Abstract
We have created a resource to rapidly map genetic traits to specific chromosomes in yeast. This mapping is done using a set of 16 yeast strains each containing a different chromosome with a conditionally functional centromere. Conditional centromere function is achieved by integration of a GAL1 promoter in cis to centromere sequences. We show that the 16 yeast chromosomes can be individually lost in diploid strains, which become hemizygous for the destabilized chromosome. Interestingly, most 2n − 1 strains endoduplicate and become 2n. We also demonstrate how chromosome loss in this set of strains can be used to map both recessive and dominant markers to specific chromosomes. In addition, we show that this method can be used to rapidly validate gene assignments from screens of strain libraries such as the yeast gene disruption collection.
THE centromere is the chromosomal element that ensures faithful segregation of duplicated chromosomes to both the mother and daughter during cell division. The yeast centromere is composed of an ∼125-bp sequence that can be subdivided into three functional elements present at all yeast centromeres (Fitzgerald-Hayes et al. 1982; Panzeri et al. 1985). These conserved DNA elements are binding sites for the centromere binding proteins Cbf1, Cse4, Mif2, and the CBF3 complex (Ndc10, Ctf13, Cep3, and Skp1) that together make up the inner kinetochore (reviewed in Cheeseman et al. 2002). The inner kinetochore associates with the central and outer kinetochore proteins to establish a link between the centromere of every chromosome and the mitotic spindle apparatus to ensure proper segregation during mitosis (Cheeseman et al. 2002).
Early experiments designed to study centromeric DNA showed that chromosomal insertion of a strong promoter adjacent to CEN DNA abrogated centromere function (Panzeri et al. 1984). This led to the construction of conditional centromeres by cloning strong regulatable promoters such as GAL1 adjacent to CEN sequences to allow controlled inactivation (Chlebowicz-Sledziewska and Sledziewski 1985; Hill and Bloom 1987). Conditional centromeres have been used to alter the stability of plasmids, YACs and whole chromosomes (Chlebowicz-Sledziewska and Sledziewski 1985; Hill and Bloom 1987; Smith et al. 1990). In a plasmid context, conditional centromeres can be used to amplify copy number since inactivating a CEN sequence relieves copy number control causing plasmid number to increase in the mother cell. At the same time, inactivation of the CEN sequence results in a higher frequency of plasmid loss, which can be selected with a counter-selectable marker. Likewise, when conditional centromeres are placed in a chromosomal context, chromosome loss can be induced to generate 2n − 1 diploids by counterselecting a marker on the conditional chromosome (Hill and Bloom 1987).
We took advantage of the Hill and Bloom (1987) observation to produce a set of 16 centromere-conditional, counterselectable chromosomes in both MATa and MATα strains. Here we show that these strains produce chromosome specific 2n − 1 monosomy and concomitant loss of heterozygosity (LOH). We show that chromosome-specific LOH can be used to map both recessive and dominant mutations using different readouts of the induced homozygous phenotype. This procedure is useful to map any unknown gene to a specific chromosome, which in turn accelerates the refinement of its genetic location. Finally, we show that this same approach can be used to verify that the phenotype of a haploid deletion library strain is indeed due to the “advertised” gene disruption.
MATERIALS AND METHODS
Reagents and yeast media:
G418 was purchased from MediaTech (Herndon, VA). Five (5)-fluoroorotic acid (5-FOA) was purchased from American Bioanalytical (Natick, MA). Taq DNA polymerase was purchased from Continental Laboratory Products (San Diego).
Yeast extract–peptone–dextrose (YPD) medium, synthetic complete (SC) medium, synthetic dropout (e.g., SC −trp) media and 5-FOA medium were prepared as described (Sherman et al. 1986) except that 60 μg/ml of leucine were added to synthetic plates where appropriate. Sporulation medium was prepared as in Klapholz and Esposito (1982). Lithium acetate (LiOAc) transformations were performed as described (Schiestl and Gietz 1989).
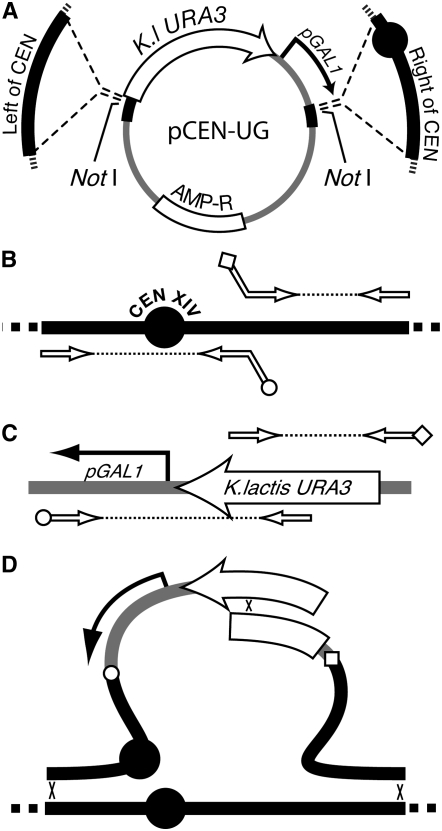
CEN-UG plasmid construction:
A PCR product with the Kluyveromyces lactis URA3 gene was cloned into the BglII and EcoRI sites in pUK21 (Vieira and Messing 1991) to generate plasmid pUK21-KlUra3 (M3739), and a PCR product with the GAL1 promoter was cloned into the BamHI and XbaI sites in plasmid pUC21 to generate plasmid pUC21-GAL (M3738). A FseI–EagI fragment from pUC21-GAL containing the GAL1 promoter was then cloned into pUK21-KlUra3 to generate pUK21-KlUGAL (M3756). A SacI–BssHII fragment from pUK21-KlUra3 was then cloned into pUC21-NotI (Voth et al. 2001) to generate pKlUGAL-NotI (M3828) containing the K. lactis URA3 gene and the pGAL promoter flanked by multiple cloning site cassettes. CEN-proximal DNA sequences (“left of CEN” and “right of CEN”) were PCR amplified from each yeast chromosome for cloning into plasmid pKlUGAL-NotI (see supplemental Table S1 for primer sequences). The PCR primers used to amplify left of CEN DNA (∼800–1200 bp DNA to the immediate left of each centromere), added terminal SacI and SfiI restriction sites. Likewise, PCR primers used to amplify right of CEN DNA (∼800–1200 bp to the right of the centromere) added BssHII and BsiWI restriction sites. The left and right DNA fragments were then digested at the primer-encoded restriction sites for cloning into pKlUGAL-NotI to generate the pCEN-UG plasmid for 15 of the yeast chromosomes (Figure 1A). These plasmids each contain a specific centromere region interrupted by a K. lactis URA3 gene and the GAL1 promoter (Figure 1A). Integrating DNA fragments were liberated from the plasmids by NotI digestion before transformation into strain W303-1A to replace the native centromere. The CEN-proximal insertion for chromosome 14 was not isolated after two attempts using the plasmid-based clone. We therefore moved the GAL1 promoter insertion to the other side of the chromosome 14 centromere using the PCR fusion method described below.
Figure 1.—
Construction of pCEN-UG plasmids and conditional centromere strains. (A) The pCEN-UG plasmids were constructed by amplifying centromere-proximal DNA (thick solid offset lines), including the CEN locus (solid circle) and cloning into plasmid pKlUGAL-NotI. This cloning generates a CEN-proximal region that is interrupted by the K. lactis URA3 gene and a GAL1 promoter. The CEN disruption fragment is liberated from the plasmid by digestion with NotI. AMP-R indicates the beta lactamase gene from the pUK21 plasmid backbone. (B) The CEN14 insertion DNA was made by PCR amplification of centromere-proximal sequences using primers indicated by the open arrows. (C) The promoter-marker cassette from A was amplified using primers indicated by the open arrows. The PCR products from B and C contain sequence tags on the primers that are reverse and complementary so that the resulting DNAs could be fused in a second round of PCR (open circle in B fuses to open circle in C and open diamond in B fuses to open diamond in C). (D) Recombination between the marker segments and genomic integration (denoted by x's) replaces the native centromere with a conditional centromere.
CEN-UG PCR fragment construction:
A conditional centromere for chromosome 14 was constructed by PCR amplification of DNA to the left and right of the chromosome 14 centromere followed by PCR-based fusion to overlapping fragments of the K. lactis URA3-GAL1 promoter cassette (Figure 1, B–D). Amplification and PCR fusions were performed as described (Reid et al. 2002). In the first round of PCR, a 418-bp left DNA fragment that included CEN14 was amplified using primer CHR14newUPR and adaptamer CHR14newUPF (see supplemental Table S1 for primer sequences). A left fragment of the K. lactis URA3-GAL1 cassette from pCEN-UG was amplified using primer Kl-GALintF and adaptamer Kl-GALR. A 567-bp fragment of chromosome 14 DNA adjacent to and right of the centromere was amplified using primer CHR14newDNF and adaptamer CHR14newDNR. A right fragment of the K. lactis URA3-GAL1 cassette was amplified using primer Kl-GALintR and adaptamer Kl-GALF. PCR products from the above reactions were fused in a second round of PCR using the complementary adaptamer tags to generate the illustrated products (Figure 1D). Transformation of the two PCR products into yeast and subsequent recombination generated a strain with a conditional centromere 14 (Figure 1D). All strains containing conditional centromeres were verified by PCR amplification using primers flanking the region of homology paired with primers internal to the K. lactis URA3 coding region. Thus correct insertions could be monitored by amplification of a correct-sized DNA molecule (data not shown).
Induction of chromosome loss:
Diploid strains containing a conditional centromere were made by crossing a CEN-conditional strain to a test strain in patches, incubating on YPD medium for 24 hr at 30°, then replica plating to the appropriate dropout medium to select for diploids. These diploids were then replica plated to rich medium containing galactose as a carbon source (YPgal) and grown for 24 hr at 30° to induce centromere destabilization. Finally the CEN-destabilized strains were streaked onto medium containing 5-FOA to select for cells that have lost the K. lactis URA3 gene and become 2n − 1.
Camptothecin treatment:
Camptothecin (CPT) (Sigma, St Louis) was dissolved in DMSO and added to synthetic media at a concentration of 5 μg/ml with 0.25% DMSO final concentration. Drug-free control plates also contained 0.25% DMSO. Strains tested for CPT sensitivity in the verification experiments contained an additional wild-type TOP1 allele on a CEN vector.
TOP1 expression plasmid:
The copper-inducible promoter from the CUP1 gene was obtained from plasmid pPW66R (Dohmen et al. 1994) after insertion of an MluI linker to replace the DHFR-CDC28 fusion. The pCUP1 promoter was excised on a 440-bp BamHI–MluI fragment and cloned into those same sites in plasmid YCpScTOP1 (Kauh and Bjornsti 1995). The resulting pCUP1-TOP1 fragment was then subcloned into plasmid pRS415 (Sikorski and Hieter 1989) to produce a LEU2-marked CEN plasmid expressing Top1.
RESULTS AND DISCUSSION
Strain construction:
Sixteen haploid yeast strains with conditional centromeres were made by integrating a GAL1 promoter into each chromosome directly adjacent to consensus centromere sequences (Figure 1 and Table 1). These conditional centromeres were constructed by cloning the GAL1 promoter along with K. lactis URA3 between segments of CEN-proximal DNA followed by integration into the genome, or by a PCR-based construction and integration (as in Figure 1). Proper integration of the GAL1 promoter and URA3 into each chromosome was assessed by PCR (see materials and methods) and the correct PCR fragment sizes were obtained for each modified CEN sequence (data not shown). In addition, each haploid strain grew normally on glucose medium but showed reduced viability on galactose medium, indicating that GAL1 promoter induction adjacent to a centromere is not tolerated in a haploid cell (data not shown).
TABLE 1.
Strains
| Strain | Genotype | Source |
|---|---|---|
| DY6281 | MATaCEN1∷pGal1-CEN1-URA3Klaaade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6297 | MATα CEN1∷pGal1-CEN1-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| W3616-3C | MATaCEN2∷pGal1-CEN2-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 RAD5 | This study |
| W3616-3A | MATα CEN2∷pGal1-CEN2-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 RAD5 | This study |
| DY6280 | MATaCEN3∷pGal1-CEN3-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6296 | MATα CEN3∷pGal1-CEN3-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6282 | MATaCEN4∷pGal1-CEN4-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6298 | MATα CEN4∷pGal1-CEN4-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6283 | MATaCEN5∷pGal1-CEN5-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6299 | MATα CEN5∷pGal1-CEN5-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6284 | MATaCEN6∷pGal1-CEN6-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6300 | MATα CEN6∷pGal1-CEN6-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6285 | MATaCEN7∷pGal1-CEN7-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6301 | MATα CEN7∷pGal1-CEN7-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6286 | MATaCEN8∷pGal1-CEN8-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6302 | MATα CEN8∷pGal1-CEN8-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6287 | MATaCEN9∷pGal1-CEN9-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6303 | MATα CEN9∷pGal1-CEN9-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6288 | MATaCEN10∷pGal1-CEN10-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6304 | MATα CEN10∷pGal1-CEN10-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6289 | MATaCEN11∷pGal1-CEN11-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6305 | MATα CEN11∷pGal1-CEN11-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6290 | MATaCEN12∷pGal1-CEN12-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6306 | MATα CEN12∷pGal1-CEN12-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6291 | MATaCEN13∷pGal1-CEN13-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6307 | MATα CEN13∷pGal1-CEN13-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| W3617-1A | MATaCEN14∷pGal1-CEN14-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 RAD5 | This study |
| W3617-1B | MATα CEN14∷pGal1-CEN14-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 RAD5 | This study |
| DY6293 | MATaCEN15∷pGal1-CEN15-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6309 | MATα CEN15∷pGal1-CEN15-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6294 | MATaCEN16∷pGal1-CEN16-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| DY6310 | MATα CEN16∷pGal1-CEN16-URA3Kl ade2-1 can1-100 his3-11,15 leu2-3,112 lys2 met17 trp1-1 ura3-1 rad5-535 | This study |
| K381-15C | MATaura3-1 ade6 arg4 aro7-1 asp5 met14 lys2-1 pet17 trp1 | S. Klapholz |
| K381-10A | MATα ura3-1 ade6 arg4 aro7-1 asp5 met14 lys2-1 pet17 trp1 | S. Klapholz |
| K393-27C | MATaura3-1 his2 leu1-12 lys1-1 met4 pet8 | S. Klapholz |
| K393-2D | MATα ura3-1 his2 lys1-1 met4 pet8 | S. Klapholz |
| K396-27B | MATaura3-1 ade1 his1 leu2 lys7 met3 trp5-d | S. Klapholz |
| K396-11B | MATα ura3-1 ade1 his1 leu2 lys7 met3 trp5-d | S. Klapholz |
| H7C4A1 | MATaura1 ade1 ade2 tyr1 his7 gal1-1 cdc7-4 | L. Hartwell |
| W3758-10B | MATaura3-1 trp1-1 leu2-3 ade2-1 HIS3 can1-100 LYS2 MET17 | This study |
| DY2877 | MATΔ∷LEU2 ho-lacZ swi5∷TRP1 ade2 ade6 can1 his3 leu2 trp1 ura3 | This study |
| DY3930 | MATaCTS1-lacZ∷LEU2 ace2∷HIS3 ade2 can1 his3 leu2 lys2 trp1 ura3 | This study |
| DY4359 | MATα ade6 his4 leu2 thr4 trp1 tyr1 ura3 | This study |
| DY4512 | MATaCTS1-lacZ∷LEU2 ace2∷HIS3 ade2 can1 his3 leu2 ura3 | This study |
| DY6715 | MATα cdc9 trp4 aro1B hom2 ade8 his1 lys11 gal2 ura3 | This study |
| WY352 | MATα CTS1-lacZ∷LEU2 ace2∷HIS3 mbp1-106 Chrom IV-nt 403846∷YIp-ADE2 ade2 can1 his3 leu2 trp1 ura3 | This study |
| W1362-17A | MATaADE2 can1-100 HIS3 LEU2 ura3-1 TRP1 | This study |
URA3Kl is the URA3 ortholog from Kluyveromyces lactis.
Analysis of conditional chromosome loss:
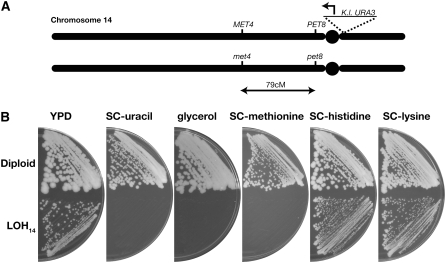
Each strain bearing a conditional chromosome was tested for its ability to lose genetic markers on that chromosome upon galactose-induced promoter activation. These experiments were performed in heterozygous diploids and chromosome loss was ascertained by LOH. In most of these experiments, the CEN-conditional strain was crossed to a test strain with one or more recessive auxotrophic markers covered by the corresponding wild-type alleles on the conditional chromosome. Chromosome loss and subsequent LOH was monitored by loss of the wild-type allele and acquisition of the auxotrophic phenotype. For example, Figure 2A shows a diagram of the relevant genetic markers on chromosome 14 after a cross of the CEN14-conditional strain (W3617-1B) to a tester strain (K393-27C). This diploid is heterozygous for the met4 and pet8 mutations on chromosome 14 and grows on SC −methionine as well as YPG (glycerol) media (Figure 2B). After induction of chromosome 14 loss (LOH14) by growth on synthetic galactose medium followed by selection on 5-FOA medium, the LOH14 strain fails to grow on SC −uracil, SC −methionine, and YPG, indicating the simultaneous loss of three different genetic markers: the CEN-linked URA3 marker, the wild-type MET4, and PET8 alleles. In contrast, this strain remained prototrophic for histidine and lysine showing no LOH for heterozygous markers on chromosomes 6 and 9.
Figure 2.—
Induction of chromosome-14 loss induces LOH for multiple alleles. (A) The conditional CEN strain W3617-1B was mated to strain K393-27C to produce a diploid with the following genotype: MATa/α CEN14/cen14∷K lactis-URA3 ura3-1/ura3-1 MET17/met17 met4/MET4 pet8/PET8 his2/HIS2 HIS3/his3-11,15 lys1-1/LYS1 LYS2/lys2. The resulting diploid contained heterozygous markers for PET8 and MET4 on chromosome 14 drawn approximately to scale. (B) The diploid strain was grown on galactose-containing medium and then 5-FOA medium to induce LOH. The diploid parent and subsequent LOH strain (LOH14) were streaked onto the indicated media to check for viability. Failure to grow signifies loss of URA3, PET8, and MET4.
Each of the 16 CEN-conditional strains was similarly tested for LOH. Table 2 lists the strains used, the markers tested for each CEN-conditional chromosome, and the LOH phenotype observed upon chromosome loss. Multiple independent diploids were produced for each experiment and 20 or more single colonies were examined for LOH. For chromosomes 3, 4, 7, and 14, multiple markers were tested for LOH events. For these and all other chromosomes, the appropriate marker loss was observed for each colony tested. At the same time, many heterozygous markers on chromosomes with unaltered centromeres were analyzed during these experiments and no LOH events were observed for those markers.
TABLE 2.
Crosses to examine chromosome-specific LOH
| Conditional CEN | Diploid genotype | LOH phenotype | Gal-CEN parent | Test parent |
|---|---|---|---|---|
| 1 | ADE1/ade1 | Adenine auxotroph | DY6297/DY6281a | K393-27B/-11B |
| 2 | LYS2/lys2 | Lysine auxotroph | W3616-3C/-3A | K381-15C/-10A |
| 3 | MATa/α | Mating competent | DY6296/DY6280 | K381-15C/-10A |
| 3 | HIS4/his4 | Histidine auxotroph | DY3930b | WY387 |
| 3 | THR4/thr4 | Threonine auxotroph | DY3930b | WY387 |
| 4 | CDC7/cdc7-4 | No growth at 37° | DY6298 | H7C4A1 |
| 4 | MBP1/mbp1 | CTS1-lacZ expression | DY4512b | WY352 |
| 4 | TRP1/trp1 | Tryptophan auxotroph | DY4512b | WY352 |
| 4 | TRP4/trp4 | Tryptophan auxotroph | DY4512b | DY6715 |
| 4 | CDC9/cdc9 | No growth at 37° | DY2877b | DY6715 |
| 4 | ARO1/aro1B | Tyrosine auxotroph | DY2877b | DY6715 |
| 4 | swi5∷TRP1/SWI5 | Tryptophan auxotroph | DY2877b | WY352 |
| 4 | ho-lacZ/ho | Loss of lacZ expression | DY2877b | DY6715 |
| 4 | ADE8/ade8 | Adenine auxotroph | DY2877b | DY6715 |
| 5 | HIS1/his1 | Histidine auxotroph | DY6299/DY6283 | K393-27B/-11B |
| 6 | HIS2/his2 | Histidine auxotroph | DY6300/DY6284 | K393-27C/-2D |
| 7 | ADE6/ade6 | Adenine auxotroph | DY6301/DY6285 | K381-15C/-10A |
| 7 | TRP5/trp5 | Tryptophan auxotroph | DY6301/DY6285 | K393-27B/-11B |
| 8 | ARG4/arg4 | Arginine auxotroph | DY6302/DY6286 | K381-15C/-10A |
| 9 | LYS1/lys1-1 | Lysine auxotroph | DY6303/DY6287 | K393-27C/-2D |
| 10 | MET3/met3 | Methionine auxotroph | DY6304/DY6288 | K393-27B/-11B |
| 11 | MET14/met14 | Methionine auxotroph | DY6305/DY6289 | K381-15C/-10A |
| 12 | ASP5/asp5 | Aspartate auxotroph | DY6306/DY6290 | K381-15C/-10A |
| 13 | LYS7/lys7 | Lysine auxotroph | DY6307/DY6291 | K393-27B/-11B |
| 14 | MET4/met4 | Methionine auxotroph | W3617-1B/-1A | K393-27C/-2D |
| 14 | PET8/pet8 | No growth on glycerol | W3617-1B/-1A | K393-27C/-2D |
| 15 | PET17/pet17 | No growth on glycerol | DY6309/DY6293 | K381-15C/-10A |
| 16 | ARO7/aro7-1 | Tyrosine auxotroph | DY6310/DY6294 | K381-15C/-10A |
Strains separated by slashes indicate congenic MATa and MATα strains.
Strains were transformed to integrate Gal-CEN insertion.
There are multiple reports of haplo-insufficiency for specific yeast genes in a diploid background (Stevens and Davis 1998; Chial et al. 1999; Deutschbauer et al. 2005) and there are also deleterious physiological effects of aneuploidy for 13 yeast chromosomes (Torres et al. 2007). We were therefore surprised to find that whole chromosome LOH events were easily obtained for all sixteen chromosomes. To further investigate these LOH events, we performed meiotic analysis for seven of the crosses listed in Table 2, plus three additional crosses using W303-derived strains. The CEN-conditional strains used for meiotic analysis are listed in Table 3 along with the tester strains that contained recessive auxotrophic marker(s). All 10 diploids were induced for chromosome loss, sporulated, and dissected. The LOH1 and LOH6 strains each showed 2:2 spore viability. All of the viable spores were uracil auxotrophs and the respective diploids also exhibited LOH for markers on chromosomes 1 or 6 (ade1 and his2), demonstrating that, in both cases, the chromosome containing the conditional centromere was lost. In contrast, LOH events for the other seven chromosomes tested in Table 3 showed a preponderance of four viable spores. In these cases, all spore products were uracil auxotrophs and showed 4:0 segregation for the marker listed in the LOH column (Table 3). The MAT locus showed 2:2 segregation as did any additional unlinked markers that were tested (fourth column in Table 3). Thus, it appears that for most whole chromosome LOH events, chromosome loss is followed by endoduplication of an entire chromosome.
TABLE 3.
Meiotic analysis of LOH in diploids
| Chromosome analyzed | No. 4-spore viable tetrads | LOH | Unlinked markers | Gal-CEN parent | Test parent |
|---|---|---|---|---|---|
| 1 | 31 (2:0)a | ade1 | 29:33 CAN1 | DY6281 | K396-27B |
| 2 | 29 | lys2-1 | 2:2 a:α | W3616-3A | K381-15C |
| 4 | 32 | trp1 | 2:2 ade2 | DY6298 | W1362-17A |
| 6 | 30 (2:0)a | his2 | 34:36 a:α | DY6300 | K393-27C |
| 7 | 22 | leu1 | 2:2 TRP1 | DY6301 | K393-27C |
| 10 | 17 | met3 | 2:2 CAN1 | DY6288 | K396-27B |
| 11 | 30 | met14 | 2:2 a:α | DY6305 | K381-15C |
| 12b | 5 | MET17 | 2:2 HIS3 | DY6306 | W3758-10B |
| 15b | 5 | HIS3 | 2:2 MET17 | DY6309 | W3758-10B |
| 16b | 13 | tyr1 | 2:2 a:α | DY6310 | K381-15C |
2:0 indicates two viable spores on rich medium.
One independent diploid analyzed.
We suspect that there is strong selective pressure for such an endoduplication event, especially for the large chromosomes, due to haplo-insufficiency (Stevens and Davis 1998; Chial et al. 1999; Deutschbauer et al. 2005). In support of this notion, we noted that during selection for LOH strains on galactose and 5-FOA media, colonies were heterogeneous in size. After additional passage, these various-sized colonies grow more uniformly, perhaps due to endoduplication. However, it is noteworthy that even after endoduplication, these diploids maintain the LOH, which permits the mapping methods presented below.
Mapping a recessive marker by chromosome LOH:
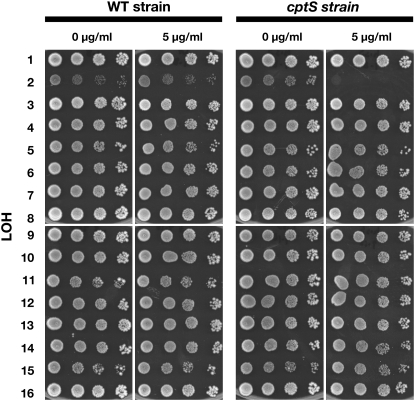
The experiments described above demonstrate that conditional chromosomes efficiently induce LOH after induction. We used conditional LOH induction to develop a method to rapidly map a recessive genetic marker to a specific chromosome. For the first test of this method, we used an unknown camptothecin-sensitive gene (cptS) in the his5Δ strain from the yeast gene disruption library (Winzeler et al. 1999) that we found during a screen for camptothecin sensitivity (R. J. D. Reid and R. Rothstein, unpublished observations). Genetic analysis showed that the cptS gene is unlinked to his5Δ. To map the unknown mutation, a cptS HIS5 segregant was isolated and crossed to each of the 16 CEN-conditional strains. Diploids were selected and induced for chromosome loss by growth on synthetic galactose medium followed by selection on 5-FOA medium. The LOH strains were then grown, serially diluted, and spotted on synthetic medium with and without camptothecin. As shown in Figure 3, the cptS LOH2 strain shows a >10,000-fold growth reduction on camptothecin compared to the no-drug control. A control cross of a wild-type strain to all of the CEN-conditional strains using the same conditions for induction of chromosome loss resulted in no increased camptothecin sensitivity in any particular LOH strain. Although the chromosome 2 heterozygous strain initially grows slowly on galactose, it is the only one that becomes CPT sensitive when the conditional centromere-bearing chromosome is lost (Figure 3). Thus, the unknown camptothecin-sensitive phenotype in the MATα his5Δ∷KanMX deletion strain is due to a recessive gene on chromosome 2.
Figure 3.—
Mapping an unknown camptothecin-sensitive mutant. An unknown gene (cptS) in the MATα library his5Δ∷KanMX strain results in camptothecin sensitivity. The cptS gene was separated from the his5Δ allele by backcross. The resulting mutant strain and a wild-type control were subsequently crossed to the 16 CEN-conditional strains, chromosome loss was induced, and the resulting LOH strains were grown, diluted, and spotted on plates with or without 0 or 5 μg/ml camptothecin. The LOH2 strain crossed to the cptS gene fails to grow in the presence of 5 μg/ml of camptothecin.
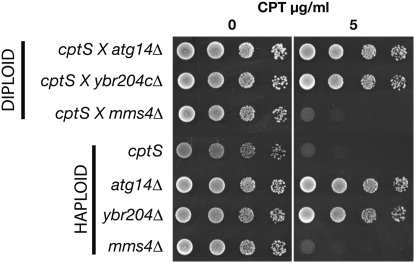
Next, we tested candidate genes on chromosome 2 for complementation of the cptS drug sensitivity. To choose the candidates, we took advantage of the fact that the cptS gene showed very high CPT sensitivity, but was not sensitive to gamma irradiation at 200 Gy (data not shown). A gene on chromosome 2, MMS4, has a similar phenotype when it is mutated (Xiao et al. 1998; Bastin-Shanower et al. 2003). Two other mutants that map on chromosome 2 (the weakly CPT-sensitive atg14Δ∷KanMX mutant and a drug-resistant mutant, ybr204cΔ∷KanMX) were also included in the complementation test (Figure 4). Diploids from the cross of the cptS strain to atg14 and ybr204c are resistant to camptothecin and thus showed complementation. However, diploids from the cross of cptS to mms4Δ fail to complement as they show camptothecin sensitivity comparable to both the mms4Δ and the cptS haploid strains indicating that the unknown cptS gene on chromosome 2 is allelic to MMS4.
Figure 4.—
Complementation analysis of a camptothecin-sensitive gene on chromosome 2. The cptS mutant strain was crossed to three strains with deletions on chromosome 2. The resulting diploids (top three strains) and the parent haploid strains (bottom four) were spotted onto plates with 0 or 5 μg/ml camptothecin to assay drug sensitivity. Failure to complement shows that cptS is allelic to mms4.
Not every case of chromosome-scale mapping will provide potential candidate genes as in the example above. As an alternative, the unknown mutant can be crossed with all of the members of the haploid gene deletion set for a particular chromosome and then be tested for complementation. This is feasible with a simple phenotypic assay such as drug sensitivity. However, even after systematic crosses, the unknown gene may not exist in the haploid deletion library (e.g., essential genes), and this approach may fail to identify the locus. Therefore, another approach to localize the unknown gene is via traditional two-point crosses. For example, chromosome 2 is 813 kb, with a physical-to-genetic distance ratio of 0.3 kb/cM (Cherry et al. 1997). Tetrad analysis can provide a reasonable measure of genetic distance up to 50 cM. Thus approximately eight equally spaced genetic markers on chromosome 2 could be used to map the genetic location of the unknown gene. Since the deletion library is marked with KanMX, appropriately spaced deletion strains can be used directly for this mapping.
Finally, the genetic location can be ascertained systematically with multiple strains from the deletion library using the synthetic genetic array method (Jorgensen et al. 2002). Using this approach, the mutant phenotype is screened after crosses with all of the markers on the chromosome of interest. The use of “magic” markers ensures that after sporulation, only haploid progeny grow, permitting selection for the KanMX marker and subsequent screening for the mutant phenotype. In this case, the gene is localized when it fails to appear in the KanMX segregants that surround its map location.
Mapping a dominant marker by chromosome LOH:
The CEN-conditional strains can also be used to map dominant alleles after LOH induction. In this case, the LOH event must be analyzed after meiosis. For example, a dominant allele on chromosome 2 that is crossed to the CEN2-conditional strain becomes homozygous after LOH2. Since chromosome 2, like most chromosomes, undergoes endoduplication, sporulation and dissection of only the LOH2 strain produces 4:0 spores for the dominant phenotype. In contrast, among the remaining CEN-conditional crosses (note, LOH3 does not sporulate), the dominant allele on chromosome 2 remains heterozygous after LOH for each chromosome and equal numbers of mutant and wild-type meiotic products are produced upon sporulation (2:2). The difference between 2:2 and 4:0 segregation can be easily determined by tetrad analysis. In LOH events where endoduplication does not occur, such as LOH1, the same analysis is made except that tetrads of LOH1 strains are 2:2 for viability. Therefore among the live spore products, an unlinked gene segregates 1:1, while a chromosome 1-linked gene segregates 2:0.
To facilitate the mapping of a dominant gene, a dissection-free protocol can be performed by random spore analysis. In the following example, we show how we confirmed the chromosome map position of a gene disruption from the yeast deletion library by scoring the KanMX dominant selectable marker. The yor364w∷KanMX4 strain from the MATα library, which contains a wild-type CAN1 gene, was crossed to each of the CEN-conditional chromosome strains, which contain the can1-100 mutation. The CAN1 gene is useful in this assay since CAN1/can1-100 diploids and CAN1 haploids are sensitive to canavanine and thus the only canavanine-resistant colonies that grow after sporulation are can1-100 haploids. Diploids were isolated and induced for chromosome loss. The LOH strains were sporulated and the asci were digested with glusulase to disrupt the tetrads and produce single spores (Sherman et al. 1986). Next, the spores were diluted and plated so that ≤300 colonies formed on synthetic medium containing canavanine. The haploid colonies were replica plated to G418 medium and the number of G418-resistant colonies was counted and compared to the total number of canavanine-resistant colonies (Table 4). For this analysis, chromosomes 3 and 5 cannot be assayed since LOH3 events homozygose the MAT locus and do not sporulate, while the can1-100 allele is lost with the conditional chromosome 5. For 13 of the 14 remaining crosses, ∼50% of the colonies in the random spore analysis are G418 resistant indicating random segregation of the KanMX-marked chromosome. In contrast, for the LOH15 strain, all of the canavanine-resistant haploids are also G418 resistant, indicating that the KanMX marker has become homozygous due to LOH. Thus the yor364Δ∷KanMX allele is mapped to chromosome 15 confirming its chromosome position.
TABLE 4.
Random spore analysis of G418 resistance in LOH diploids
| LOH | Can-R | G418-R | G418/Can |
|---|---|---|---|
| 1 | 79 | 39 | 0.49 |
| 2 | 122 | 72 | 0.59 |
| 3 | ND | ND | ND |
| 4 | 124 | 67 | 0.54 |
| 5 | ND | ND | ND |
| 6 | 93 | 55 | 0.59 |
| 7 | 194 | 102 | 0.53 |
| 8 | 28 | 13 | 0.46 |
| 9 | 95 | 52 | 0.55 |
| 10 | 58 | 34 | 0.59 |
| 11 | 63 | 36 | 0.57 |
| 12 | 208 | 124 | 0.60 |
| 13 | 35 | 20 | 0.57 |
| 14 | 276 | 156 | 0.57 |
| 15 | 60 | 60 | 1.00 |
| 16 | 81 | 44 | 0.54 |
There are some limitations to mapping procedures using the CEN-conditional strains described here. For example, any analyses that require sporulation, such as mapping a dominant gene, fail for chromosome 3. Therefore, we assign a gene to chromosome 3 by process of elimination. However, this limitation can be overcome by providing a plasmid-borne (or nonchromosome 3) copy of the same mating-type locus in the CEN3-conditional strains. Thus, when the native chromosome 3 is lost, the additional MAT locus provides mating-type function for sporulation. Another problem occurs during the random spore procedure for meiotic LOH analysis. The can1-100 allele is used to select haploid spore clones. Since LOH5 results in loss of that recessive drug-resistant mutation, we are unable to select haploid strains during plating for chromosome 5. This limitation can be overcome by introducing lyp1 or cyh2 (rpl28) mutations on chromosomes 14 or 7 into the chromosome 5 strain, since these alleles cause recessive drug resistance to thiol-lysine (Grenson 1966) and cycloheximide (Kaufer et al. 1983), respectively.
Comparison to other mapping methods:
Several methods have previously been developed to map a gene to a specific chromosome. In some of these methods, chromosome loss is randomly induced in certain genetic backgrounds, for example by sporulation of a triploid strain as in Wickner (1979). Behavior of the unknown gene is then compared to chromosome-specific markers to narrow the map position to a single chromosome. Another mapping method employs a spo11-1 mutation to suppress recombination during sporulation, thus limiting the segregation of an unknown marker vs. chromosome-specific markers in predictable ways (Klapholz and Esposito 1982). In these random chromosome loss and rec− methods, a mutation must first be crossed into appropriate strain backgrounds before mapping occurs. In contrast, no strain construction is required for our LOH mapping procedure. The unknown mutation need only be in a ura3 strain.
A mapping method that induces specific chromosome loss to allow analysis by LOH was also developed on the basis of 2μ-integration into all 16 yeast chromosomes in cir0 strains (Wakem and Sherman 1990). However in this method of chromosome loss, LOH events can routinely occur for only one chromosome arm depending on the site of 2μ-integration (R. Rothstein and G. Chrebet, unpublished observations). Thus, many more than 16 strains are required for crosses to an unknown to cover the entire genome. On the other hand, the method of chromosome loss induction described here produces LOH for all markers on a given chromosome and thus greatly simplifies the LOH analysis compared to the 2μ-integrations.
Rapid validation of library screens:
The methods described here rely on lack of complementation after an LOH event to identify the chromosome containing a given recessive marker. This principle can be used to validate a screen and verify that the gene responsible for a phenotype is likely linked to the corresponding gene disruption. As an example we used the centromere-conditional strains described in this report to verify all of our “hits” in a recently completed screen of the yeast gene disruption library. In that screen, we expressed a TOP1 allele (top1-T722A) that acts as a CPT mimetic and damages DNA (Megonigal et al. 1997). The full results of this screen will be described elsewhere (R. J. D. Reid and R. Rothstein, unpublished results). In brief, the mutant TOP1 allele was expressed from the copper-inducible CUP1 promoter on a CEN plasmid that had been transferred into each strain in the deletion library. Heterologous expression of the mutant allele induces DNA damage and most of the deletion mutants identified as sensitive to expression of the top1-T722A are also CPT sensitive. Approximately 140 strains showed increased sensitivity to expression of the top1-T722A mutant compared to wild type. To assure that DNA damage sensitivity was due to the specific KanMX-marked mutations, we performed a secondary screen by crossing each of the 140 strains to the conditional centromere strain corresponding to the deletion allele's chromosome. These hybrid diploids also contain an additional wild-type copy of TOP1 on a plasmid, which effectively increases the sensitivity of yeast strains to CPT (Reid et al. 1998). All 140 strains were retested for CPT sensitivity after LOH induction and an example of the retest results from four different chromosome-8 mutants isolated in our screen are shown in Figure 5. These four mutant strains were crossed to the CEN8-conditional strain and induced for LOH8. Any spurious recessive factors residing on another chromosome that might result in DNA damage sensitivity would be complemented in the LOH8 strain. None of the heterozygous diploids, including a wild-type control, exhibits CPT sensitivity before LOH. We found that 3 of the 4 mutant strains are drug sensitive after LOH8 induction, indicating that for those 3 strains the gene disruption on chromosome 8 was the likely cause of drug sensitivity. On the other hand, the dia4 mutant showed no CPT sensitivity in LOH8 diploids and thus we could eliminate further study of this strain. Approximately 70% of the mutants isolated in the top1-T722A mutant screen validated after rescreening by LOH.
Figure 5.—
LOH method to verify camptothecin sensitivity. Four yeast strains with gene deletions on chromosome 8 were identified in a screen for sensitivity to expression of the camptothecin mimetic top1-T722A. These strains were crossed to a CEN8-conditional strain that was transformed with plasmid-borne TOP1 expressed from the CUP1 promoter. The diploids and subsequent LOH8 strains were grown and diluted for spot assays on media with 0 or 5 μg/ml camptothecin (CPT) and 0 or 50 μm copper to induce TOP1 expression and increase camptothecin sensitivity. Only the dia4 disruption shows CPT resistance after LOH8 indicating that the sensitivity of the haploid strain is not due to dia4.
Spurious secondary mutations unlinked to the KanMX insertions have been identified in the deletion library (Lehner et al. 2007) and we showed one such example above (cptS/mms4). In principle, such a mutation can occur on the same chromosome as a library gene disruption and would not be identified as a false positive using the method described here. A hypothetical worst case occurs when verifying chromosome-4 deletion strains. But even though chromosome 4 contains ∼13% of the total coding DNA in yeast, our LOH rescreen would remove 87% of the cases of secondary mutants assuming that these would be distributed randomly on all chromosomes. We assume that the contribution of spurious secondary mutations in these screens is relatively small, so removing ∼90% of such events is quite good.
Summary:
We have produced a set of 16 centromere-conditional chromosomes that can be used to induce LOH. The systematic LOH induction described here provides a simple and effective means of mapping both recessive and dominant alleles to specific chromosomes. In addition, hybrid diploid formation and LOH can be used to complement spurious genetic variation in strains isolated in library screens. Our simple rescreening procedure effectively eradicates such variation and confirms the results from any library screen.
Acknowledgments
We thank Peter Thorpe and David Alvaro for helpful discussions and critical reading of the manuscript. This work was funded in part by National Institutes of Health grants GM57254, GM39067, and GM48624 awarded to D.S.; CA125520, GM50237, and HG01620 awarded to R.R.; and HG00193 awarded to R.J.D.R.
References
- Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen and S. J. Brill, 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M., D. G. Drubin and G. Barnes, 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J. M., C. Ball, S. Weng, G. Juvik, R. Schmidt et al., 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387 67–73. [PMC free article] [PubMed] [Google Scholar]
- Chial, H. J., T. H. Giddings, Jr., E. A. Siewert, M. A. Hoyt and M. Winey, 1999. Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication gene, NDC1, leads to aneuploidy and polyploidy. Proc. Natl. Acad. Sci. USA 96 10200–10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowicz-Sledziewska, E., and A. Z. Sledziewski, 1985. Construction of multicopy yeast plasmids with regulated centromere function. Gene 39 25–31. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer et al., 2005. Mechanisms of haplo-insufficiency revealed by genome-wide profiling in yeast. Genetics 169 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R. J., P. Wu and A. Varshavsky, 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263 1273–1276. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke and J. Carbon, 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29 235–244. [DOI] [PubMed] [Google Scholar]
- Grenson, M., 1966. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. II. Evidence for a specific lysine-transporting system. Biochim. Biophys. Acta 127 339–346. [DOI] [PubMed] [Google Scholar]
- Hill, A., and K. Bloom, 1987. Genetic manipulation of centromere function. Mol. Cell. Biol. 7 2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., B. Nelson, M. D. Robinson, Y. Chen, B. Andrews et al., 2002. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufer, N. F., H. M. Fried, W. F. Schwindinger, M. Jasin and J. R. Warner, 1983. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 11 3123–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauh, E. A., and M. A. Bjornsti, 1995. SCT1 mutants suppress the camptothecin sensitivity of yeast cells expressing wild-type DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 92 6299–6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1982. A new mapping method employing a meiotic rec-mutant of yeast. Genetics 100 387–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, K. R., M. M. Stone, R. A. Farber and T. D. Petes, 2007. Ninety-six haploid yeast strains with individual disruptions of open reading frames between YOR097C and YOR192C, constructed for the Saccharomyces genome deletion project, have an additional mutation in the mismatch repair gene MSH3. Genetics 177 1951–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megonigal, M. D., J. Fertala and M. A. Bjornsti, 1997. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J. Biol. Chem. 272 12801–12808. [DOI] [PubMed] [Google Scholar]
- Panzeri, L., I. Groth-Clausen, J. Shepard, A. Stotz and P. Philippsen, 1984. Centromeric DNA in yeast, pp. 46–58 in Chromosomes Today, edited by M. D. Bennett, A. Gropp and U. Wolf. Allen & Unwin, London.
- Panzeri, L., L. Landonio, A. Stotz and P. Philippsen, 1985. Role of conserved sequence elements in yeast centromere DNA. EMBO J. 4 1867–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, R. J., P. Benedetti and M. A. Bjornsti, 1998. Yeast as a model organism for studying the actions of DNA topoisomerase-targeted drugs. Biochim. Biophys. Acta 1400 289–300. [DOI] [PubMed] [Google Scholar]
- Reid, R. J. D., I. Sunjevaric, M. Keddache and R. Rothstein, 2002. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast 19 319–328. [DOI] [PubMed] [Google Scholar]
- Schiestl, R. H., and R. D. Gietz, 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16 339–346. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. R., A. P. Smyth and D. T. Moir, 1990. Amplification of large artificial chromosomes. Proc. Natl. Acad. Sci. USA 87 8242–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, R. C., and T. N. Davis, 1998. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J. Cell Biol. 142 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli et al., 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317 916–924. [DOI] [PubMed] [Google Scholar]
- Vieira, J., and J. Messing, 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100 189–194. [DOI] [PubMed] [Google Scholar]
- Voth, W. P., J. D. Richards, J. M. Shaw and D. J. Stillman, 2001. Yeast vectors for integration at the HO locus. Nucleic Acids Res. 29 E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakem, L. P., and F. Sherman, 1990. Chromosomal assignment of mutations by specific chromosome loss in the yeast Saccharomyces cerevisiae. Genetics 125 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R. B., 1979. Mapping chromosomal genes of Saccharomyces cerevisiae using an improved genetic mapping method. Genetics 92 803–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Xiao, W., B. L. Chow and C. N. Milo, 1998. Mms4, a putative transcriptional (co)activator, protects Saccharomyces cerevisiae cells from endogenous and environmental DNA damage. Mol. Gen. Genet. 257 614–623. [DOI] [PubMed] [Google Scholar]