Abstract
Heritable diseases are caused by germ-line mutations that, despite tissuewide presence, often lead to tissue-specific pathology. Here, we make a systematic analysis of the link between tissue-specific gene expression and pathological manifestations in many human diseases and cancers. Diseases were systematically mapped to tissues they affect from disease-relevant literature in PubMed to create a disease–tissue covariation matrix of high-confidence associations of >1,000 diseases to 73 tissues. By retrieving >2,000 known disease genes, and generating 1,500 disease-associated protein complexes, we analyzed the differential expression of a gene or complex involved in a particular disease in the tissues affected by the disease, compared with nonaffected tissues. When this analysis is scaled to all diseases in our dataset, there is a significant tendency for disease genes and complexes to be overexpressed in the normal tissues where defects cause pathology. In contrast, cancer genes and complexes were not overexpressed in the tissues from which the tumors emanate. We specifically identified a complex involved in XY sex reversal that is testis-specific and down-regulated in ovaries. We also identified complexes in Parkinson disease, cardiomyopathies, and muscular dystrophy syndromes that are similarly tissue specific. Our method represents a conceptual scaffold for organism-spanning analyses and reveals an extensive list of tissue-specific draft molecular pathways, both known and unexpected, that might be disrupted in disease.
Keywords: proteomics, systems biology, computational biology
Pathology caused by defects in human genes is usually highly tissue-specific (1–4). In heritable diseases, this suggests that specific spatiotemporal functions of the implicated genes are disrupted due to germ-line mutations. Research on tissue specificity of human diseases has focused on the analysis of single-disease genes in affected tissues (5, 6), and although it has been shown that disease genes generally tend to be expressed in a limited number of tissues (2), it is still unclear in many cases how the tissue-specific expression patterns of disease genes correlate with their pathological manifestations.
Proteomics approaches have established that most gene products exert their function as members of one or more protein complexes (7–11), and that mutations in different proteins participating in the same complex, such as cellular machines, rigid structures, dynamic signaling networks, and posttranslational modification systems, generally lead to similar phenotypes (8, 12, 13). A next logical step is to model entire disease complexes and to analyze the link between tissue specificity of the complexes and the pathological manifestations with which they are associated when defective. However, such efforts are hampered by the lack of adequate coverage on experimental proteomic data in humans and of strategies for systematically analyzing hundreds of diseases, and their related genes and protein complexes, across multiple tissues of the human organism.
Here, we describe a strategy (Fig. 1) for systematically correlating pathological manifestations of diseases with expression patterns of implicated genes and protein complexes across many human tissues. For this analysis we created and validated a number of datasets, including >1,500 disease-associated protein complexes, and to these added tissue and subcellular localization. Then, a method for systematically associating diseases to affected tissues was developed. Across all diseases in the Online Mendelian Inheritance in Man (OMIM) (14) database to which causative genes could be mapped, we analyze the correlation between tissue-specific expression and pathological manifestation both at the cellular level of single-disease genes and for entire disease-associated protein complexes. Finally, we systematically compared the tissue-specific pattern of expression and pathology in cancer-initiating genes and complexes, causing familial cancers, with that of non-cancer disease genes and complexes.
Fig. 1.
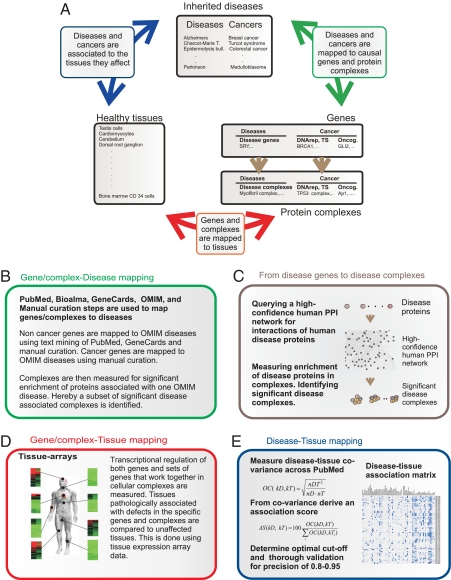
Overview of the study. (A) The different analyses and how they relate to each other. (B) 59 inherited cancers and >1,000 other Mendelian disorders are mapped to 2,227 causative genes and 1,524 complexes by using a combination of automated parsing of OMIM and PubMed. Genes and complexes are stratified into 3 major categories, noncancer disease, cancer gain of function, and cancer loss of function. This stratification is done by a combination of manual curation and semiautomated steps. (C) A unique set of 1,524 protein complexes associated with disease are generated by querying the proteins of disease genes for direct interaction partners in a human protein interaction network followed by several quality control steps. (D) Transcriptional regulation of both genes and sets of genes that work together in cellular complexes are analyzed across tissues of the human organism. (E) Diseases are mapped to relevant tissues by using association degree of particular diseases and tissues across PubMed. Steps are taken to reduce errors in word recognition and handle synonyms accurately. These steps are followed by determination of an optimal cutoff and rigorous quality control. Hereby, we produced a matrix where diseases are mapped to tissues relevant to the pathology with a precision of >0.8. Cancers are mapped to tissues that are the primary origin of tumor formation with a precision >0.95.
Results
Systematic Generation of an Atlas of Disease-Associated Protein Complexes with Tissue Resolution.
By mining the GeneCards (15) resource for genes associated with diseases, we generated a list of 2,227 unique disease-related proteins. Similar to the method that we reported earlier (13), an in silico approach for generating disease-associated protein complexes based on an inferred human protein–protein interaction network was used [see supporting information (SI) Text and Fig. S1]. Following this strategy, we generated 1,524 raw complexes comprising 45,662 unique interactions between 5,202 unique proteins. The quality of the complexes was validated by measures identical to the ones reported in major experimental screens in Saccharomyces cerevisiae, Escherhichia coli, and Homo sapiens (7–11, 16, 17), showing that the quality of our data matches the reproducibility, average probabilistic interaction scores, accuracy, and coverage reported in these studies and that the complexes are true biological entities (see SI Text, Table S1, and Fig. S2). Finally, the complexes were mapped to tissues by using the expression data from 73 nondiseased tissues from the Novartis Research Foundation Gene Expression Database (GNF) (18). The expression level of a complex in a tissue was calculated by averaging over the expression levels of all genes represented in the complex.
Mapping Complexes to Diseases.
To map complexes to diseases we systematically identified the proteins that had been associated to each of the diseases mentioned in OMIM. This was done by using the protein to OMIM mapping displayed in GeneCards (http://www-bimas.cit.nih.gov/cards/) database. We then measured the overlap between proteins in complexes and proteins associated with the diseases and calculated the significance of this overlap. Because a number of complexes are known to be involved in different diseases we allowed for a complex to be associated with more than one disease. In total the 1,524 raw complexes were mapped to 1,054 OMIM diseases. In the further text we refer to these as disease complexes.
Disease–Tissue Association Matrix.
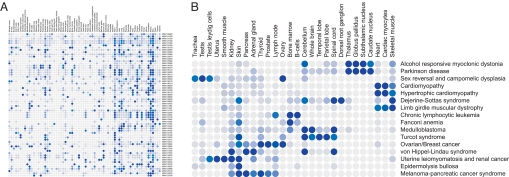
To our knowledge no systematic mapping of diseases to affected tissues exists. We determined the covariance of a disease with a tissue by identifying the number of publications comentioning the disease and tissue (and synonyms thereof), relative to the number of publications mentioning the disease or tissue alone (19). We transformed the covariance into an association score between a tissue and a disease by calculating the fraction of covariance that a given tissue–disease pair constituted, of the total covariance for a given disease. Calculating an association score for the 73 tissues used in the GNF tissue atlas (18) versus 1,054 OMIM diseases yielded a disease–tissue association matrix (Fig. 2). By manually validating the associations we determined a cutoff where tissues associated with the pathology of a given disease could be determined with a precision of >80% (see SI Text, Table S2, and Fig. S3), meaning that above this threshold tissues relevant to the pathology of a given disease can be accurately identified among the GNF atlas tissues in >80% of the cases. High confidence associations scoring above this threshold are blue to dark blue in Fig. 2. Tissues associated with the pathology of a given diseases are in the further text defined as disease–tissue associations scoring above this cutoff.
Fig. 2.
Disease–tissue association matrix. The color range goes from light gray, which corresponds to no association of disease and tissue, to dark blue at 12% association. Only high confidence associations scoring above 8% (blue to dark blue) are used in the further analysis. The percent association is the proportion of a disease's association to a particular tissue in the Novartis Research Foundation Gene Expression Database (GNF) atlas, out of the cumulative association to all tissue in the atlas. (A) The first 100 diseases mapped to the 73 tissues in the GNF atlas. A more detailed view of the matrix can be seen by using the zoom tool. (B) A subset of the disease–tissue associations.
Mapping Complexes to Cancers.
A large number of genes have been associated with cancers, due to aberrant expression or somatic mutations in tumors. However, few of these genes have actually been proven to play a role in the initiation of the tumor. Hence, an automated mapping of cancer genes to complexes would include many genes that are mutated in tumors but do not cause the cancer. Because we are interested in studying the tissue distribution of disease-initiating genes and complexes, we manually created an exhaustive list of heritable cancer genes that initiate tumors through germ-line mutations. These genes were mapped to OMIM diseases describing the cancers manually (Table S3). For this subset of genes, there is compelling evidence that defects are the primary cause of the cancer. In total we extracted a subset of 51 genes in which mutations lead to heritable cancers and mapped them to 59 cancers. Because most cancer mutations are either loss or gain of function that could influence the mechanisms of disease progression and have bearing on the mechanisms of tissue specificity, we further stratified the cancer genes into loss or gain of function as defined in Vogelstein et al. (4). Examples of loss-of-function genes are tumor suppressor or DNA repair genes that become defective when mutated, and examples of gain of function are kinases that become constitutively activated by mutations (Table S4). Cancer-associated complexes were identified as complexes enriched for this subset of genes. In the further text we refer to these as cancer complexes.
Generating a Disease–Tissue Association Matrix for Cancers.
Cancer to tissue association mapping is not straightforward. In this study we were interested in exclusively studying the tissues in which tumors are initiated through germ-line mutations of particular genes. Because cancers generally affect many tissues through downstream effects such as metastases, associations to noninitiating tissues had to be filtered out. Furthermore, many cancer syndromes, arising from germ-line mutations in cancer genes, also include nonmalignant pathology, for which disease–tissue association had to be disregarded in this analysis. For this reason, we manually analyzed the complete subset of tissues associated with heritable cancer syndromes resulting in a precision approximating 100% for the cancer–tissue associations (SI Text and Table S5).
Correlation Between Pathology and Tissue-Specific Expression.
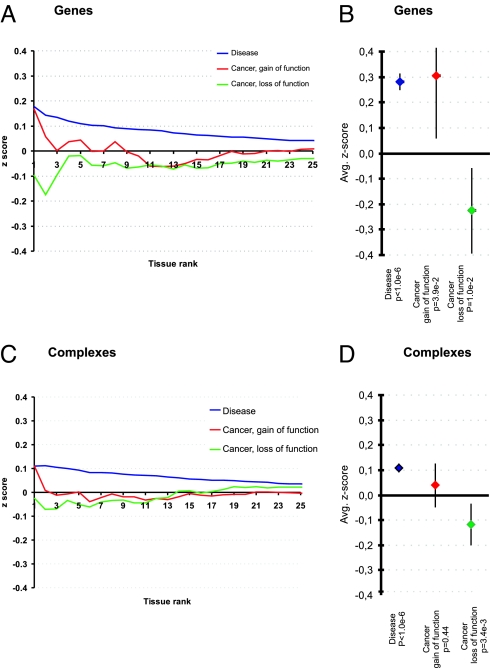
First, we analyzed the expression of disease genes in the tissue with the highest disease association in the disease–tissue matrix (rank 1). This analysis was repeated for the 2nd to 25th highest associated tissues (rank 2 to 25) and the average z score at each rank level was plotted as a curve (Fig. 3A). For example, myosin heavy chain 6 (MYH6) is involved in hypertrophic cardiomyopathy and the tissues from the GNF atlas ranked first and second in relation to hypertrophic cardiomyopathy are heart and cardiac myocytes. We determined the z score of MYH6 in heart (tissue rank 1), the average z score of MYH6 in the 2 highest ranked tissues, heart and cardiac myocytes (tissue rank 2). This procedure is repeated for ranks 3 to 25. This gives a set of rank-dependent z scores for MYH6. This procedure is repeated for every disease gene in every disease yielding rank-dependent z scores for every gene–disease combination, which is plotted in Fig. 3A. This figure shows the clear tendency of overexpression for disease genes in tissues with the highest rank (blue curve). The curves for cancer genes show 2 different trends. Although gain-of-function genes are overexpressed in tissues with the highest rank (red curve), loss-of-function genes are underexpressed (green curve).
Fig. 3.
Expression levels of disease genes and complexes in pathologically associated tissues. (A) The expression level of genes associated with diseases and cancers in the tissues most associated with the particular disease caused by the genes. Tissues are ranked with the most associated tissue at the intersection with the y axis and in declining order from left to right. This plot shows the trend of overexpression for disease genes and gain-of-function cancer genes in tissues with the highest rank. Loss-of-function cancer genes are generally underexpressed in the tissues with the highest rank. (B) The average disease gene expression in associated tissues is shown. Disease genes are overexpressed with an average z score of 0.28 (P < 10E-6). The cancer-associated genes show 2 different trends: gain-of-function genes follow the trend of all disease genes, with an average z score of 0.30 (P = 3.9E-2), but loss-of-function genes have a tendency to be underexpressed in the tissues associated with tumor formation, with an average z score of −0.21 (P = 1.0e-2). (C and D) The same analysis is shown at the level of protein complexes, where the trend is conserved.
To see whether the observed expression trends were significant, we averaged the z scores in the tissues associated with the disease and compared the scores with their expression levels in nonaffected tissues (Fig. 3B). For non-cancer disease genes we observed a significant tendency of overexpression (P < 1.0E-6), which is also the case for gain-of-function cancer genes (P = 3.9E-2), but with less significance. Loss-of-function cancer genes show the converse trend of underexpression (P = 1.0E-2).
We carried out the same analysis for the protein complexes which showed that the expression trend observed for disease genes is conserved at the level of disease protein complexes (see Fig. 3 D and C). These disease complexes display a significant tendency to be overexpressed in tissues where they are involved in pathology (P < 10E-6, blue curve). While protein complexes significantly enriched for gain-of-function cancer genes follow the tendency of overexpression (P = 0.44, red curve), complexes enriched for loss-of-function cancer genes are underexpressed (P = 3.4E-3, green curve).
Because the z scores were lower for the cancer genes and complexes compared with the more robust values of the non-cancer disease genes and complexes, we tested whether this result was influenced by the dataset and normalization method. We replicated the analysis by using a different robust multiarray (RMA)-based normalization scheme (20). Expression data normalized with this algorithm still showed a significant overexpression of disease genes and complexes, but both the over and underexpression trends for the cancer genes and complexes decreased in significance. To test whether a few diseases or tissues were driving the observed trend, we analyzed the expression trend broken down into single tissues (Fig. S4a) and by bootstrapping the dataset both on disease and tissue level. This analysis shows that most tissues and diseases contribute to the observed results and they are robust to bootstrapping of the dataset (Fig. S4b).
Examples of Disease Complexes with Tissue and Phenotype Correlation.
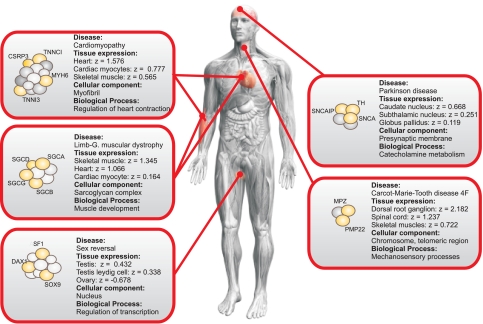
Examples of the correlations found between tissue expression and pathology or phenotype reported are provided in Fig. 4. Also, the most significant gene ontology (GO) subcelluar and functional categories for the complex in question are indicated followed by the significance with which the complex can be assigned to this GO category (21). Tissue names are as defined in the GNF atlas. The full sets of proteins in each complex can be seen in Fig. S5.
Fig. 4.
Representative examples of disease complexes are displayed. Diseases are associated with tissues by using our disease–tissue matrix, and expression data are from the GNF dataset. The expression levels of complexes are shown as z scores. If a disease is associated with more than 3 tissues, only the 3 most associated tissues are shown for clarity. In a given complex, proteins relevant to the disease in question are yellow. The figure shows the general tendency of overexpression of the complexes in the tissues in which they are involved in pathology compared with their expression level in other tissues. All members of the complexes can be seen in Fig. S5.
XY sex reversal can be caused by mutations in the transcription factors SRY (Sex determining Region Y) (22), SOX 9 (the SRY sex determining region Y-box 9 gene) (23), NR5A1 (the nuclear receptor subfamily 5A1), more commonly known as SF1 (24, 25); and NR0B1 (nuclear receptor subfamily 0B1), more commonly known as DAX1 (26). Additionally, SOX 9 is associated with campomelic dysplasia, a bone disorder that leads to a number of associated skeletal and cartilaginous deformities (27). SF1 is needed for gonad and adrenal differentiation (25, 28) and for proper steroidogenesis as well as for Mullerian Inhibiting Substance (MIS) ligand and MIS receptor expression (28, 29). DAX1, which leads to XY sex reversal both when overexpressed, by inhibiting SF1 (26), and when inactivated, as it is required for testis differentiation by regulating expression of SOX9 (30). Although the activity of SF1, DAX1, and SOX9 is required for testis differentiation, development, and maintenance, none of these genes are essential for ovarian development and maintenence (30–33). Here, we identify a transcriptional regulation complex (GO:0006355: P = 1.9E-8) containing DAX1, SF1, and SOX9, all of which are known to be associated with sex reversal (P = 6.9E-6). Furthermore, the complex contains SOX8 that is closely related to SOX9 and implicated in regulating the expression of testis-specific genes (34). Whereas the complex is overexpressed in testis cells, it is underexpressed in ovaries (Fig. 4), which coincides with the known biology of the most well characterized of its components. Our method thus has predictive value because it can (i) detect interactions between molecules that, by themselves, are known to be important in sex differentiation and determination by producing sex reversal, (ii) validate these findings by demonstrating dimorphic tissue-specific expression that correlates with the pathology resulting from inactivation of several members of the complex, and (iii) reveal the importance of new interactors worthy of further study.
Four other complexes, where tissue-specific overexpression correlates with pathological manifestations, are depicted in Fig. 4 (see SI Text and Fig. S5 for more details on these 4 complexes and for examples of cancer-related complexes). These include (i) a complex involved in Charcot–Marie–Tooth disease type 4F and overexpressed in spinal cord, dorsal root ganglion, and skeletal muscles; (ii) a sarcoglycan complex involved in Limb–girdle muscular dystropy overexpressed in skeletal muscle, cardiac myocytes, and heart; (iii) a myofibril complex involved in familial cardiomyopathy overexpressed in several tissues associated with the disease such as heart and cardiac myocytes; (iv) and a complex involved in catechol metabolism and Parkinson disease, overexpressed in a number of relevant brain tissues including the caudate nucleus, subthalamic nucleus, and globus pallidus. Although the overexpression of the sarcoglycan and myofibril complex in muscle tissues is well known, the ovarian–testes dimorphic expression pattern of the sex-reversal complex, and the overexpression of a Parkinson complex in several relevant brain tissues of the basal ganglia are suggestive of the underlying tissue-specific biology of these disorders. Across all examples the tissue-specific expression patterns correlate with the pathological changes observed when one or several members of the complex are defective.
Discussion
The complex dataset reported here is >3 times larger than our reported set of complexes (13) and contains approximately 7 times more interactions than the only reported experimental screen for human complexes (35). To our knowledge, this dataset comprises a unique set of systematically generated complexes with tissue, phenotype, and subcellular resolution in any mammalian organism. The entire atlas is made available online at (http://www.cbs.dtu.dk/suppl/dgf/).
A theoretical limitation of our approach is that we use gene expression data to map complexes to tissues because of the lack of good coverage of quantitative proteomics expression data. Early studies of the relationships between mRNA expression and protein abundance levels have consistently reported modest correlations (36–38). Recent work, which uses a probabilistic framework to model the relationship between the experimentally recorded protein and mRNA patterns, has confirmed that in 75% of all genes tissue mRNA expression patterns linearly correlate with protein abundance, and this overall good correlation is shown for the dataset we use in this work (39). However, to test how a lack of correlation for 25% of the genes affects our results, we randomized 25% of the data points and found that the results achieved for disease genes and complexes, and for loss-of-function cancer genes and complexes, were robust (P < 1.0E-3, see SI Text). Furthermore, the tissue resolution of our complexes is supported by the observation that they are significantly enriched in proteins cooccurring in tissue samples that are analyzed by using manually curated immunohistochemistry data (SI Text and Fig. S2).
Our results support the notion that known disease genes generally are tissue specific (1, 2), by being selectively overexpressed in the tissues in which specific gene defects cause pathology. Alternatively high levels of gene expression may be needed for the functional activity of the tissue. Moreover, we show that this trend is conserved also at the level of the protein complexes in which the disease genes carry out their biological function.
Most known genes that initiate cancer are involved in ubiquitous processes such as DNA repair, cell cycle regulation, and apoptosis (4, 40–42). And it remains a key puzzle in oncology to determine how germ-line mutations in general genes initiate tissue-specific tumors (40). To investigate this contradiction, we also analyzed the expression patterns of cancer genes and complexes involved in heritable cancer syndromes. The gain-of-function cancer genes and complexes follow the trend of non-cancer disease genes and are generally overexpressed in tissues where they initiate tumors, conversely complexes enriched for loss-of-function genes are underexpressed in the tissues where mutations cause neoplastic transformation. Our results for cancer genes and complexes were not robust when different algorithms were used to normalize the expression data. There could be a number of reasons for the lack of a tissue-specificity signal for the analyzed cancer genes and complexes. The current concepts of cancer indicate that some tumors are initiated by a small subset of stem cells (43) whose specific expression levels would be impossible to detect in tissue samples with the resolution used here. Another hypothesis is that tumor initiation is caused by a combination of mutations in a key gene, exposure to mutagenic substances or ionizing radiation, and high proliferation rates of specific cell populations in a tissue (40), a combination we do not analyze here. However, our results highlight the fundamental difference between the tissue specificity of cancers and other diseases, and shows that this difference is consistent on both gene and complex level.
Functional genomics and sequencing have been extremely useful tools for identifying the complete sets of genes in humans and model organisms, and deducing how disruption of different genes in a common molecular pathway can lead to similar phenotypic pathologies. These results indicate how the function of genes is organized in space and time. The next step is to analyze entire systems that are significantly associated with human diseases. This has proven difficult in humans because of experimental limitations and ethical issues, suggesting that other strategies must be considered. Using data integration and systems biology we take a step toward this goal by integrating and refining existing data, and by creating new data sets. Hereby we identify a comprehensive list of functional modules that are associated with pathological processes in humans. We analyze their spatial tissue-specific and subcellular patterns and correlate this information with the diseases that are the result of defects in the modules. As such, our dataset and the scaffold of the analysis presented could be useful in disease systems biology of humans, and provides draft mechanistic pathways that can serve as potential molecular drug targets.
Materials and Methods
Mapping Genes and Complexes to Tissues.
We used the GNF tissue atlas (18) that includes reproduced RNA expression experiments from 79 human tissues. Six tissues were removed because they were derived from cancer tissues. We chose the GNF dataset because it displays high reproducibility (44), and the transcript levels show generally a linear relationship with protein abundance (39). We log-transformed hybridization levels and normalized within each tissue (to ensure equal weight), followed by a normalization across all tissues, thereby ensuring that expression levels represented the relative presence of a transcript in one tissue compared with the other 72 healthy tissues in the dataset. For complexes, the normalized expression levels of all genes in a complex were averaged for each tissue. To test the effect of different normalization methods on our results, we prepared the same dataset with Eklund and Szallasi's normalization method (20) and compared the results.
A Curated Set of Genes in Which Mutations Lead to Tumor Formation.
We curated a set of genes in which mutations had been shown to lead to heritable tumor formation and mapped them to OMIM diseases (see Table S3). By following the definitions introduced by Vogelstein et al. (4) we also noted whether the genes were oncogenes or nononcogenes (such as tumor suppressors or DNA repair proteins) (see Table S4).
Mapping of Complexes to OMIM Diseases.
We calculated the enrichment of proteins involved in the same OMIM disease by using the annotations in GeneCards, which has previously been shown to be an accurate way of mapping genes to diseases (13). We calculated the significance of an enrichment by using a hypergeometric test.
Under- and Overexpression Significance.
We averaged the expression z score over all disease genes in the most disease-associated tissue as determined from the disease–tissue matrix. For each rank from 1 through 25, we calculated the average z score yielding a curve. In Fig. 3A, this curve is plotted as the average z scores of all gene–disease pairs in tissues with a particular rank. This procedure was repeated for gain-of-function and loss-of-function cancer genes. Again this approach was repeated on a protein complex level. All reported significances are 2-tailed using the Student's t test.
Disease-Tissue Association Matrix.
To identify the tissues most affected by diseases described in the OMIM database (14), we used comentioning of a given disease with a given tissue across PubMed (19). The tissue names from the Novartis Research Foundation Gene Expression Database (GNF) (18) were manually curated and translated to corresponding medical subject heading (MeSH) terms (to reduce errors in word recognition and handle synonyms properly). Similarly, the disease names were determined by using disease titles provided in OMIM. Also, these titles were manually curated and translated to the relevant MeSH terms. We used Ochiai's coefficient (OC) as a measure of similarity derived from the cooccurrences (45–47), and calculated an association score (see below), as the percentage of the total normalized cooccurrence of a given disease that could be attributed to a given tissue. Validation was carried out as described in SI Text.
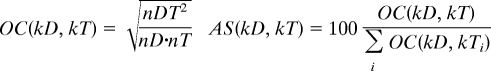 |
Supplementary Material
Acknowledgments.
We thank Matthias Mann, Jiri Bartek, Gert-Jan B. van Ommen, Barbara Pober, and Jonathan Rosand for valuable input on the manuscript and project, Anders Lendager and Lene Hep from MAPT for help with the figures, Zenia Marian Størling for assistance wtih the initial analyses, Kasper Fugger and Christopher Workman for helpful discussions, and Olga Rigina for curating the PPI databases. This work was supported by Villum Kann Rasmussen Foundation, the Simon Spies Foundation, National Institute of Child Health and Development Grant CD RO1 HD0551-50, and the National Institutes of Health
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810772105/DCSupplemental.
References
- 1.Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goh KI, et al. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao EC, Lipkin SM. Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucleic Acids Res. 2006;34:840–852. doi: 10.1093/nar/gkj489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 5.Beyer K, et al. Identification and characterization of a new alpha-synuclein isoform and its role in Lewy body diseases. Neurogenetics. 2008;9:5–23. doi: 10.1007/s10048-007-0106-0. [DOI] [PubMed] [Google Scholar]
- 6.Kim KY, Kee MK, Chong SA, Nam MJ. Galanin is up-regulated in colon adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:2373–2378. doi: 10.1158/1055-9965.EPI-06-0740. [DOI] [PubMed] [Google Scholar]
- 7.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 8.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 9.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 10.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 11.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 12.van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA. A text-mining analysis of the human phenome. Eur J Hum Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 13.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 14.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safran M, et al. GeneCards 2002: Towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18:1542–1543. doi: 10.1093/bioinformatics/18.11.1542. [DOI] [PubMed] [Google Scholar]
- 16.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 17.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 18.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korbel JO, et al. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005;3:e134. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eklund AC, Szallasi Z. Correction of technical bias in clinical microarray data improves concordance with known biological information. Genome Biol. 2008;9:R26. doi: 10.1186/gb-2008-9-2-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camon E, et al. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32(Database issue):D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polanco JC, Koopman P. Sry and the hesitant beginnings of male development. Dev Biol. 2007;302:13–24. doi: 10.1016/j.ydbio.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 23.Patel M, et al. Primate DAX1, SRY, and SOX9: evolutionary stratification of sex-determination pathway. Am J Hum Genet. 2001;68:275–280. doi: 10.1086/316932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker KL. The roles of steroidogenic factor 1 in endocrine development and function. Mol Cell Endocrinol. 1998;140:59–63. doi: 10.1016/s0303-7207(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 25.Park SY, Tong M, Jameson JL. Distinct roles for steroidogenic factor 1 and desert hedgehog pathways in fetal and adult Leydig cell development. Endocrinology. 2007;148:3704–3710. doi: 10.1210/en.2006-1731. [DOI] [PubMed] [Google Scholar]
- 26.Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R. Dax1 antagonizes Sry action in mammalian sex determination. Nature. 1998;391:761–767. doi: 10.1038/35799. [DOI] [PubMed] [Google Scholar]
- 27.Pop R, Zaragoza MV, Gaudette M, Dohrmann U, Scherer G. A homozygous nonsense mutation in SOX9 in the dominant disorder campomelic dysplasia: A case of mitotic gene conversion. Hum Genet. 2005;117:43–53. doi: 10.1007/s00439-005-1295-y. [DOI] [PubMed] [Google Scholar]
- 28.MacLaughlin DT, Donahoe PK. Sex determination and differentiation. N Engl J Med. 2004;350:367–378. doi: 10.1056/NEJMra022784. [DOI] [PubMed] [Google Scholar]
- 29.Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 30.Meeks JJ, Weiss J, Jameson JL. Dax1 is required for testis determination. Nat Genet. 2003;34:32–33. doi: 10.1038/ng1141. [DOI] [PubMed] [Google Scholar]
- 31.Notarnicola C, Malki S, Berta P, Poulat F, Boizet-Bonhoure B. Transient expression of SOX9 protein during follicular development in the adult mouse ovary. Gene Expr Patterns. 2006;6:695–702. doi: 10.1016/j.modgep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Bouma GJ, Washburn LL, Albrecht KH, Eicher EM. Correct dosage of Fog2 and Gata4 transcription factors is critical for fetal testis development in mice. Proc Natl Acad Sci USA. 2007;104:14994–14999. doi: 10.1073/pnas.0701677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biason-Lauber A, Schoenle EJ. Apparently normal ovarian differentiation in a prepubertal girl with transcriptionally inactive steroidogenic factor 1 (NR5A1/SF-1) and adrenocortical insufficiency. Am J Hum Genet. 2000;67:1563–1568. doi: 10.1086/316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schepers G, Wilson M, Wilhelm D, Koopman P. SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem. 2003;278:28101–28108. doi: 10.1074/jbc.M304067200. [DOI] [PubMed] [Google Scholar]
- 35.Ewing RM, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mootha VK, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 37.Griffin TJ, et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 38.Le Roch KG, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kislinger T, et al. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 40.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrocca F, et al. Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res. 2006;66:10287–10291. doi: 10.1158/0008-5472.CAN-06-2289. [DOI] [PubMed] [Google Scholar]
- 42.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 43.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 44.Huminiecki L, Lloyd AT, Wolfe KH. Congruence of tissue expression profiles from Gene Expression Atlas, SAGEmap and TissueInfo databases. BMC Genomics. 2003;4:31. doi: 10.1186/1471-2164-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochiai A. Zoogeographic studies on the soleoid fishes found in Japan and its neighbouring regions. Bull Jpn Soc Sci Fish. 1957;22:526–530. [Google Scholar]
- 46.Jackson DA, Somers KM, Harvey HH. Similarity measures: Measures of co-occurrence and association or simply measures of co-occurrence? Am Nat. 1989;133:436–453. [Google Scholar]
- 47.Udoh E, Rhoades J. Third International Conference on Information Technology: New Generations. Washington, DC: IEEE Computer Society; 2006. pp. 490–494. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.