Abstract
During mitosis different types of cells can have differential requirements for chromosome segregation. We isolated two new alleles of the separation anxiety gene (san). san was previously described in both Drosophila and in humans to be required for centromeric sister chromatid cohesion (Hou et al., 2007; Williams et al., 2003). Our work confirms and expands the observation that san is required in vivo for normal mitosis of different types of somatic cells. In addition, we suggest that san is also important for the correct resolution of chromosomes, implying a more general function of this acetyltransferase. Surprisingly, during oogenesis we cannot detect mitotic defects in germ line cells mutant for san. We hypothesize the female germ line stem cells have differential requirements for mitotic sister chromatid cohesion.
Keywords: Drosophila, germ line, oogenesis, mitosis, sister chromatid cohesion, separation anxiety
Introduction
Drosophila embryonic development starts with thirteen nuclear divisions without cytokinesis (Foe et al., 1993). The nuclei migrate outward to the egg periphery during nuclear division 8 and 9, with most nuclei arriving at the surface of the embryo during interphase 10. After four additional nuclear divisions, the cortical nuclei arrest mitosis during interphase 14. Once arrested, the nuclei become synchronously encased by polarized invaginations of the plasma membrane and a monolayer of epithelial cells is formed de novo.
Sister chromatid cohesion is crucial for chromosome alignment during metaphase (Losada, 2007; Nasmyth and Haering, 2005). The evolutionarily conserved multisubunit cohesin complex is required for sister chromatid cohesion. This complex contains four core subunits: Smc1, Smc3, Scc3, and Scc1/Mcd1/Rad21 (Losada, 2007). In vertebrates, Scc3 has two isoforms: SA1 and SA2 (Losada et al., 2000; Sumara et al., 2000). Another protein, Pds5, is weakly associated with the cohesin complex and may regulate the dynamic interaction of cohesin with chromatin (Hartman et al., 2000; Panizza et al., 2000)
In yeast cohesion is established in multiple steps: before S-phase the Scc2 and Scc4 proteins regulate the chromosomal loading of cohesin at centromeres and at regularly spaced intergenic regions along chromosome arms (Ciosk et al., 2000; Glynn et al., 2004; Tomonaga et al., 2000). During ensuing DNA replication, sister chromatid cohesion is established (Uhlmann and Nasmyth, 1998). The Eco1/Ctf7 protein is the main regulator of this event (Skibbens et al., 1999; Toth et al., 1999). Yeast mutants lacking the acetyltransferase Eco1/Ctf7 (or Eso1 in fission yeast) exhibit defective cohesion despite cohesins continuing to localize to the chromosomes (Skibbens et al., 1999; Tanaka et al., 2000; Toth et al., 1999). Recent studies support the idea that this protein makes a direct connection with the replication fork when establishing cohesion (Lengronne et al., 2006; Moldovan et al., 2006). The cohesin links will then remain until anaphase when they are removed through proteolytic cleavage of Scc1 (Uhlmann et al., 1999).
Meiotic and mitotic sister chromatid cohesion are distinct. Meiotic cells contain specific cohesin subunits, including the α-kleisin Rec8 (Nasmyth and Haering, 2005). Rec8 is crucial for meiotic cohesion and for synaptonemal complex (SC) formation in all organisms studied (Klein et al., 1999; Molnar et al., 1995; Pasierbek et al., 2001). In Drosophila, meiotic cohesion depends on the protein Orientation Disruptor (Ord) (Bickel et al., 1996; Bickel et al., 1997; Miyazaki and Orr-Weaver, 1992). In ord mutants, sister chromatids segregate randomly through both meiotic divisions (Bickel et al., 1997; Webber et al., 2004). Ord is enriched at the centromeres of meiotic chromosomes in both males and females (Bickel et al., 1997; Webber et al., 2004). Smc1 and Smc3 subunits colocalize with Ord at centromeres of ovarian germ line cells and in flies lacking Ord activity, cohesin SMCs fail to accumulate at oocyte centromeres (Khetani and Bickel, 2007).
In Drosophila, the separation anxiety (san) gene encodes an acetyltransferase known to be required for mitotic sister chromatid cohesion in neuroblasts and S2 cells (Williams et al., 2003). san function was associated with sister chromatid cohesion since mutant cells showed loss of the cohesin Scc1 specifically at the centromeres. Recently, san was shown to have a homologue in humans (Arnesen et al., 2006). RNAi experiments depleting SAN in HeLa cells caused defects in sister chromatid cohesion and cohesin SMC1 was no longer detected at the centromeres (Hou et al., 2007).
In this study we isolated two new loss-of-function alleles of san in a forward genetic screen for maternal mutants defective in blastoderm cellularization. We confirm and expand the observation that during mitosis san is required in vivo and in different types of somatic cells for chromosome segregation. In addition, our work suggests that san is also important for chromosome resolution. This implies a more general function of this acetyltransferase and a possible interplay between cohesion and chromosome condensation/resolution. Surprisingly, during oogenesis we cannot detect mitotic defects in germ line cells mutant for san. We hypothesize the female germ line stem cells have differential requirements for mitotic sister chromatid cohesion.
Results
atado is required maternally for the correct segregation of chromosomes during syncytial blastoderm
To identify new genes involved in Drosophila blastoderm cellularization and germ-band extension, we took advantage of a previously reported maternal screen (Barbosa et al., 2007). This screen used the FLP-FRT/ovoD system (Chou and Perrimon, 1992) to generate germ line mutant clones. The screen was carried out in the right-arm of the second chromosome (2R) and 137 independent mutant lines within the “germ cells only” class of mutants were isolated on the basis of an extremely abnormal soma but where the germ cells were formed normally at the posterior pole of the embryo. The secondary screen involved isolation of mutants defective in cuticle production. Absence of cuticle is a good marker for defects in apicobasal polarization of epithelial cells. Secondary screening allowed the isolation of 47 of the initial 137 lines. Complementation studies of these mutants identified 9 complementation groups by zygotic lethality or sterility. Complementation group 2 contained two alleles, which we will initially refer to as atado1 and atado2. These two alleles will later be renamed san3 and san4, respectively.
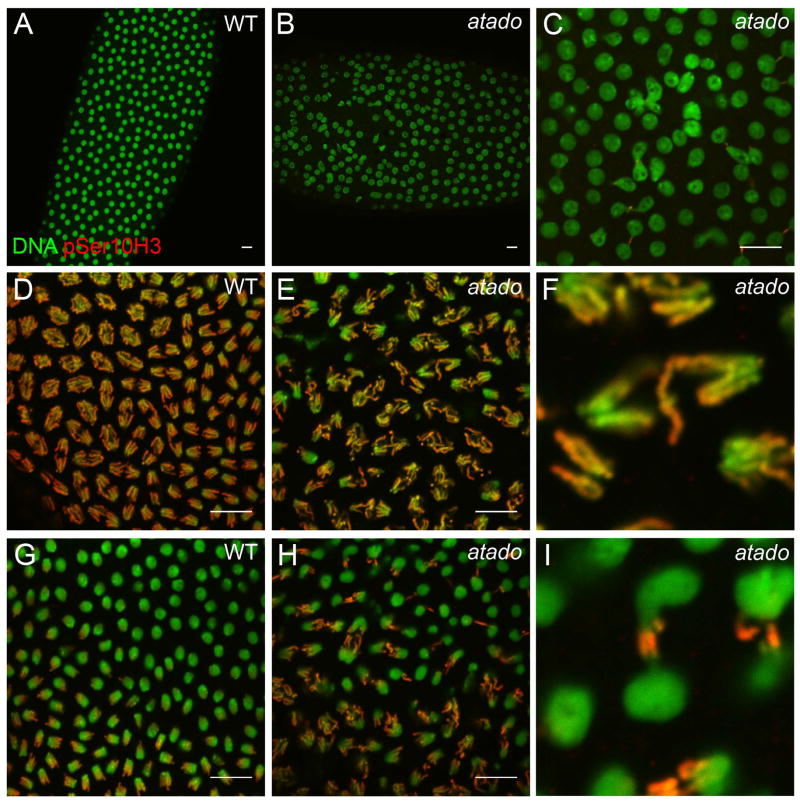
To characterize the role of the atado gene during Drosophila early embryonic development, we examined germ line clone embryos of both alleles of atado. Both mutant alleles had similar maternal phenotypes, with 87% (n=39) of atado2 embryos exhibiting nuclear division abnormalities during syncytial blastoderm (2.5%, n=40, in wild-type embryos) (Supplementary Fig. 1). During syncytial blastoderm development wild-type nuclei divide synchronously and are evenly distributed throughout the embryo (Fig. 1A, D, G). We observed that the atado embryos frequently showed nuclei division asynchrony (data not shown) and irregular distribution of the nuclei (Fig. 1B). Yet, the most striking phenotype in atado embryos was chromosome segregation defects during mitosis (Fig. 1). Anaphase, whilst being mainly bipolar in atado embryos, appeared significantly more disorganized in atado than in wild-type embryos (Fig. 1D, E). atado embryos showed a high frequency of chromosome lagging (Fig 1F) and formation of chromosome bridges (Fig. 1H, I). We also observed interphase nuclei fused together or attached by chromosome bridges (Fig. 1C, Supplementary Fig. 1). Due to these chromatin bridges we refer to our mutant as atado, “tied-up” in Portuguese.
Figure 1. atado is required maternally for chromosome segregation during syncytial blastoderm.
Embryos mutant for atado2 show abnormal anaphases (D, E), with a high frequency of chromosome bridges (A–C, G–I) and chromosome lagging (F). All panels show syncytial blastoderm embryos. atado2 mutant embryos were obtained after the induction of germ line clones (maternal mutants). All embryos were stained for DNA (green) and pSer10 Histone H3 (red). Low magnification of a wild type (A) and atado2 (B) embryo during interphase. (C) Shows a detail of the atado2 embryo shown in B. High magnification of a wild type (D) and atado2 (E) embryo during anaphase. (F) Shows a detail of the atado2 embryo shown in E. High magnification of a wild type (G) and atado2 (H) embryo during late anaphase/telophase. (I) Shows a detail of the atado2 embryo shown in H. pSer10 histone H3 staining marks condensed chromosomes. For quantitative data on the observed chromosome bridges, refer to Figure S1. Scale bars equals 10μm.
Chromosome lagging in atado embryos may be explained by kinetochore abnormalities, but we failed to detect any obvious defects in the localization of Centromere identifier (Cid) during metaphase or anaphase (Supplementary Fig. 2A, B). Chromosome lagging could also be explained by centrosome/mitotic spindle defects. Yet, we did not detect any obvious defects in the localization of Centrosomin (Cnn) (Supplementary Fig. 2C–F), and the mitotic spindle was bipolar and correctly attached to chromosomes and centrosomes (Supplementary Fig. 2E, F).
atado is necessary zygotically for neuroblasts mitosis and imaginal discs development
atado mutations were zygotically lethal. To better characterize the zygotic lethality of atado alleles, we followed the development of transheterozygote atado1/atado2 mutants. All isolated transheterozygote atado larvae reached the third instar larval stage, pupated and died at the pupa stage (n=14). All isolated heterozygous larvae (atado/+) were viable (n=16). Therefore, the maternal contribution of atado was sufficient for development to larval stage, but not for pupa and metamorphosis to the adult.
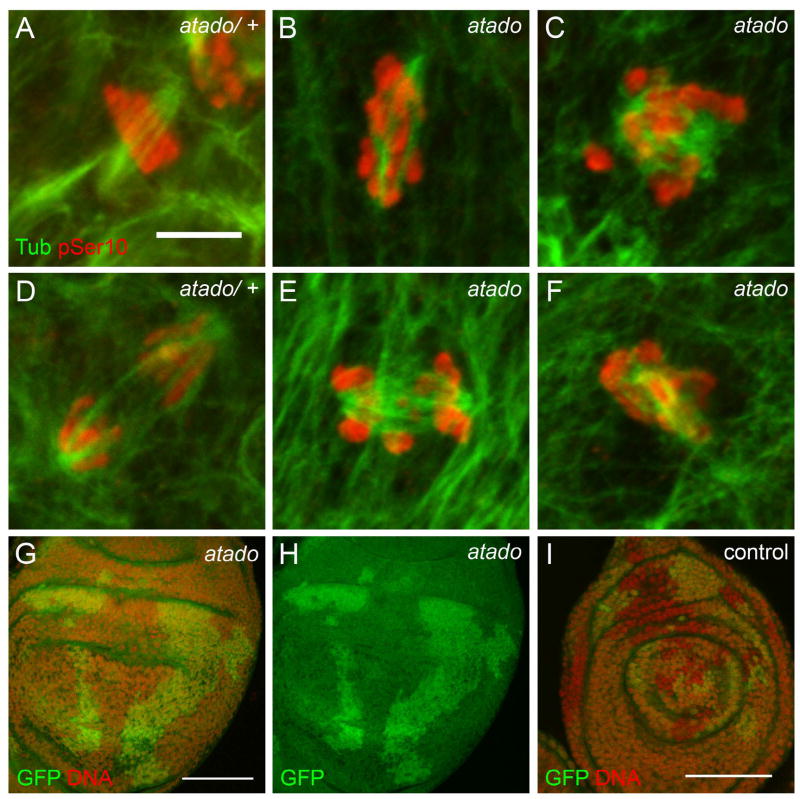
We expected the lethality to be associated with mitotic defects, and hence we analyzed the brains and imaginal discs of transheterozygote third instar larvae. We observed that atado mutant larvae had smaller brains and extremely small imaginal discs (Supplementary Fig. 3, data not shown). This is the typical zygotic phenotype of several cell cycle mutants (Gatti and Baker, 1989; Krause et al., 2001). atado neuroblasts showed chromosome congression defects during metaphase (Fig. 2A, B) and abnormal segregation of chromosomes during anaphase (Fig. 2D, E). To further confirm that atado is important for cell proliferation, we induced atado mutant clones in imaginal discs of an otherwise heterozygote larva (one copy GFP). Imaginal disc clones mutant for atado2 (marked by the absence of GFP) were absent or significantly smaller than the twin-spot wild-type clones (marked by two copies of GFP) (Fig. 2G, H). In contrast, control clones also marked by absence of GFP had a similar size to the twin-spot clones (Fig. 2I). This suggested that atado is required for normal development of larvae imaginal discs.
Figure 2. atado is required zygotically for normal mitosis of larvae neuroblasts and imaginal discs development.
Larvae zygotically mutant for atado2 contain smaller brains (refer to Figure S3) and the neuroblasts show mitotic defects (A–F). Neuroblasts mutant for atado2 show chromosome congression defects during metaphase (A, B) and chromosome lagging during anaphase (D, E). In larvae imaginal discs, clones with two copies of GFP (control clones) are significantly bigger than the twin-clones without GFP (clones mutant for atado2) (G–H). In control imaginal discs both types of clones have equivalent sizes (I). (A–F) Neuroblasts were stained for α-Tubulin (green) and pSer10 Histone H3 (red). (G–I) Imaginal discs were stained for DNA (red). Scale bars equals (A–F) 5μm, (G–H) 50μm, and (I) 30μm.
atado is allelic to separation anxiety (san), a gene required for mitotic sister chromatid cohesion
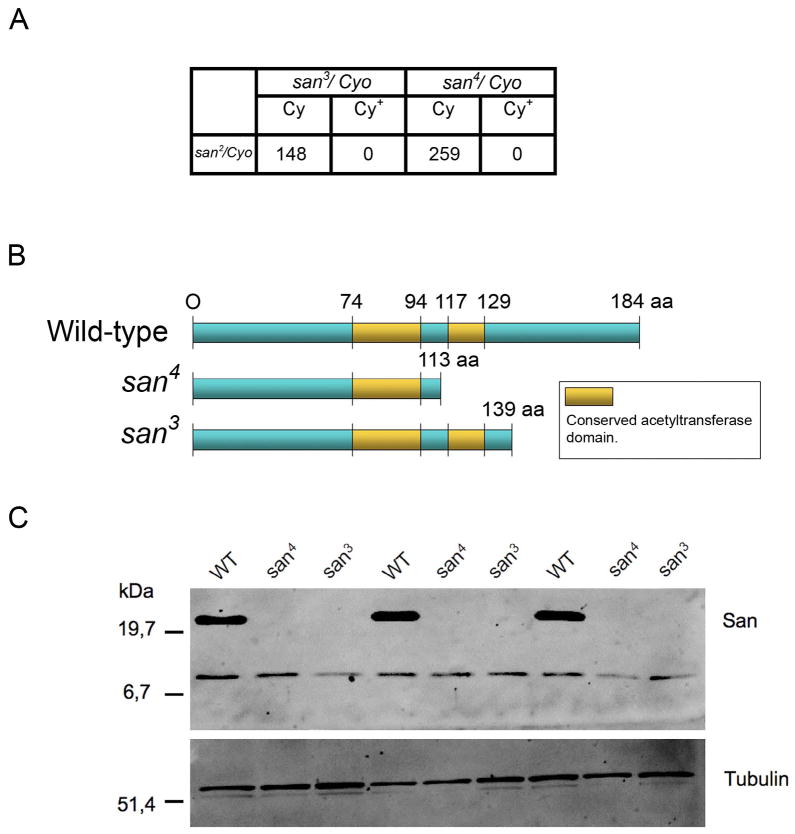
To identify the gene responsible for the atado mutant phenotypes, we mapped both atado alleles using the Bloomington 2R deficiency kit (See Materials and Methods), defining a cytological interval comprising 47 genes. By a candidate gene approach we concluded that atado was most likely allelic to the gene separation anxiety (san) since both atado alleles failed to complement san2 (Fig. 3A), a lethal P-element of the san gene (Williams et al., 2003). Furthermore, san2 germ line clones produced mutant embryos phenotypically indistinguishable from atado1 and atado2 (Supplementary Fig. 4). The san gene was previously predicted to encode an acetyltransferase, which transfers acetyl groups to the N-terminus of other proteins (Williams et al., 2003). San protein contains 184 amino acids and analysis of San primary sequence revealed an acetyltransferase domain composed by two acetyltransferase subdomains from amino acids 74–94 and 117–129, respectively (Fig. 3B) (Williams et al., 2003). Sequencing both alleles of atado confirmed that atado was allelic to san, as both alleles contained distinct nonsense mutations within san open reading frame (ORF). These two nonsense mutations were predicted to cause severe truncations of the San protein (Fig. 3B). We failed to detect San from embryonic total protein extracts (Fig. 3C). The San antibody is polyclonal and it was raised against most of San protein (Williams et al., 2003). It was nevertheless possible that it did not recognize the truncate proteins encoded by the atado alleles. We expressed in bacteria the smallest truncated protein, which was predicted to be encoded by atado2/san4. The San antibody used in this work was able to recognize the recombinant protein in total extracts from bacteria (Supplementary Fig. 5). We concluded that the isolated atado alleles are loss-of-function alleles of san. We therefore renamed atado1 and atado2, respectively to san3 and san4 alleles.
Figure 3. atado is allelic to separation anxiety (san).
Both alleles of atado contain nonsense mutations within san open-reading frame and are protein null for San. The mutations of both alleles of atado were mapped to a small cytological interval using the 2R deficiency kit. Using a candidate gene approach it was observed that both alleles of atado failed to complement a loss-of-function allele of the gene separation anxiety (san2) (A). The atado alleles (atado1 and atado2) were therefore respectively renamed as san3 and san4. Both isolated alleles of san contain nonsense mutations within san open-reading frame, which will putatively lead to the truncation of San protein (B). Total protein extracts from embryos mutant for both alleles of san show absence or undetectable levels of San protein (C).
san is required for chromosome resolution
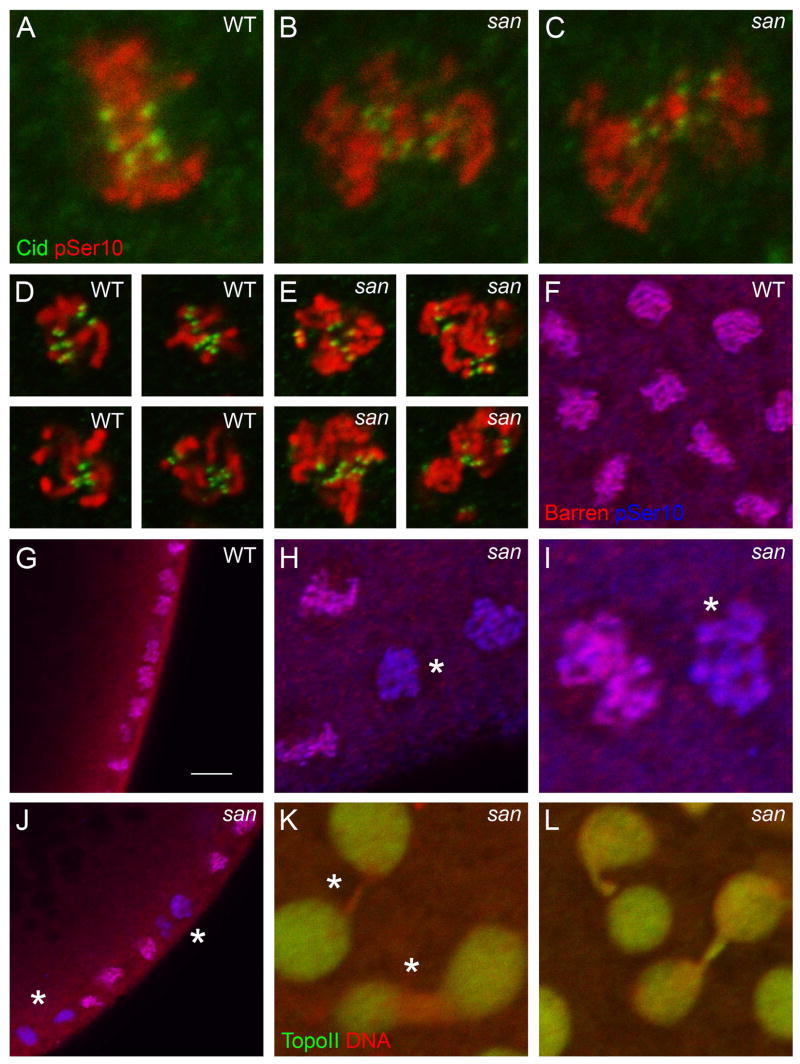
Syncytial blastoderm embryos mutant for san showed dramatic chromosome segregation defects during anaphase (Fig. 1). We observed that during metaphase the defects in chromosome congression and alignment were comparatively mild (Fig. 4A–C and Supplementary Fig. 4). This is in contrast to the dramatic defects in chromosome congression and alignment recently reported in Scc1-depleted embryos (Pauli et al., 2008). Additionally, we only detected a minor separation of the sister chromatid kinetochores (due to the loss of centromeric cohesion) in san mutant embryos arrested in metaphase (Fig. 4D, E). Given this evidence we decided to investigate if San had additional functions that could explain the observed phenotypes.
Figure 4. A subset of nuclei from syncytial blastoderm embryos mutant for san show reduced levels of Barren and Topoisomerase II.
Embryos mutant for san4 show mild defects in chromosome congression and alignment during metaphase (A–C). san4 embryos incubated with colchicine arrest the cell cycle with condensed chromosomes and show a minor separation of the sister chromatid kinetochores (D, E). Subsets of mitotic nuclei in san4 embryos show a dramatic reduction in Barren localization (F–J; asterisks indicates reduction in Barren localization; see text for quantification). Subsets of interphasic nuclei in san4 embryos show reduced levels of TopoII localization specifically in the chromosome bridges (K, L; asterisk indicates reduction of TopoII). All panels show syncytial blastoderm embryos. (A–E) Embryos were stained for Cid (green) and pSer10 Histone H3 (red). (F–J) Embryos were stained for Barren (red) and pSer10 Histone H3 (blue). (K, L) Embryos were stained for Topoisomerase II (green) and DNA (red). (D, E) Embryos were incubated with colchicine for 15 min at room temperature. Scale bar equals 10μm.
Cohesion defects can explain the lagging chromosomes and the high frequency of chromosome bridges observed in san mutant embryos, but we noticed that a large proportion of these bridges did not involve centromeric regions of the chromosome (negative for Cid staining). Of the scored bridges between nuclei in late-mitosis/interphase, 55.6%± 18.2 were negative for Cid staining, 32%±11.5 were positive for Cid staining, and in 12.4%±10 of the cases the result was inconclusive (n= 104 bridges) (Supplementary Fig. 6). Bridges involving distal chromosome regions were previously described in mutants defective in chromosome condensation and/or resolution (Bhat et al., 1996; Steffensen et al., 2001). Consistent with the hypothesis that san could be involved in chromosome resolution/condensation; we observed that during mitosis a subset of nuclei showed a dramatic decrease in the levels of Barren (Fig. 4F–J) (number of cortical nuclei with a detectable reduction of Barren localization, wild-type: 0±0 nuclei, n=780, 11 embryos, san4: 19.7%±12.9 nuclei, n=980, 11 embryos; Student’s t test, 95% confidence interval, p<3×10−5). Embryos mutant for barren are defective in chromosome segregation with the formation of chromosome bridges (Bhat et al., 1996). Although the reduction of Barren localization suggests defects in chromosome resolution and/or condensation, we did not detect reduced levels of this condensin specifically at chromosome bridges (data not shown). Consistent with chromosome resolution defects, during interphase we observed a subset of chromosome bridges showing reduced levels of Topoisomerase II (Fig. 4K, L). TopoII is important for DNA decatenation and chromosome resolution (Holm et al., 1989; Uemura et al., 1987). Chromosomal localization of TopoII is condensin-dependent (Coelho et al., 2003). We did not detect any change in the levels of TopoII in embryo total extracts (data not shown).
During oogenesis san is not required for germ line mitosis
Our data implied san as being important for mitosis of distinct types of somatic cells. This is consistent with previous published work that showed that san is required in vivo in neuroblasts, ex vivo in S2 cells and human HeLa cells, for mitotic sister chromatid cohesion (Hou et al., 2007; Williams et al., 2003). At this point it is important to emphasize that the maternal screen from which we isolated both san3 and san4 alleles was designed to exclude mutants with mitotic defects during oogenesis as egg laying had to be normal. To investigate if egg laying from females with a germ line mutant for san was normal, we induced germ line clones using the FLP-FRT/ovoD system (Chou and Perrimon, 1992). We compared the number of eggs laid by san and control females (with an identical FRT chromosome), and concluded that egg laying between these females was identical for more than 15 days after pupa eclosion (Supplementary Fig. 7) (See Materials and Methods). Since clones were induced at larvae stages by heat-shock, this suggested that san and control germ line stem cells divided continuously and at similar rates for almost 20 days after clone induction.
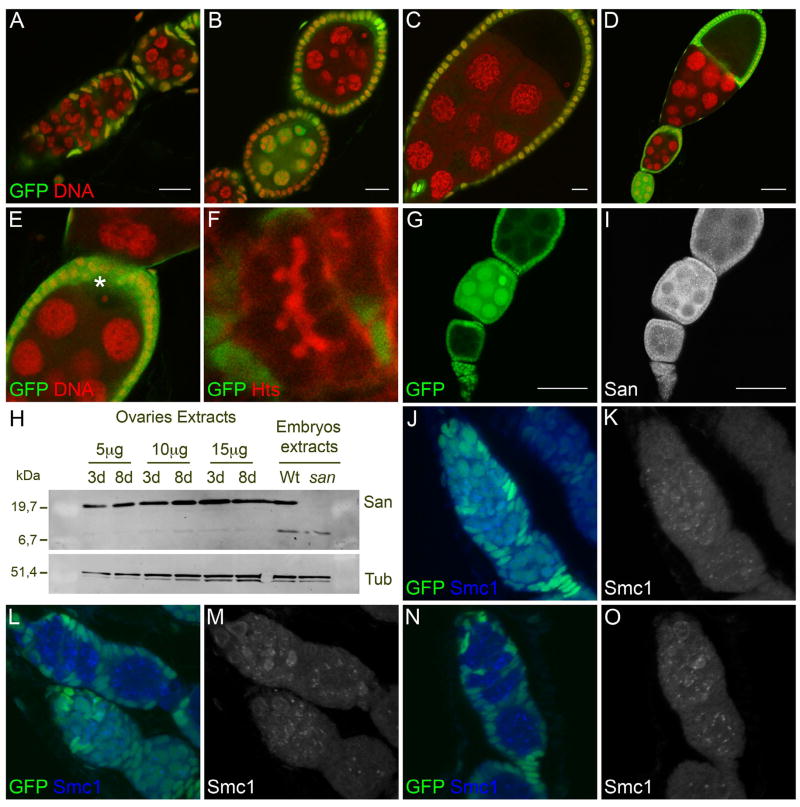
To test the hypothesis that san mitotic function is not required during oogenesis, we generated females with genetically mosaic ovaries using the FLP-FRT-mediated mitotic recombination and a nuclear GFP clone marker. Absence of GFP (green) indicates that the cells are homozygous for san mutations. After the induction of clones at larvae stages, we observed a consistently high frequency of san clones in the adult ovaries (data not shown). This suggested that the proliferating larvae primordial germ cells mutant for san could efficiently compete for the adult germ line stem cell niche. Confirming that san is not required during oogenesis for germ line mitosis, egg chambers mutant for san from females with 7/8 or 15 days old after pupa eclosion showed a normal determination of the oocyte (Fig. 5B–D), a normal condensed karyosome (Fig. 5E - asterisks indicates a condensed karyosome), a normal number of nurse cells (15 nurse cells, n=16), a normal fusome (Fig. 5F), and a normal eggshell without fused dorsal appendages (spindle phenotype) (data not shown). Similar results were obtained with a previously isolated loss-of-function allele of san (san2) (Supplementary Fig. 8A–E).
Figure 5. During oogenesis san is not required for germ line mitosis.
Absence of endogenous GFP (green) indicates that the cells are homozygous for san mutations. Germ line mutant clones of san4 were induced at larvae stages using heat-shock-controlled flipase. Ovaries were dissected from adult females that were 7/8 (A–C, F, G, I–O) or 15 days (D, E) old after hatching (pupal eclosion). Drosophila germarium whose germ line is mutant for san4 develops normally (A). Germ line stem cells mutant for san4 divide normally since egg-laying is identical to the one observed in control females (refer to Figure S7) and egg-chambers mutant for san4 develop normally (A–D), with a normal condensed karyosome (E; asterisk indicates karyosome) and a normal fusome (F). San is expressed in the ovaries (H) and no obvious difference in the expression levels of San could be detected between young (3 days) and older females (8 days). San is expressed within the ovaries germ line (G, I). Germarium whose germ line is mutant for san4 (L–O) show Smc1 expression identical to the one observed in wild-type germarium (J, K). Absence of GFP indicates that the germ-line is mutant for san4. (A–E) Ovaries were stained for DNA (red). (F) Ovary was stained for fusome marker Hts (red). (G, I) Ovary was stained for San (gray). (J–O) Ovaries were stained for Smc1 (blue or gray). (H) Total ovaries and embryonic protein extracts were analyzed by SDS-PAGE and western blot using an anti-San polyclonal antibody. An anti-α-tubulin antibody was used as a loading control. Scale bars equals (A–C) 10μm, (D, G, I) 50μm. (E) Is a detailed view of the image shown in (D).
Since germ line clones mutant for san did not show mitotic defects during oogenesis, we investigated if San is required within the somatic follicle cells. Each Drosophila egg chamber contains 16 germ line cells surrounded by a follicle cell epithelium of somatic origin. Follicle cell clones were induced by heat-shock at larvae stages (as described for germ line clones - See Material and Methods). Whereas we were able to isolate large clones (negative for GFP) using a control chromosome (Supplementary Fig. 8G–L), in the case of san mutants (FRT san2 and FRT san4), the isolated clones were both significantly smaller and less frequent (Supplementary Fig. 8A–F); suggesting a requirement of san function within follicle cells.
We were able to recover san clones within the follicle cells if they were induced in adult flies and if the ovaries were dissected two/three days after induction (data not shown). This was most likely due to San protein perdurance within the follicle cells.
We also tested whether San was expressed in wild-type ovaries. Western blots of total protein extracts showed that San was expressed in the ovaries (Fig. 5H), with no obvious differences in the expression levels detected between younger (3 days) and older females (8 days). Additionally, San immunostaining of mosaic ovaries confirmed San expression within the ovaries germ line, as we detected a specific cytoplasmic staining between the control (presence of GFP) and mutant egg chambers (absence of GFP) (Fig. 5G, I).
Previous studies with san homologue in HeLa cells showed that the inactivation of san expression by RNAi induced an abnormal expression of cohesin SMC1 (Hou et al., 2007). To check if integrity of the cohesin complex during oogenesis was affected in san mutants, mosaic ovaries were stained for Smc1. Consistent with the absence of defects during oogenesis, we could not detect any change in Smc1 localization in germaria whose germ-line was mutant for san (Fig 5J–O), with bright foci and a diffuse staining present as was previously described for wild-type germarium (Khetani and Bickel, 2007). We failed to detect specific immunostaining for Scc1 in wild-type germarium (data not shown). We also did not detect specific immunostaining for Scc1 and Smc1 in the syncytial blastoderm embryo (data not shown).
Discussion
We isolated two new alleles of the san gene: san3 and san4. san was previously described to be required for centromeric sister chromatid cohesion in Drosophila (neuroblasts and ex vivo in S2 cells) and in humans (ex vivo in HeLa cells) (Hou et al., 2007; Williams et al., 2003).
Our work confirms and extends the observation that san is required in vivo for mitosis of different types of somatic cells. It also suggests that in addition to the previously described role in sister chromatid cohesion, san is also important for chromosome resolution. We also propose that female germ line stem cells have differential requirements for sister chromatid cohesion, since we could not detect mitotic defects when they were mutant for san.
san is required in vivo for mitosis of different types of somatic cells
Since the mitotic function of san had only been studied in vivo in Drosophila neuroblasts (Williams et al., 2003), we decided to characterize its function during different stages of Drosophila development. Our analysis shows that san is important for chromosome segregation during syncytial blastoderm nuclei division (maternal phenotype) and, as previously shown, in larvae neuroblast mitosis (zygotic phenotype). We also observe that imaginal discs from larvae mutant for san are extremely small. Clonal analysis confirms san function is required within larvae imaginal discs and adult ovary follicle cells.
san is required for chromosome resolution
Defects in mitotic sister chromatid cohesion in yeast and higher eukaryotes have similar defects in chromosome alignment during metaphase and chromosome lagging during anaphase (Dorsett et al., 2005; Sonoda et al., 2001; Toyoda and Yanagida, 2006; Vass et al., 2003). Drosophila embryos depleted for Scc1 have dramatic defects in chromosome congression and alignment during metaphase (Pauli et al., 2008). In contrast, san mutant embryos have clear defects in chromosome segregation, but only show remarkably mild abnormalities during metaphase. These phenotypic discrepancies suggest that loss of san function causes other problems beyond sister chromatid cohesion defects.
Embryos mutant for san show a high frequency of chromosome bridges during anaphase. Sister chromatid cohesion defects can cause a high frequency of anaphase bridging (Pauli et al., 2008; Vass et al., 2003). Yet a large proportion of chromosome bridges in san embryos are negative for Cid staining. Bridges involving distal regions of the chromosomes were previously described in mutants with chromosome condensation and/or resolution defects (Bhat et al., 1996; Cobbe et al., 2006; Dej et al., 2004; Steffensen et al., 2001).
Chromosome condensation during mitosis relies on the condensin multisubunit protein complex (Hirano, 2005). Together with topoisomerase II, this complex has an important role in organizing the individual axes of sister chromatids prior to their segregation during anaphase (Hirano, 2006). Mutants for the condensin complex SMC4 or gluon and Barren show chromosome segregation defects with the formation of chromosome bridges during anaphase. After depletion of SMC4, TopoII fails to localize to a clearly defined chromatid axial structure and there is a significant decrease in TopoII DNA decatenation activity (Coelho et al., 2003). TopoII is important for chromosome resolution, and inactivation of topoII in Drosophila embryos and S2 cells results in chromosome bridges (Buchenau et al., 1993; Chang et al., 2003).
Given the phenotypic similarities between san, topoII, and barren mutant embryos we investigated if Barren and TopoII expression was abnormal in san mutant embryos. In san mutant embryos a subset of mitotic nuclei show a dramatic decrease in Barren levels, suggesting defects in chromosome condensation. Interestingly, we also observe a subset of the chromosome bridges connecting the interphase nuclei showing reduced levels of TopoII. We hypothesize that san is important not only for sister chromatid cohesion but also for chromosome resolution, which implies a more general function of this acetyltransferase.
Several lines of evidence argue against a cross talk between the cohesin and condensin complexes: 1) cohesins and condensins were isolated as separate complexes in solution (Losada et al., 1998). 2) Condensin DmSMC4 depletion did not alter the localization or removal of cohesins from mitotic chromatin in Drosophila S2 cells (Coelho et al., 2003). 3) In yeast has been shown that although sister chromatid separation did not occur normally in condensin Ycs4 mutants (Cap-D2 in Drosophila), cohesin MCD1/SCC1 was released normally from chromosomes at the metaphase-anaphase transition (Bhalla et al., 2002). 4) In higher eukaryotes cohesion depletion did not appear to affect chromosome condensation (Losada et al., 1998; Sonoda et al., 2001; Vass et al., 2003).
Nevertheless, previous reports in budding yeast have suggested mechanistic interactions between cohesins and condensins (Castano et al., 1996; Lavoie et al., 2002). We show that san is genetically upstream of both sister chromatid cohesion and chromosome resolution/condensation. Our current interpretation is that San acetyltransferase activity is necessary for sister chromatid cohesion and chromosome resolution/condensation without being a core component of these processes. To what extent we are uncovering a cross talk is still unclear, but this is a plausible possibility.
During oogenesis san is not required for germ line mitosis
The female ovary is composed of 16–20 ovarioles (Gilboa and Lehmann, 2004; Spradling, 1993). The germarium is at the anterior tip of each ovariole and is responsible for egg chamber formation. Two to three germ line stem cells are positioned at the anterior tip of each germarium. These cells divide asymmetrically: the most anterior daughter cell keeps a germ line stem cell fate whereas the most posterior daughter cell (the cystoblast) starts a differentiation program that ultimately produces an egg. The cystoblast divides mitotically four times in order to form a cyst of 16 cells connected by ring-canals and a highly branched cytoskeletal structure called fusome. One of these cells becomes the oocyte, the other 15 nurse cells.
In accordance with previously published work (Williams et al., 2003), we clearly demonstrate that san is necessary in vivo for normal mitosis in Drosophila melanogaster. It was therefore surprising that analysis of germ line clones mutant for san did not detect any obvious mitotic defects during oogenesis. 1) The mitotic divisions of the germ line stem cells and cysts mutant for san are normal based on morphological analysis of the DNA, number of nurse cells, and fusome organization. 2) Egg-laying from females whose germ line is mutant for san is equivalent to the control females, even two weeks after pupa eclosion (hatching). This suggests that loss of san function does not reduce the mitotic rate or impair the viability of the germ line stem cells. 3) Egg chambers mutant for san show a normal determination and positioning of the oocyte, and a normal condensed karyosome. 4) Drosophila eggs mutant for san have a normal size and a normal dorsal ventral (DV) patterning (absence of spindle phenotype). If san activity were necessary within the germ line for chromosome segregation we would expect defects in the subsequent stages of oogenesis. Consistent with this expectation, defects in the repair of DNA double strand breaks (DSB) during meiosis activates the DNA damage checkpoint (Ghabrial and Schupbach, 1999), delaying meiosis, inhibiting karyosome condensation, and mutant females lay eggs with DV patterning defects.
A possible explanation for the lack of mitotic abnormalities in females whose germ line is mutant for san is that San protein is extremely stable. We do not favor this hypothesis for four separate reasons: 1) we induce germ line clones for san during larvae development (2nd and 3rd instar larvae) and analyze their effect at least 12 days later in adult females that were 7/8 and 15 days old (after pupa eclosion). 2) We consistently detect mitotic defects in embryos laid by females whose germ line is mutant for san. 3) san zygotic mutants reaching larvae stage is a typical phenotype associated with several previously characterized cell cycle mutants (Gatti and Baker, 1989; Krause et al., 2001). 4) san mutant clones in larvae imaginal discs show defects only three days after clone induction.
The isolated alleles of san contain nonsense mutations within the open reading frame (ORF) and since they are protein-null alleles, the lack of oogenesis defects is not the result of a putative hypomorphic nature of the isolated alleles. Similar results were also obtained with san2, a previously isolated loss-of-function allele of san (Williams et al., 2003). We conclude that during oogenesis san is not required for germ line mitosis.
san is possibly not required during oogenesis due to a functional redundancy with deco acetyltransferase. This is improbable since these two proteins are thought to have different substrates: San was predicted to be an N-acetyltransferase (Williams et al., 2003), and Deco was predicted to acetylate internal lysines (Ivanov et al., 2002). Additionally, San was shown to localize to the cytoplasm during the interphase of Drosophila Kc cells and human HeLa cells (Hou et al., 2007; Williams et al., 2003), whereas members Eco1/Ctf7 family (that includes Drosophila Deco) localize to the nucleus (Hou and Zou, 2005; Skibbens et al., 1999; Toth et al., 1999). Nevertheless, even if the observed results are a consequence of a redundancy between san and deco specifically within the germ line, this is consistent with the hypothesis that during oogenesis female germ line cells have differential requirements for mitotic sister chromatid cohesion (see bellow).
Female germ line cells have differential requirements for mitotic sister chromatid cohesion
Here we have shown that during mitosis different types of cells can have differential requirements for chromosome segregation. We cannot discard the possibility that some somatic cells might not require the mitotic function of san, but our data suggest that female germ line cells have differential requirements for mitotic sister chromatid cohesion. Consistent with this hypothesis a previous analysis of Smc1 germ line clones (focused primarily on prophase I cysts) has not reported mitotic defects during oogenesis (Khetani and Bickel, 2007). Identification of the in vivo substrates of san acetyltransferase should help a better understanding of the molecular nature of these differences.
Materials and Methods
Fly Work and Genetics
atado/san alleles were isolated from a maternal screen previously done in the laboratory of Ruth Lehmann (Barbosa et al., 2007). From this screen we identified 9 complementation groups on the right arm of the second chromosome. These mutants fail to form embryonic cuticle or have scraps of cuticle, however the primordial germ cells are formed properly at the posterior pole of the embryo. Complementation Group 2 contained two alleles that initially were named atado1 and atado2. Later on these alleles were respectively renamed to san3 and san4. All flies were raised at 25°C unless otherwise indicated, using standard techniques.
Germ line clones were induced using the FLP/FRT ovoD system (Chou and Perrimon, 1992). Germ line clones of san3 and san4 were made by crossing FRT42B san/CyO virgins to hs flp; FRT42B ovoD/CyO males and heat shocking the progeny once at 37°C for 1 hour during second and third larval instar.
Mosaic ovaries with the nuclear GFP clone marker were generated by FLP-FRT-mediated mitotic recombination as described (Caceres and Nilson, 2005; Chou and Perrimon, 1992). FRT42B san4/CyO virgins were crossed with yw P[w+, hsFLP]1; P[mini- w+, FRT42B] P[w+; ubi-nls-GFP]/CyOhshid males. The P[w+; ubi-nls-GFP] FRT chromosomes bear a polyubiquitin promoter that drives ubiquitous nuclear green fluorescent protein (GFP) expression. Recombination was induced by a 1 hour heat shock at 37°C during the second and third instar stage. Adult ovaries were harvested from females with 7–8 days and 15 days old, and were subsequently processed for immunofluorescence. Germ line and follicle cell clones were identified by the absence of nuclear endogenous GFP.
To generate clones marked by the absence of GFP in imaginal discs, FRT42B san4/CyOhshid males were crossed with yw P[w+, hsFLP]1; P[mini- w+, FRT42B] P[w+; ubi-nls-GFP]/CyOhshid virgins. The offspring were heat-shocked for 2 hours at 37°C at both 24 and 48 hours after a 24 hour egg collection, corresponding to the first and second larval instar.
To test the egg laying, 1 to 3 days old females from both hs flp; FRT 42B san4/FRT42B ovoD germ line clones and control hs flp; FRT42B/ovoD FRT 42B, where germ line clones had been induced by heat-shock during the second and third larval instar, were mated in parallel to 30 wild-type (Oregon R) males. Egg laying was counted every 24 hours during 15 days.
Both san3 and san4 alleles were balanced over a CyO Actin-GFP to enable isolation of transheterozygous mutant larvae. Mutant larvae were harvested on the basis of lack of GFP and transferred to fresh tubes. Development was followed until pupae stage where san mutant larvae died.
In order to compare san mutant phenotypes with a known lethal P-element insertion on the separation anxiety gene (san) (Williams et al., 2003), we recombined san2 allele with a FRT42B chromosome. This allowed us to generate san2 germ line clones and confirm that they are phenotypically indistinguishable from san3 and san4 germ line clones.
Cloning atado
Both alleles of atado/san gene were mapped using the Bloomington 2R deficiency kit. Deficiencies were crossed with both atado/san alleles and F1 progeny scored for zygotic lethality. The following seven deficiencies failed to complement atado alleles: Df(2R)en-A, Df(2R)en-B, Df(2R)E3363, Df(2R)Exel6060, Df(2R)ix[87i3], Df(2R)ED2219 and Df(2R)ED2155. This allowed us to map atado/san mutation to the cytological interval 47E3-47F5, comprising 47 genes. By a candidate gene approach we concluded that both alleles of atado/san failed to complement a known lethal P-element of the san gene, san2 (Williams et al., 2003).
To molecularly characterize the isolated san mutations, genomic PCR was carried out from heterozygous mutants of san gene (san/Cyo). As a control, and in order to detect DNA polymorphisms, we used a mutant from a different complementation group isolated in the same screen. Two independent genomic PCR fragments from each allele were sequenced and compared with each other, and with the control. Both san alleles have distinct nonsense mutations within san open reading frame that are predicted to cause a truncation of the San protein.
Immunohistochemistry
For phenotypic analysis of san embryos, at 2–3 hours of age, embryos were collected and fixed (after dechorionation in 50% bleach for 5 min) by gentle shaking for 1 hour in 4 mL heptane, 0.125 mL 37% formaldehyde and 0.875 mL PEMS. Fixation was followed of devitellinization by addition of 4 mL methanol and shaking vigorously during 1 minute. Following rehydration, embryos were blocked in phosphate buffered saline (PBS, pH 7.4) containing 0.1% Tween-20, 0.1% bovine serum albumin and 5% serum (BBS), at 4°C overnight. To analyze nuclei arrested in metaphase, san embryos were incubated 15 minutes with 250 μM colchicine, PBS and heptane, prior to fixation.
Primary antibody incubations were carried in BBS at 4°C overnight. Antibodies used were anti-Cid at 1:500 kindly provided by David Glover’s laboratory, anti-Neurotactin at 1:133 (BP106 Hybridoma Bank), anti-pSer10-Histone H3 at 1:1000 (Upstate Biotechnology), anti-Topoisomerase II at 1:400 (Buchenau et al., 1993), anti-Barren at (1:2000) (Bhat et al., 1996), anti-Cnn at 1:1000 kindly provided by Jordan Raff, Anti-α Tubulin clone YL1/2 at 1:50 (Serotec UK). The embryos were washed extensively in PBS containing 0.1% Tween-20 (PBT), reblocked in BBS and incubated with the appropriate secondary antibody for two hours at room temperature (RT). Secondary antibodies were Cy3- and Cy5-conjugated at 1:1000 (Jackson ImmunoResearch Laboratories, West Grove, PA). After extensively washed in PBT, DNA was stained with OliGreen (Molecular Probes, Eugene, OR) at 1:5000 with the addition of 5 μg/mL RNAse A. For scoring the number of cortical nuclei with reduced levels of Barren we focused in embryos where the remaining nuclei (positive for Barren) were at metaphase or early anaphase stages. These are the stages when Barren localization in the chromosomes is the highest.
Ovaries were processed for immunofluorescence as described (Navarro et al., 2004), with exception of DNA staining. Primary antibody was rabbit polyclonal anti-San (Williams et al., 2003) at 1:1000 and secondary antibody was Cy5-conjugated at 1:1000. For DNA staining, ovaries were incubated with 100μg of RNAse/mL for 30 min following incubation with 0.17 μg/mL propidium iodide. Ovaries were 2 times 5-min washed in PBT and 2 rinses in PBS, following mounting. Endogenous GFP was used to distinguish mutant clone egg chambers from chambers that did not have clones.
For ovary staining with anti-Smc1 antibody, ovaries were processed as described (Song et al., 2002). Primary antibody was anti-Smc1 at 1:2000 (Khetani and Bickel, 2007) and anti-Hts at 1:50 (1B1 Hybridoma Bank). Secondary detection was done using rodamin red at 1:1000 (Jackson ImmunoResearch Laboratories) and Cy5-conjugated at 1:1000.
Imaginal discs were dissected in PBS from crawling late third instar larvae. Discs were fixed in 4% formaldehyde with PEMS during 30 minutes on ice, following a 15 min wash in PBS with 0,2% Triton X-100. DNA was stained with propidium iodide as previously described.
Whole brains from transheterozygous mutants of san (san3/san4) were dissected from third instar larvae in PBS and fixed for 20 minutes in 3.7% formaldehyde, 2 mM EGTA in PBS. Briefly, brains were three times 5-min washed with PBS, permeabilized for 10 min in PBS + 0,3% Triton X-100 and blocked in PBS containing 0.1% Triton X-100 and 1% BSA for 1 hour at RT. Anti-α Tubulin at 1:50 and anti-pSer10 Histone H3 at 1:300 were incubated with PBS + 0.1% Triton X-100 and 1% BSA overnight at 4°C. After three 5-min washes, Cy3- and Cy5-conjugated secondary antibodies were incubated for 4 hours at RT. Brains were rinsed with PBS and DNA was stained with propidium iodide as previously described.
Embryos and all tissues were mounted in Fluorescent Mounting Medium (DakoCytomation, Inc) and immunostainings were visualized using a Leica SP5 confocal microscope. All images are confocal sections, with the exception of Fig. 5J–O, Sup. Fig. 4, and Sup. Fig. 6, which are Z-stacks. The Z-stacks projections were obtained using Image J program (Grouped ZProjector, maximum pixel intensity).
Western Blotting
Embryos were collected on apple juice agar after 3 hour egg laying. Each protein sample was collected by lysing 10 embryos in SDS-PAGE sample buffer. Samples were boiled for 5 minutes and loaded on 5×8 cm 12.5% polyacrylamide gels.
Ovaries were dissected from 3- to 8-day-old Oregon females. Ovaries were homogenized in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 0,1% NP-40, 2 mM DTT, 10 mM NaF and protease inhibitor (Roche). A Bio–Rad™ Bradford protein microassay ensured loading of 5, 10 and 15 μg of protein onto 12 % SDS polyacrylamide gels. As for embryo extracts, samples were boiled for 5 minutes in SDS-PAGE sample buffer before loading.
Proteins were then transferred onto Hybond-ECL membranes (Amersham) and western blotting was performed with standard procedure. Briefly, the membrane was blocked in 5% non-fat milk/PBT (0,1% Tween-20, 1xPBS) overnight at 4°C. Primary antibodies were incubated with the membrane overnight at 4°C. Following washes with PBT secondary antibodies were incubated for 4 hours at RT. After washes with PBT, the membranes were detected with an ECL Plus western blotting detection system (GE Healthcare). Primary antibodies used were anti-Cid (1:2000), rabbit polyclonal anti-San (1:1000), rabbit anti-Smc4 (1:1000) (Steffensen et al., 2001), rat anti-Tubulin (1:250). Secondary detection was performed with rabbit and rat HRP-conjugated antibodies used at a final concentration of 1:5000.
Recombinant San4 protein expression and detection
We expressed in bacteria a recombinant C-terminal polyhistidine (6xHis)-tagged fragment of San corresponding to residues 1–113 by cloning a cDNA EcoRI-XhoI fragment into the pET22b vector (Novagene) and expressing it in E. coli BL21. This is the truncated protein predicted to be encoded by atado2/san4. Since the San antibody was generated against a GST-San fusion protein, we cloned san4 cDNA in a His-tag expression vector to avoid cross-reactivity with GST. The San4-6xHis protein was induced with 10mM IPTG in E. coli BL21. Bacterial samples were collected at 90, 120 and 180 minutes from both induced and non-induced cultures. The bacterial pellets from each time point were ressuspended in SDS-sample buffer, boiled for 5 minutes and loaded on two 15% polyacrylamide gels. One gel was stained with brilliant coomassie and the other gel was used for western blotting using the San antibody.
Supplementary Material
Embryos mutant for atado2 show high frequency of chromosome bridges during syncytial blastoderm. (A) Syncytial blastoderm is abnormal in 87% of the embryos mutant for atado2 and 2.5% of the control embryos. (B) Chromosome bridges during late-mitosis and interphase can be observed in 12.3%±20.5 of the nuclei mutant for atado2 (n= 1033 nuclei, 12 embryos) and only 0.7%±1.8 of the control nuclei (n=2081 nuclei, 12 embryos) (Student’s t test, 95% confidence interval, p<0.03). Nuclei were only scored when pSer10 histone H3 staining was absent (or almost absent) from the syncytial nuclei. This allows the exclusion of most nuclei going through mitosis.
Embryos mutant for atado2 apparently have normal kinetochores, centrosomes, and mitotic spindles. Embryos mutant for atado2 show normal localization of Cid (A, B), Cnn (C, D), and α-Tubulin (E, F). All panels show syncytial blastoderm embryos. (A, B) Embryos were stained for Cid (green), DNA (red), and pSer10 H3 (blue). (C, D) Embryos were stained for DNA (green) and Cnn (red). (E, F) Embryos were stained for α-Tubulin (green), DNA (red), Cnn (blue). Scale bars equals (A, B) 10μm and (C, D) 5μm.
Larvae mutant for atado (zygotic mutants) show smaller and morphological abnormal brains. Transheterozygotic atado1/atado2 third-instar larvae show smaller and morphologically abnormal brains (br) when compared to the ones from control larvae (atado/Cyo Actin-GFP). Control larvae were identified using an Actin-GFP transgene. Central ganglia (gl) from atado and control larvae have similar sizes.
Embryos mutant for san2 (germ-line clones) are phenotypically indistinguishable from atado mutant embryos. Embryos mutant for san2 show abnormal anaphases (C, D), with a high frequency of chromosome bridges (E). Chromosome congression during metaphase occurs normally, yet a subset of chromosomes although positive for pSer10 H3 (B) appear to have chromosome condensation problems (A). All panels show syncytial blastoderm embryos. The same embryo was stained for DNA (A) and pSer10 H3 (B). Embryos (C–F) were stained for DNA (green) and pSer10 Histone H3 (red). (D, E) show details of the embryo shown in C. Embryo shown in (F) is wild type. Embryos shown in (A–E) are mutant for san2 (germ-line clones crossed with wild type males). Images shown are Z-stacks obtained using Image J program (Grouped ZProjector, maximum pixel intensity).
San antibody is able to recognize a truncated protein predicted to be encoded by atado2/san4. Western blot from bacteria protein extracts from induced (with IPTG) and mock induced (no IPTG) bacteria show that San antibody is able to recognize a truncated protein predicted to be encoded by atado2/san4. An embryonic extract from wild type embryos is used as a positive control. Since the San antibody was raised against a GST-San recombinant protein, the truncate protein was tagged with a 6xHis-tag in order to avoid GST cross-reactivity.
In san2 embryos, a large proportion of the bridges between interphase nuclei are negative for Cid staining. All panels show interphase nuclei from syncytial blastoderm embryos mutant for san2. A large proportion of bridges were negative for Cid staining (C–F). Yet some were positive for Cid (G, H). Of the scored bridges between nuclei in late-mitosis/interphase: 55.6%± 18.2 were negative for Cid staining, 32%±11.5 were positive for Cid staining, and in 12.4%±10 of the cases the result was inconclusive (n= 104 bridges). All embryos were stained for DNA (green) and Cid (red). Panels (C–I) show details of the embryos shown in (A, B). Images shown are Z-stacks obtained using Image J program (Grouped ZProjector, maximum pixel intensity).
san is not required within the germ line for normal egg laying. The egg-laying of san4 and control females (with an identical FRT chromosome) is identical. Germ line clones using the FRT-ovoD system were induced by heat-shock at larvae stages. For more experimental information please see MATERIALS AND METHODS.
During oogenesis san is required within somatic follicle cells but is not required within the germ line for normal mitosis. san2 is a previously characterized loss-of-function allele of san. Absence of endogenous GFP (green) indicates that the cells are homozygous for san2 (A–F) or control FRT chromosome (G-L). Germ line and follicle cell clones of san2 (A–F) or control FRT chromosome (G–L) were induced at larvae stages using heat-shock-controlled flipase and the ovaries were dissected from adult females that were 7/8 old after hatching (pupal eclosion). Drosophila germarium whose germ line is mutant for san2 developed normally (A–F). Follicle cell clones mutant for san2 were rare and small (B, D, E, F). Follicle cell clones from control FRT chromosome were more frequent and significantly larger than the ones observed for san2 (G–L). Ovaries are stained for DNA (red). Scale bars equal (A, C) 25μm, (E) 50μm, (G) 40μm, and (J) 50μm. The images shown in (A, B), (C, D), (E, F), (G–I), and (J, K) correspond to the same egg chambers.
Acknowledgments
We thank Monica Bettencourt-Dias, Alvaro Tavares, Miguel Godinho, Raquel Oliveira, Richard Hampson, Nicolas Malmanche, and Claudio Sunkel for suggestions that greatly improved the manuscript; the Martinho lab, Monica Bettencourt-Dias, Alvaro Tavares, Gonçalo Costa, and IGC fly community for discussions. We thank A. Arkov, Y. Arkova, N. Kimm, P. Kunwar, A. Renault, H. Sano and H. Zinszner for help in performing the mutagenesis screen from which the two atado alleles were originally isolated. This work was supported by a Marie Curie International Reintegration Grant (029165) and grants from Fundação para a Ciência e Tecnologia [Grants PPCDT/DG/BIA/82013/2006 and PTDC/BIA-BCM/69256/2006]. A.R.M. has a fellowship from Fundação para a Ciência e Tecnologia [SFRH/BD/28767/2006].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnesen T, Anderson D, Torsvik J, Halseth HB, Varhaug JE, Lillehaug JR. Cloning and characterization of hNAT5/hSAN: an evolutionarily conserved component of the NatA protein N-alpha-acetyltransferase complex. Gene. 2006;371:291–5. doi: 10.1016/j.gene.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Barbosa V, Kimm N, Lehmann R. A maternal screen for genes regulating Drosophila oocyte polarity uncovers new steps in meiotic progression. Genetics. 2007;176:1967–77. doi: 10.1534/genetics.106.069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell. 2002;13:632–45. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–14. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Bickel SE, Wyman DW, Miyazaki WY, Moore DP, Orr-Weaver TL. Identification of ORD, a Drosophila protein essential for sister chromatid cohesion. Embo J. 1996;15:1451–9. [PMC free article] [PubMed] [Google Scholar]
- Bickel SE, Wyman DW, Orr-Weaver TL. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 1997;146:1319–31. doi: 10.1093/genetics/146.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchenau P, Saumweber H, Arndt-Jovin DJ. Consequences of topoisomerase II inhibition in early embryogenesis of Drosophila revealed by in vivo confocal laser scanning microscopy. J Cell Sci. 1993;104(Pt 4):1175–85. doi: 10.1242/jcs.104.4.1175. [DOI] [PubMed] [Google Scholar]
- Caceres L, Nilson LA. Production of gurken in the nurse cells is sufficient for axis determination in the Drosophila oocyte. Development. 2005;132:2345–53. doi: 10.1242/dev.01820. [DOI] [PubMed] [Google Scholar]
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 1996;10:2564–76. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Chang CJ, Goulding S, Earnshaw WC, Carmena M. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J Cell Sci. 2003;116:4715–26. doi: 10.1242/jcs.00797. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–53. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–54. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Cobbe N, Savvidou E, Heck MM. Diverse mitotic and interphase functions of condensins in Drosophila. Genetics. 2006;172:991–1008. doi: 10.1534/genetics.105.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PA, Queiroz-Machado J, Sunkel CE. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J Cell Sci. 2003;116:4763–76. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- Dej KJ, Ahn C, Orr-Weaver TL. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168:895–906. doi: 10.1534/genetics.104.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–53. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Odell GM, Edgar BA. The development of Drosophila melanogaster. Vol. 1. C. s. h. l. press; 1993. Mitosis and Morphogenesis in the Drosophila embryo: point and counterpoint; p. 149. [Google Scholar]
- Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–53. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Schupbach T. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat Cell Biol. 1999;1:354–7. doi: 10.1038/14046. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 2004;131:4895–905. doi: 10.1242/dev.01373. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–26. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensins: organizing and segregating the genome. Curr Biol. 2005;15:R265–75. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–68. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Chu CW, Kong X, Yokomori K, Zou H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J Cell Biol. 2007;177:587–97. doi: 10.1083/jcb.200701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–18. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–8. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- Khetani RS, Bickel SE. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J Cell Sci. 2007;120:3123–37. doi: 10.1242/jcs.009977. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Krause SA, Loupart ML, Vass S, Schoenfelder S, Harrison S, Heck MM. Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol Cell Biol. 2001;21:5156–68. doi: 10.1128/MCB.21.15.5156-5168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156:805–15. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–99. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Losada A. Cohesin regulation: fashionable ways to wear a ring. Chromosoma. 2007;116:321–9. doi: 10.1007/s00412-007-0104-x. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–97. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–16. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid misbehavior in Drosophila ord mutants. Genetics. 1992;132:1047–61. doi: 10.1093/genetics/132.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–32. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6:427–35. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–64. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–60. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-Type-Specific TEV Protease Cleavage Reveals Cohesin Functions in Drosophila Neurons. Dev Cell. 2008;14:239–51. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–19. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–7. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, Earnshaw WC, Takeda S. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell. 2001;1:759–70. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- Spradling AC. The development of Drosophila melanogaster. Vol. 1. C. S. H. L. Press; 1993. Development genetics of oogenesis; p. 1. [Google Scholar]
- Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MM, Sunkel CE. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr Biol. 2001;11:295–307. doi: 10.1016/s0960-9822(01)00096-3. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–62. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–69. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–70. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–33. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol Biol Cell. 2006;17:2287–302. doi: 10.1091/mbc.E05-11-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–25. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–18. doi: 10.1016/s0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Webber HA, Howard L, Bickel SE. The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol. 2004;164:819–29. doi: 10.1083/jcb.200310077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–36. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Embryos mutant for atado2 show high frequency of chromosome bridges during syncytial blastoderm. (A) Syncytial blastoderm is abnormal in 87% of the embryos mutant for atado2 and 2.5% of the control embryos. (B) Chromosome bridges during late-mitosis and interphase can be observed in 12.3%±20.5 of the nuclei mutant for atado2 (n= 1033 nuclei, 12 embryos) and only 0.7%±1.8 of the control nuclei (n=2081 nuclei, 12 embryos) (Student’s t test, 95% confidence interval, p<0.03). Nuclei were only scored when pSer10 histone H3 staining was absent (or almost absent) from the syncytial nuclei. This allows the exclusion of most nuclei going through mitosis.
Embryos mutant for atado2 apparently have normal kinetochores, centrosomes, and mitotic spindles. Embryos mutant for atado2 show normal localization of Cid (A, B), Cnn (C, D), and α-Tubulin (E, F). All panels show syncytial blastoderm embryos. (A, B) Embryos were stained for Cid (green), DNA (red), and pSer10 H3 (blue). (C, D) Embryos were stained for DNA (green) and Cnn (red). (E, F) Embryos were stained for α-Tubulin (green), DNA (red), Cnn (blue). Scale bars equals (A, B) 10μm and (C, D) 5μm.
Larvae mutant for atado (zygotic mutants) show smaller and morphological abnormal brains. Transheterozygotic atado1/atado2 third-instar larvae show smaller and morphologically abnormal brains (br) when compared to the ones from control larvae (atado/Cyo Actin-GFP). Control larvae were identified using an Actin-GFP transgene. Central ganglia (gl) from atado and control larvae have similar sizes.
Embryos mutant for san2 (germ-line clones) are phenotypically indistinguishable from atado mutant embryos. Embryos mutant for san2 show abnormal anaphases (C, D), with a high frequency of chromosome bridges (E). Chromosome congression during metaphase occurs normally, yet a subset of chromosomes although positive for pSer10 H3 (B) appear to have chromosome condensation problems (A). All panels show syncytial blastoderm embryos. The same embryo was stained for DNA (A) and pSer10 H3 (B). Embryos (C–F) were stained for DNA (green) and pSer10 Histone H3 (red). (D, E) show details of the embryo shown in C. Embryo shown in (F) is wild type. Embryos shown in (A–E) are mutant for san2 (germ-line clones crossed with wild type males). Images shown are Z-stacks obtained using Image J program (Grouped ZProjector, maximum pixel intensity).
San antibody is able to recognize a truncated protein predicted to be encoded by atado2/san4. Western blot from bacteria protein extracts from induced (with IPTG) and mock induced (no IPTG) bacteria show that San antibody is able to recognize a truncated protein predicted to be encoded by atado2/san4. An embryonic extract from wild type embryos is used as a positive control. Since the San antibody was raised against a GST-San recombinant protein, the truncate protein was tagged with a 6xHis-tag in order to avoid GST cross-reactivity.
In san2 embryos, a large proportion of the bridges between interphase nuclei are negative for Cid staining. All panels show interphase nuclei from syncytial blastoderm embryos mutant for san2. A large proportion of bridges were negative for Cid staining (C–F). Yet some were positive for Cid (G, H). Of the scored bridges between nuclei in late-mitosis/interphase: 55.6%± 18.2 were negative for Cid staining, 32%±11.5 were positive for Cid staining, and in 12.4%±10 of the cases the result was inconclusive (n= 104 bridges). All embryos were stained for DNA (green) and Cid (red). Panels (C–I) show details of the embryos shown in (A, B). Images shown are Z-stacks obtained using Image J program (Grouped ZProjector, maximum pixel intensity).
san is not required within the germ line for normal egg laying. The egg-laying of san4 and control females (with an identical FRT chromosome) is identical. Germ line clones using the FRT-ovoD system were induced by heat-shock at larvae stages. For more experimental information please see MATERIALS AND METHODS.
During oogenesis san is required within somatic follicle cells but is not required within the germ line for normal mitosis. san2 is a previously characterized loss-of-function allele of san. Absence of endogenous GFP (green) indicates that the cells are homozygous for san2 (A–F) or control FRT chromosome (G-L). Germ line and follicle cell clones of san2 (A–F) or control FRT chromosome (G–L) were induced at larvae stages using heat-shock-controlled flipase and the ovaries were dissected from adult females that were 7/8 old after hatching (pupal eclosion). Drosophila germarium whose germ line is mutant for san2 developed normally (A–F). Follicle cell clones mutant for san2 were rare and small (B, D, E, F). Follicle cell clones from control FRT chromosome were more frequent and significantly larger than the ones observed for san2 (G–L). Ovaries are stained for DNA (red). Scale bars equal (A, C) 25μm, (E) 50μm, (G) 40μm, and (J) 50μm. The images shown in (A, B), (C, D), (E, F), (G–I), and (J, K) correspond to the same egg chambers.