
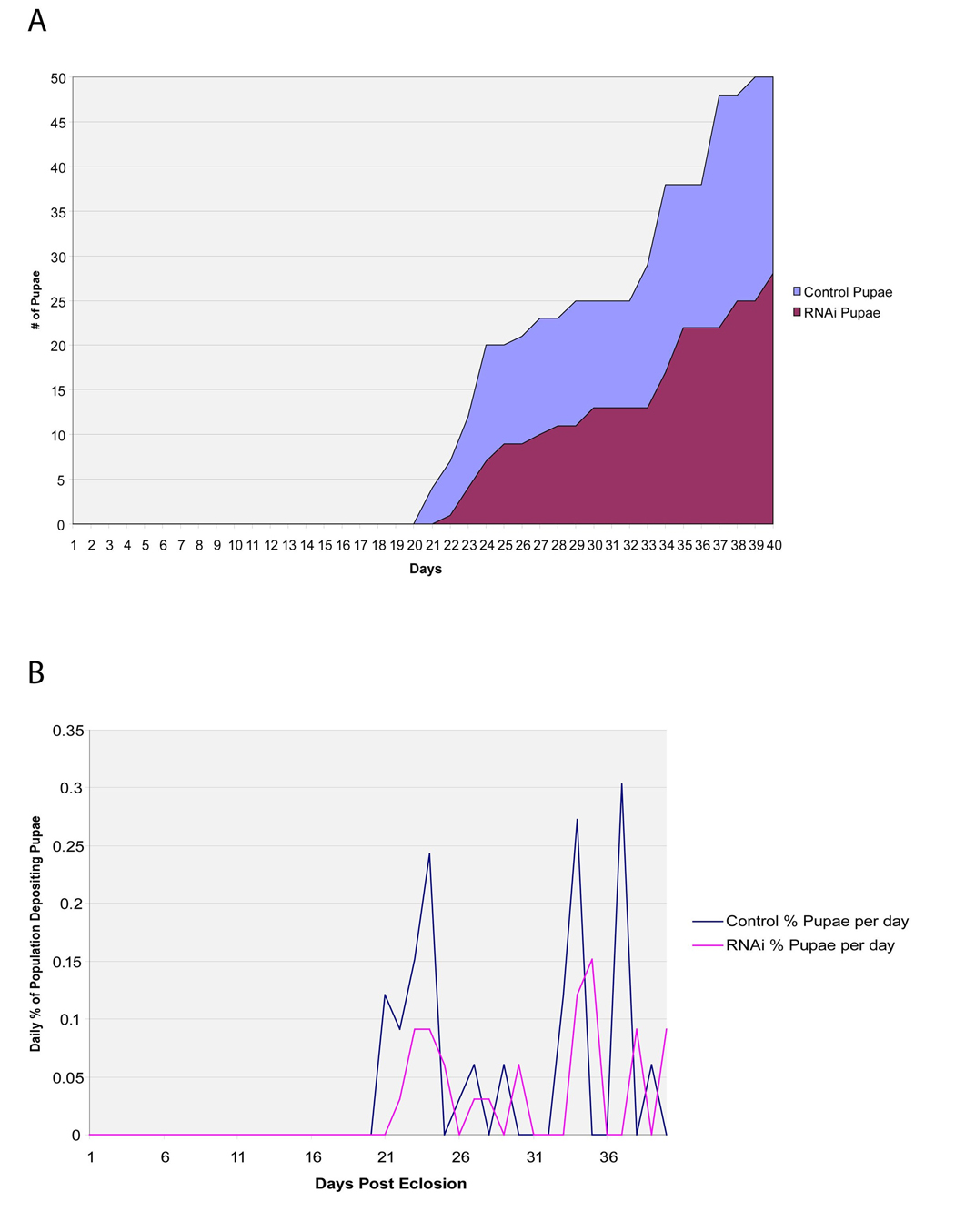
Functional analysis of GmmMGP was performed by dsRNA injection into pregnant females. Two cages of females were treated with either dsMGP or dsGFP (control) at five days of age and again at 13 days of age. The gmmmgp knockdown group had a lower rate of pupal deposition relative to the control group as measured by monitoring the percentage of females depositing pupae per day (Fig. 3a). The rate of pupal deposition by the knockdown flies is roughly half that of the control flies with 4% of the control flies depositing daily versus 2% of the knockdown flies. The cumulative number of pupae deposited by the two cages was 28 in the dsMGP group as opposed to 50 by the dsGFP group during the 40 days of observation (Fig. 3b).
Figure 3. Fecundity effects on female flies treated with dsRNA against gmmmgp.
Two cages of 30 female flies were treated with either gmmmgp or gfp dsRNAs at 5 and 13 days post eclosion. Numbers of offspring and female mortality were monitored daily. A. Measurement of cumulative pupal deposition per day for each cage. B. Measurement of percentage of surviving females depositing pupa per day
In situ localization of symbionts within the milk gland
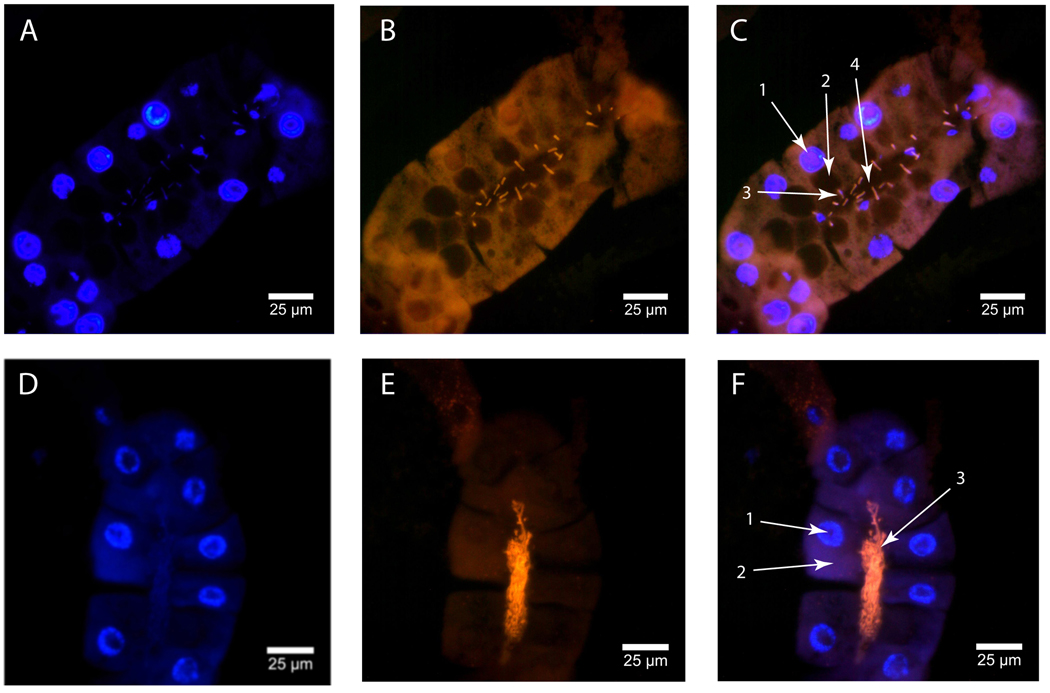
Specific identification of the bacterial fauna residing in the milk gland was accomplished by in situ analysis of milk gland using fluorescently labeled oligonucleotide probes specific to either Wigglesworthia or Sodalis 16S ribosomal RNA. Staining with the Sodalis specific probe reveals large numbers of Sodalis within the lumen of the milk gland (Fig. 4c). In addition, Sodalis are also detectable in the cytoplasm of the milk gland secretory cells.
Figure 4. In situ staining of Sodalis in pregnant female milk gland.
Tissue sections were stained with DAPI and Sodalis specific DIG-labeled 16S ribosomal RNA probe. (A–C: Longitudinal milk gland section). A: DAPI staining of milk gland cell nuclei, B: In situ staining of Sodalis. The staining pattern shows concentrated Sodalis in the extracellular space of the lumen. Bacteria can also be seen intracellularly within the cytoplasm of the secretory cells. C: Merged images: C1- nucleus, C2- cytoplasm (Sodalis), C3- lumen (Sodalis).
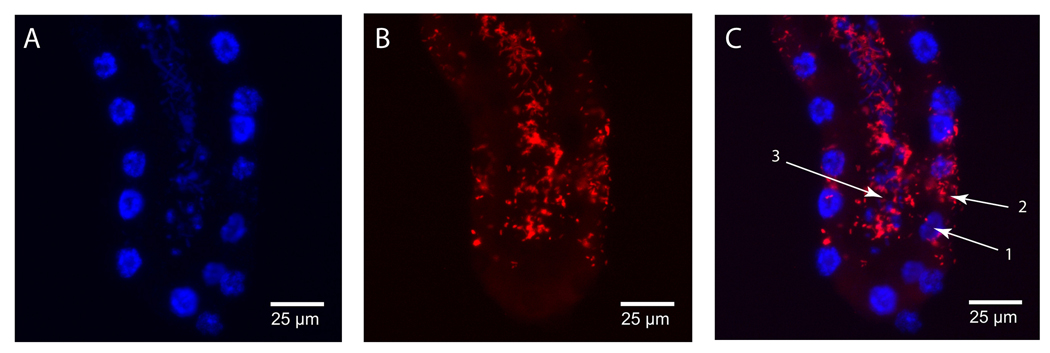
The staining pattern for Wigglesworthia differs from Sodalis. Wigglesworthia staining is exclusive to the lumen of the milk gland (Fig. 5c). Individual Wigglesworthia are observed within the canals in the intima leading to the secretory reservoirs of the secretory cells (Fig. 5f). This observation was made by (Denlinger and Ma 1974) in their transmission electron microscopy study. The species specific labeling technique used here confirms the identify of these bacteria as Wigglesworthia.
Figure 5. In situ staining of Wigglesworthia in pregnant female milk gland.
Tissue sections were stained with DAPI and Wigglesworthia specific DIG-labeled 16S ribosomal RNA probe. (A–F: Longitudinal milk gland sections). A+D: DAPI staining of milk gland cell nuclei, B+E: In situ staining of Wigglesworthia. Bacteria are concentrated within the lumen and can be seen positioned within the canals leading to the secretory reservoirs. C+F: Merged images: C1- nucleus, C2- secretory reservoir, C3- bacteria within the canal leading to the reservoir (Wigglesworthia), C4- lumen with bacteria (Wigglesworthia), F1- nucleus, F2 – cytoplasm, F3- lumen with concentrated bacteria (Wigglesworthia), F4- lumen.
Discussion
Data presented confirm the synthesis and secretion of GmmMGP and GmmTsf by the milk gland secretory cells for use in larval nutrition. Functional evidence of GmmMGP’s role in larvigenesis is demonstrated by the negative impact on fecundity resulting from its knockdown. Bacterium specific FISH analysis indicate the presence of both tsetse symbionts, Sodalis and Wigglesworthia, in the milk gland. Sodalis resides both in the milk gland lumen and within the milk gland secretory cells, while Wigglesworthia is only detected in the lumen. This data confirm the identity of the previously visualized bacteria as a population of extracellular Wigglesworthia living in the milk gland lumen.
The milk gland is a unique tissue in the Dipteran order (found only in species within the superfamily Hippoboscoidea). It is responsible for the conversion of nutrients into the sole source of nourishment for the fly’s intrauterine larvae. The milk gland also appears to harbor the flies symbiotic flora and act as the conduit for their transfer from mother to offspring. The origin of this tissue is a reproductive accessory gland that has adapted to fulfill the role of milk production in tsetse. The evolutionary biology of the milk gland requires further study. Tsetse accessory glands differ from those in other insects in structure and function. Function of insect accessory glands is diverse and is usually associated with secretion of adhesive material to secure and protect eggs (Romoser 1981). However, one viviparous species of cockroach generates a milk secretion from its accessory glands (Williford et al. 2004).
In many flies, including mosquitoes, the fat body is responsible for the majority of protein synthesis associated with oogenesis (Raikhel 1992). Gene expression in tsetse is different in that the major vitellogenic protein (GmmYP1) is synthesized by the reproductive tract and the two characterized milk proteins (GmmMGP and GmmTsf) by the milk gland (Attardo et al. 2006). These observations suggest changes have occurred in the transcriptional regulation of genes such as the tsetse yolk protein and transferrin orthologs. In other flies these genes are expressed by the fat body. Future comparative analysis of the regulatory regions of these genes with those of orthologs from other flies will illustrate the conservation and divergence of regulatory elements that define the temporal and tissue specificity of these genes. The shift away from fat body specific expression to other tissues suggests that the physiology of the fat body in tsetse may be different from that observed in other insects. Previous work demonstrated that the fat body acts as a store for lipids that are mobilized to the milk gland and integrated into the milk (Langley et al. 1981). It is possible that the role of the fat body in protein synthesis has been shifted to other tissues to allow for enhanced lipid storage function. Fat body function in tsetse requires further study to determine its role in nutrient storage, metabolism, and transference of lipids to the milk gland.
GmmMGP is a protein from the lipocalin family. Lipocalins are known for their ability to bind small hydrophobic molecules (Flower et al. 2000). They are also associated with lactation secretions. Lipocalins are found in the lactation secretions of marsupials (Collet and Joseph 1993). These proteins are also a primary component of the secretion generated by the viviparous cockroach species (Williford et al. 2004). These parallels with tsetse suggest functional conservation in these proteins that may be a result of not only their nutritional properties but their functional properties as a carrier of lipid and/or sterol type molecules. The possibility that GmmMGP may perform dual roles suggests it plays an important role in the delivery of nutrients from mother to larva. The adverse effect on fecundity of females in which GmmMGP has been knocked down supports this hypothesis.
The transfer of nutrients and bacteria are essential for larval development and the fertility of the resulting adult flies. Given the abundant yet incomplete nutritional value of blood, tsetse is dependent upon its symbionts to maintain its lifecycle. Clearing bacterial fauna results in an infertility phenotype in tsetse (Nogge 1976). Microscopic analysis shows aposymbiotic flies are dysfunctional in their reproductive cycle. Some flies have been observed to reabsorb completely developed oocytes, while other aposymbiotic flies undergo ovulation but, abort the offspring either during embryogenesis or early larvigenesis (Attardo, unpublished observations). This suggests that the symbionts are providing a factor(s) essential for larval development. It remains to be determined what this factor is and whether it is affecting ovulation, embryogenesis and/or larval development.
It is likely that the symbiotic flora, Wigglesworthia and Sodalis are transmitted into the developing larva as a component of the milk secretions as predicted in (Denlinger and Ma 1975). Our evidence suggests that the fly has two separate populations of Wigglesworthia; one that lives intracellularly in the midgut bacteriome and one that lives extracellularly in the lumen of the milk gland. These two populations of Wigglesworthia may fulfill different physiological functions in host biology. It remains to be seen when and where symbiont colonization takes place in the larva and if bacterial sub populations are generated during larval development.
Details of the bacterial interaction with the milk gland remain to be investigated as Sodalis are also observed living within the milk gland cells. Furthermore, Wigglesworthia are often observed localized within the channels heading in to the secretory vacuoles. This observation suggests the possibility the bacteria are responding to chemotaxic stimuli from a product of the secretory cells.
These investigations generate new questions regarding the function of milk gland proteome in larvigenesis and the nature of their tissue/cell specific regulation. The biology of the bacteria in the milk gland, their vertical transmission to offspring and colonization of tissues in the developing larva demands further study.
Acknowledgments
We would like to acknowledge Severine Balmand (UMR203 Biologie Fonctionnelle Insectes et Interactions, IFR41, INRA, INSA-Lyon, F-69621 Villeurbanne, France) for her technical assistance and training in FISH techniques. This work was generously funded by grants from the NIGMS (069449) and Ambrose Monell Foundation to S.A. as well as the NIH Ruth Kirshstein Postdoctoral Training Award F32 GM077964 to Geoffrey M. Attardo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aksoy S. Tsetse - A haven for microorganisms. Parasitology Today. 2000;16:114–118. doi: 10.1016/s0169-4758(99)01606-3. [DOI] [PubMed] [Google Scholar]
- Anselme C, Vallier A, Balmand S, Fauvarque MO, Heddi A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Applied Environmental Microbiology. 2006;72:6766–6772. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Guz N, Strickler-Dinglasan P, Aksoy S. Molecular aspects of viviparous reproductive biology of the tsetse fly (Glossina morsitans morsitans): Regulation of yolk and milk gland protein synthesis. Journal of Insect Physiology. 2006;52:1128–1136. doi: 10.1016/j.jinsphys.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Aksoy S. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Molecular Biology. 1999;8:125–132. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- Cmelik SHW, Bursell E, Slack E. Composition of Gut Contents of Third-Instar Tsetse Larvae (Glossina Morsitans Westwood) Comparative Biochemistry and Physiology. 1969;29 447-&. [Google Scholar]
- Collet C, Joseph R. A novel member of the lipocalin superfamily: tammar wallaby late-lactation protein. Biochimica et Biophysica Acta. 1993;1167:219–222. doi: 10.1016/0005-2760(93)90165-6. [DOI] [PubMed] [Google Scholar]
- Denlinger DL, Ma WC. Dynamics of pregnancy cycle in tsetse Glossina-morsitans. Journal of Insect Physiology. 1974;20:1015–1026. doi: 10.1016/0022-1910(74)90143-7. [DOI] [PubMed] [Google Scholar]
- Denlinger DL, Ma WC. Maternal nutritive secretions as possible channels for vertical transmission of microorganisms in insects: the tsetse fly example. Annals of the New York Academy of Sciences. 1975;266:162–165. doi: 10.1111/j.1749-6632.1975.tb35097.x. [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochimica et Biophysica Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Guz N, Attardo GM, Wu Y, Aksoy S. Molecular aspects of transferrin expression in the tsetse fly (Glossina morsitans morsitans) Journal of Insect Physiology. 2007;53:715–723. doi: 10.1016/j.jinsphys.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley PA, Bursell E, Kabayo J, Pimley RW, Trewern MA, Marshall J. Hemolymph Lipid Transport from Fat-Body to Uterine Gland in Pregnant Females of Glossina-Morsitans. Insect Biochemistry. 1981;11:225–231. [Google Scholar]
- Ma WC, Denlinger DL, Jarlfors U, Smith DS. Structural modulations in the tsetse fly milk gland during a pregnancy cycle. Tissue and Cell. 1975;7:319–330. doi: 10.1016/0040-8166(75)90008-7. [DOI] [PubMed] [Google Scholar]
- Meier R, Kotrba M, Ferrar P. Ovoviviparity and viviparity in the Diptera. Biological Reviews of the Cambridge Philosophical Society. 1999;74:199–258. [Google Scholar]
- Moloo SK. An artificial feeding technique for Glossina. PARASITOLOGY. 1971;63:507–512. doi: 10.1017/s0031182000080021. [DOI] [PubMed] [Google Scholar]
- Nogge G. Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia. 1976;32:995–996. doi: 10.1007/BF01933932. [DOI] [PubMed] [Google Scholar]
- Raikhel AS. Advances in Disease Vector Research. New York: Springer; 1992. Vitellogenesis in mosquitos; pp. 1–39. [Google Scholar]
- Riddiford LM, Dhadialla TS. Protein-Synthesis by the Milk Gland and Fat-Body of the Tsetse-Fly, Glossina-Pallidipes. Insect Biochemistry. 1990;20:493–500. [Google Scholar]
- Romoser WSS, J G. The Science of Entomology. Prentice Hall. (2nd edn.) 1981:575. [Google Scholar]
- Tobe SS, Davey KG, Huebner E. Nutrient transfer during the reproductive cycle in Glossina austeni Newst.: histology and histochemistry of the milk gland, fat body, and oenocytes. Tissue and Cell. 1973;5:633–650. doi: 10.1016/s0040-8166(73)80050-3. [DOI] [PubMed] [Google Scholar]
- Tobe SS, Langley PA. Reproductive physiology of Glossina. Annual Review of Entomology. 1978;23:283–307. doi: 10.1146/annurev.en.23.010178.001435. [DOI] [PubMed] [Google Scholar]
- Williford A, Stay B, Bhattacharya D. Evolution of a novel function: nutritive milk in the viviparous cockroach, Diploptera punctata. Evolution and Development. 2004;6:67–77. doi: 10.1111/j.1525-142x.2004.04012.x. [DOI] [PubMed] [Google Scholar]