Abstract
A proteome-wide mapping of interactions between hepatitis C virus (HCV) and human proteins was performed to provide a comprehensive view of the cellular infection. A total of 314 protein–protein interactions between HCV and human proteins was identified by yeast two-hybrid and 170 by literature mining. Integration of this data set into a reconstructed human interactome showed that cellular proteins interacting with HCV are enriched in highly central and interconnected proteins. A global analysis on the basis of functional annotation highlighted the enrichment of cellular pathways targeted by HCV. A network of proteins associated with frequent clinical disorders of chronically infected patients was constructed by connecting the insulin, Jak/STAT and TGFβ pathways with cellular proteins targeted by HCV. CORE protein appeared as a major perturbator of this network. Focal adhesion was identified as a new function affected by HCV, mainly by NS3 and NS5A proteins.
Keywords: functional analysis, hepatitis C, interactome, virus–host cell
Introduction
Hepatitis C virus (HCV) infection is characterized by a high rate of chronicity and concerns 170 millions of individuals worldwide. Chronically infected patients present liver injury essentially mediated by immune mechanisms and metabolic disorders associated with hepatic steatosis, fibrogenesis and insulin resistance to various extent (Negro, 2006; Moradpour et al, 2007). Long-term-infected patients have a high risk of developing cirrhosis and hepatocarcinoma, but despite considerable efforts, molecular basis of HCV pathology remains poorly understood. HCV genome is a positive-strand RNA of 9.6 kb encoding a polyprotein that is post-translationally processed into structural (CORE, E1, E2 and p7) and non-structural (NS2, NS3, NS4A, NS4B, NS5A and NS5B) proteins (Appel et al, 2006). HCV variants have been classified into six genotypes with biological and antigenic differences. Whereas infection by all genotypes is associated with insulin resistance and fibrosis, a correlation between hepatic steatosis severity and viral replication is preferentially observed for genotype 3. Genotypic differences also correlate with interferon sensitivity, with genotypes 2 and 3 responding better to combined interferon and ribavirine therapy. We focused here on HCV genotype 1b, which is associated with insulin resistance, fibrosis, mild steatosis and poor sensitivity to treatment (Lonardo et al, 2004; Strader et al, 2004).
The rapidly growing knowledge of protein–protein interaction (PPI) networks (interactome) for human, model organisms and host–pathogen begins to provide network-based models for diseases. In a network approach, viral pathogenesis can be viewed as the expression of new constraints on the protein network imposed by the virus when connecting to the cellular interactome. Identification of topological and functional properties that are lost or deregulated, or that emerged in the ‘infection network', becomes a major challenge for a systems understanding of viral infection (Tan et al, 2007).
High-throughput yeast two-hybrid (Y2H) screens of human cDNA library (Calderwood et al, 2007) and computation-based analysis (Uetz et al, 2006) have been used previously to study Epstein–Barr virus (EBV), Kaposi sarcoma herpes virus and varicella zoster virus interactions with host cell factors. Analysis of virus–human protein inter-interactome network revealed that host interactors tend to be enriched in proteins that are highly connected in the cellular network (Calderwood et al, 2007; Dyer et al, 2008). These hub proteins are thought to be essential for the normal cell functioning and during pathogenesis.
Several laboratories have joined their efforts to develop infection mapping project (I-MAP). The goal of I-MAP is to provide a comprehensive view of viral infections at the protein level by mapping the interactions of a large number of viral proteins with host proteins. Screening and mapping have been designed to address specific questions, such as virulence/attenuation, species barrier, identification of therapeutic targets, chronicity and the risk of cancer development.
Here, a proteome-wide mapping approach of interactions between HCV and cellular proteins was performed to provide a comprehensive view of viral infection (Figure 1A). A viral ORFeome was first generated that included ORFs encoding all full-length mature proteins and several protein domains of genotype 1b strain (Supplementary Figure S1). These viral baits were screened against human cDNA libraries using a highly stringent Y2H assay (IMAP Y2H data set). Together with interactions extensively mined and curated from the literature (IMAP LCI data set), this comprehensive host–virus infection network was integrated into a reconstructed human protein–protein interactome. Analysis of the ‘infection network' (V-HHCV; Figure 1A) revealed topological features of cellular interactors and identified functional pathways related to viral biology and pathogenesis.
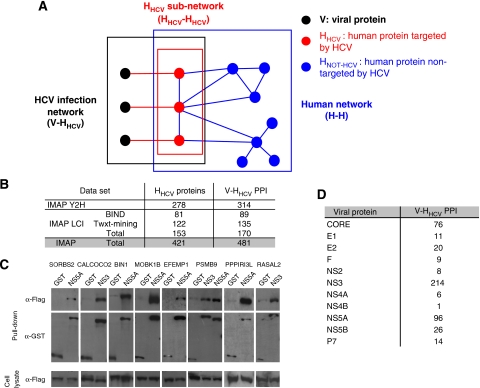
Figure 1.
The HCV interaction network. (A) Nomenclature. V: viral protein (black node). HHCV: human protein interacting with HCV proteins (red node). HNot-HCV: human protein not interacting with HCV proteins (blue node). V-HHCV: HCV–human protein interaction (red edge). HHCV–HHCV: interaction between HCV-interacting human proteins (blue edge). H–H: human–human protein interaction (blue edges). V-HHCV represents the interactions between HCV and human proteins (black box). HHCV–HHCV is composed of human proteins interacting with viral proteins (red box). H–H network represents interactions between human proteins (blue box). (B) Number of proteins and interactions in HCV–human interaction network. Number of human proteins interacting with HCV proteins (HHCV) and corresponding number of protein–protein interactions (V-HHCV PPI). Data are given for our yeast two-hybrid screens (IMAP Y2H) and for literature-curated interactions (IMAP LCI). (C) Validation of Y2H interactions by co-affinity purification assay. Nine out of 22 positive co-AP assays are shown, representing the following: NS5A-SORBS2, NS3-CALCOCO2, NS5A-BIN1, NS5A-MOBK1B, NS5A-EFEMP1, NS3-PSMB9 and NS5A-PSMB9, NS5A-PPPIRI3L, NS3-RASAL2. After pull-down with GST-tagged viral baits or with negative-control GST alone cellular preys are identified with anti-Flag antibody. Anti-GST antibody identifies either GST-alone or GST-tagged viral baits. Expression of cellular preys in cell lysate is controlled by anti-Flag (bottom panel). (D) Number of interactions by viral protein.
Results and discussion
Construction of an HCV–human interactome map
A comprehensive interactome map between HCV and cellular proteins was generated by Y2H screens. Twenty-seven constructs encoding full-length HCV mature proteins or discrete domains were cloned using a recombination-based cloning system (Walhout et al, 2000) (Supplementary Figure S1). Four independent screens were performed with each HCV bait protein, probing two distinct human cDNA libraries, either by mating (IMAP1 screens) or by transformation (IMAP2 screens; see Materials and methods). Fetal brain and spleen cDNA libraries were used instead of a liver library because the liver is known to overexpress a large number of secreted proteins, which could interfere with the quality of the screens. Comparing EST data from fetal brain and spleen with EST data from liver revealed that 87% of genes expressed in the liver are also expressed in brain or spleen (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene). A total of 314 HCV–human PPIs were identified, involving 278 human proteins (Figure 1B, IMAP Y2H data set in Supplementary Table SI). More than 90% of the cellular interactors identified are expressed in the liver. Pairwise interactions between HCV and human proteins were also extracted from the literature by automatic text mining and checked by expert curation (see Materials and methods; IMAP LCI data set in Supplementary Table SI). A total of 135 PPI were extracted from Pubmed and 89 were extracted from BIND database (Bader et al, 2003) (Figure 1B). The resulting HCV–human interactome is thus composed of 481 PPIs with 65% new interactions, involving 11 HCV proteins and 421 distinct human proteins (Figure 1B). IMAP1 and IMAP2 screens share 22 interactions (7% of IMAP Y2H data set). This overlap is in the range of previously reported data using two Y2H high-throughput screening methods (Lim et al, 2006) and suggests that despite screens characteristics, summarized in Supplementary Table SII, saturation has not been reached. Differences in the screening methods, such as sensitivity of the different yeast strains to selective drugs, differential growth rate of colonies and low penetrance of interaction phenotype, could account for this observation. The low redundancy between IMAP Y2H and IMAP LCI data sets may also emphasize a high false-negative rate of the Y2H system, which would be in agreement with recent studies (Rual et al, 2005; Huang et al, 2007). An interesting hypothesis is that different methods of screening may lead to the exploration of different spaces of the HCV–human interactome. As false-positives may also contribute to the weak overlap of IMAP1 and IMAP2, two validation methods were used to assess the confidence of the IMAP Y2H data set. Two-thirds of the data set was retested by direct Y2H between viral protein baits and cellular protein preys identified by our Y2H screens (Y2H pairwise matrices). From the remaining interactions, 26 PPIs (25%) were retested by co-affinity purification and 22 PPIs could be validated (validation rate: 85%; Figure 1C and Supplementary Table SI). This Y2H data set was thus of very high confidence for further analysis at the topological and functional levels. In Table I, the top 21 new interactions validated by GST pull-down experiments and identified in one or two screens are shown. Analysis of the HCV-infection network (V-HHCV, Figure 1A) showed that NS3, NS5A and CORE are the most connected proteins, with 214, 96 and 76 cellular partners, respectively, highlighting the potential multi-functionality of these proteins during infection (Supplementary Table SI, Figure 1D). Highly interacting proteins are known to be significantly more disordered than low-degree (LD) proteins (Haynes et al, 2006). Interestingly, NS3, NS5A and CORE are the only HCV proteins predicted to contain at least one intrinsic disordered region, according to DISOPRED2 (Ward et al, 2004) (prediction of protein disorder server; data not shown). This correlates well with the high degree (HD) of these proteins. In addition, 45 cellular proteins are targeted by more than one viral protein, suggesting their essentiality for virus biology (Calderwood et al, 2007) (Supplementary Table SIII).
Table 1.
Top 21 HCV–human protein–protein interactions
| HCV protein | Human gene name | Human protein official full name | Biological process |
|---|---|---|---|
| NS3 | CALCOCO2 | Calcium binding and coiled-coil domain 2 | May have a function in viral life cycles |
| NS3 | EIF4ENIF1 | Eukaryotic translation initiation factor 4E nuclear import factor 1 | Protein nuclear import |
| NS3 | FRS3 | Fibroblast growth factor receptor substrate 3 | FGF receptor signalling pathway |
| NS3 | CCDC21 | Coiled-coil domain containing 21 | Unknown |
| NS3 | BCKDK | Branched-chain ketoacid dehydrogenase kinase | Branched-chain family amino-acid catabolic process |
| NS3 | GBP2 | Guanylate-binding protein 2, interferon-inducible | Immune response |
| NS3 | KPNA1 | Karyopherin alpha 1 (importin alpha 5) | NLS-bearing substrate import to the nucleus |
| NS3 | PSMB9 | Proteasome subunit, beta type, 9 | Ubiquitin-dependent protein catabolic process |
| NS3 | RASAL2 | RAS protein activator-like 2 | Regulation of small GTPase-mediated signal transduction |
| NS3 | SMURF2 | SMAD-specific E3 ubiquitin protein ligase 2 | Regulation of TGFβ receptor signalling pathway |
| NS5A | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | Integrity of fascia connective tissues |
| NS5A | GOLGA2 | Golgi autoantigen, golgin subfamily a, 2 | Vesicular transport |
| NS5A | GPS2 | G protein pathway suppressor 2 | JNK cascade |
| NS5A | ITGAL | Integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) | Integrin-mediated signalling pathway |
| NS5A | MOBK1B | MOB1, Mps one binder kinase activator-like 1B (yeast) | Cell cycle progression |
| NS5A | NAP1L2 | Nucleosome assembly protein 1-like 2 | Nucleosome assembly |
| NS5A | PPP1R13 L | Protein phosphatase 1, regulatory (inhibitor) subunit 13-like | Regulation of transcription, DNA-dependent |
| NS5A | PSMB9 | Proteasome subunit, beta type, 9 | Ubiquitin-dependent protein catabolic process |
| NS5A | SORBS2 | Sorbin and SH3 domain containing 2 | Cytoskeletal adaptor activity |
| NS5A | TXNDC11 | Thioredoxin domain containing 11 | Cell redox homeostasis |
| NS5A | VPS52 | Vacuolar protein sorting 52 homolog (S. cerevisiae) | Vesicular transport |
HCV proteins are referenced according to their NCBI mature peptide product name (column 1). Human proteins are referenced with their cognate NCBI gene name or their protein official full name with their main biological function (column 2, 3 and 4, respectively). The 21 interactions have been validated by GST pull-down. In the first three entries, interactions were identified in two screens. In the rest of the entries, interactions were identified in one screen. Interaction Bin1–NS5A has been removed from this list as already described in literature.
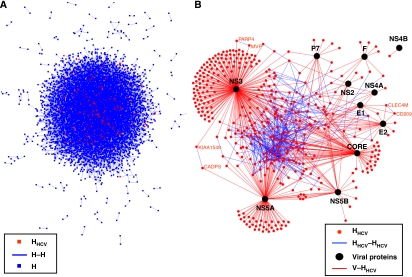
A human PPI network (H–H network; Figure 1A) was reconstructed from eight databases (Gandhi et al, 2006) (see Materials and methods). This network is composed of 44 223 non-redundant PPIs between 9520 different proteins (Figure 2A, complete list of PPIs in Supplementary Table SIV), corresponding to 30% of the human proteome (the remaining proteins have no known cellular partners and can therefore not be included in this network). Interestingly, human proteins targeted by HCV (HHCV) are clearly over-represented in this H–H network (IMAP Y2H data set: 76%; and IMAP LCI data set: 88%, exact Fisher test, P-value <2.2 × 10−16). This suggests that HCV preferentially targets host proteins already known to be engaged in protein–protein interactions (Rual et al, 2005; Stelzl et al, 2005). For the IMAP LCI data set, the higher percentage of HHCV integrated in the human interactome may be explained by inspection bias of well-studied proteins and biological pathways. Analysis of HHCV–HHCV subnetwork (all connected HHCV proteins) showed that cellular proteins interacting with HCV are significantly more interconnected than expected for random subnetworks (Figures 1A and 2B, Supplementary methods). Indeed, the 338 HHCV integrated into the human interactome are distributed into 131 connected components (versus 276 expected by random subnetworks; z-score-based test P-value <10−10, Supplementary Table SV). The largest one is composed of 196 HHCV (versus 18 expected by random subnetworks; z-test, P-value <10−10) and 127 are disconnected proteins. The three remaining connected components comprised two proteins. Two contained functionally related proteins (CLEC4M and CD209 are lectins involved in viral entry (Lozach et al, 2003); MVP and PARP4 are involved in Vault complex (Kedersha and Rome, 1986)) and one contained proteins not known to be functionally linked (KIAA1549 and CADPS).
Figure 2.
Graphical representation of the HCV–human interaction network. (A) Graphical representation of H–H network. Each node represents a protein and each edge represents an interaction. Red and blue nodes are HHCV and HNot-HCV, respectively. (B) Graphical representation of V–HHCV interaction network. Black node: viral protein; red node: human protein; red edge: interaction between human and viral proteins (V–HHCV); blue edge: interaction between human proteins (HHCV–HHCV). The largest component containing 196 proteins is represented in the middle of the network. Names of cellular proteins belonging to the three other connected components are also represented.
Topological analysis of the HCV–human interaction network
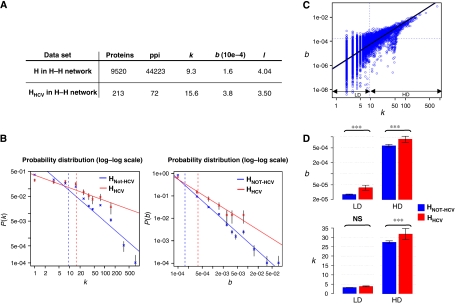
To assess how HCV proteins interplay with the cellular protein network, we next focused on the centrality measures of HHCV proteins integrated into the H–H interactome. Local (degree) and global (shortest path length and betweenness) centrality measures were calculated. Briefly, the degree (k) of a protein in a network corresponds to its number of direct partners and is therefore a measure of local centrality. Betweenness (b) is a global measure of centrality, as it measures the number of shortest paths (the minimum distance between two proteins in the network, l) that pass through a given protein. To provide an unbiased analysis, calculations were done on the basis of the 213 HHCV from the IMAP Y2H data set integrated in the human interactome. The average degree, betweenness and shortest path length of the H–H network are 9.3, 1.6 × 10−4 and 4.04, respectively, which is in good agreement with previous reports (Ramirez et al, 2007) (Figure 3A). As the distribution of properties such as node degree and node betweenness in PPI networks appear to follow a power law, summarizing values by their distributions appears more appropriated for comparative analysis (Goh et al, 2002; Joy et al, 2005). The degree distribution of HHCV and of the human interactome are significantly distinct (U-test P-value <10−3), with an average degree of HHCV higher than the average degree of the human interactome (15.6 versus 9.3). The comparison of degree probability distribution reveals that HHCV are preferentially represented in all class above the mean degree (Figure 3B, left). This indicates that HCV proteins have a strong tendency to interact with highly connected cellular proteins. However, as degree measures only local connectivity of proteins, global characteristics that could reflect information exchange and propagation in the network were investigated (Hernandez et al, 2007). At a global scale, the betweenness distribution of HHCV and of the human interactome are significantly distinct (U-test P-value <10−3), with an average betweenness of HHCV higher than the average betweenness of the human interactome (3.8 × 10−4 versus 1.6 × 10−4). As for the degree, the comparison of betweenness probability distribution shows an excess of HHCV in all class above the mean betweenness (Figure 3B, right). In addition, the shortest path length distribution of HHCV and of the human interactome were found significantly distinct (U-test P-value <10−5), with an average shortest path length of HHCV lower than average shortest path length of the human interactome (3.50 versus 4.04) revealing the topological proximity of HHCV. Both local and global centrality of HHCV from the IMAP LCI data set were higher than for the IMAP Y2H data set, emphasizing the problem of literature inspection bias and reinforcing the unbiased approach of Y2H screening (Supplementary Table SV). To ensure that the preferential attachment to central HHCV was not due to inherent bias associated to false positive in the H–H interactome, we performed the same analysis with a high-confidence, but less comprehensive, human interactome (Supplementary Table SV). This trend was maintained with this data set, confirming that HHCV are highly central within the human interactome, both locally and globally, and appear relatively close to each other in this network. For comparative analysis of HCV and EBV, the centrality measures were also computed for HEBV (data set from Calderwood et al (2007). Degree, betweenness and shortest path followed the same tendency with HEBV proteins (Supplementary Table SV and Supplementary Figure S2) and were in good agreement with a previous report (Calderwood et al, 2007). These results indicate that preferential attachment on central proteins may be a general hallmark of viral proteins as recently suggested by analysis of the literature (Dyer et al, 2008). The high centrality of proteins was previously shown to correlate with their functional essentiality for the yeast model organism (Jeong et al, 2001; Ekman et al, 2006). In mammals, lethal and disease-related proteins were found enriched in central proteins (Wachi et al, 2005; Stark et al, 2006; Goh et al, 2007; Hernandez et al, 2007). This suggests that HCV proteins interacted with essential proteins in the cell.
Figure 3.
Topological analysis of the HCV–human interaction network. (A) Topological analysis of H and HHCV in H–H network. Average values of degree (k), betweenness (b) and shortest path length (l) for all human proteins and for HHCV from the IMAP Y2H data set. (B) Degree and betweenness distribution of H and HHCV proteins in H–H network. P(k) is the probability of a node to connect k other nodes in the network. P(b) is the probability of a node to have a betweeness equal to b in the network. Normalized log degree (left) and log betweenness (right) distribution of H (blue) and HHCV proteins (red). Solid line represents linear regression fit. Vertical dashed lines give mean degree and betweenness values. Each class is represented with conventional standard error. (C) Degree and betweenness correlation of H in H–H network. Normalized log degree (x axis) and log betweenness (y axis) of H proteins into H–H network. Black solid line represents the linear regression fit (R2=0.56). Horizontal and vertical dashed lines give the mean degree and betweenness values, respectively. Low-degree (LD) and high-degree (HD) classes were defined by using the average degree cutoff. (D) Mean degree and betweenness of HNot-HCV and HHCV for LD and HD proteins. Top: mean betweenness (log scale) of HNot-HCV (blue) and HHCV (red) is given for LD and HD classes. Bottom: mean degree of HNot-HCV (blue) and HHCV (red) is given for LD and HD classes. The conventional standard error threshold and the U-test P-value are represented (***P-value <10−10, NS: not significant).
To determine which of the degree or the betweenness most influences the probability of interaction between viral and cellular proteins, we used a generalized linear model to test the separate and additive effects of both measures (Supplementary methods). This analysis revealed that betweenness better explains the probability of interaction between viral and human proteins (ANOVA P-value <10−3). Figure 3C shows a partial correlation between k and b centrality measures (R2=56%, P-value <10−16), explained by the high variability of betweenness at LD values. We thus asked whether this high variability observed at LD could explain the preponderant effect of betweenness. For this purpose, the data sets were split in LD and HD protein classes according to the average degree of the human interactome. For cellular proteins included in LD class, HCV interacts preferentially with proteins of high-betweenness independently of their degree property (Figure 3D). Within the HD class, interaction with HCV proteins is dependent on both betweenness and degree of cellular proteins. On the basis of a recent study in yeast (Joy et al, 2005), it can be extrapolated that LD high-betweenness HHCV proteins could exert an effect as connectors or bottlenecks between cellular modules and may thus be essential for the infection.
Functional analysis of the HCV–human interaction network
To better understand biological functions targeted by HCV, we next tested the enrichment of specific pathways for all interactors of a given viral protein. This was done by analysing the HHCV proteins with regard to the KEGG functional annotation pathways (Table II, Materials and methods). Although this approach is not totally unbiased because functions have not yet been attributed to all proteins, it remains a powerful way of incorporating conventional biology in system-level data sets. This analysis showed enrichment for three pathways associated with HCV clinical syndromes (insulin, TGFβ and Jak/STAT pathways) and identified focal adhesion as a novel pathway affected by HCV.
Table 2.
KEGG pathway enrichment for HHCV
| KEGG name | CORE | E1 | E2 | NS3 | NS5A | NS5B |
|---|---|---|---|---|---|---|
| Cell–cell and cell–ECM interactions | ||||||
| Adherens junction | 5 | 6 (5) | ||||
| Cell communication | 6 (2) | 8 (8) | ||||
| Cell adhesion molecules (CAMs) | 3 (1) | |||||
| ECM–receptor interaction | 2 (1) | 6 (6) | ||||
| Focal adhesion | 10 (9) | 8 (2) | ||||
| Gap junction | 4 | |||||
| Tight junction | 2 | |||||
| Signalling pathways | ||||||
| TGFβ signalling pathway | 4 | |||||
| Jak-STAT signalling pathway | 6 | 3 | ||||
| Adipocytokine signalling pathway | 5 | |||||
| MAPK signalling pathway | 3(2) | |||||
| Phosphatidylinositol signalling system | 2 | 4 | ||||
| Wnt signalling pathway | 7 (3) | |||||
| Insulin signalling pathway | 3 | |||||
| B cell receptor receptor signalling pathway | 3 | |||||
| T cell receptor signalling pathway | 5 | |||||
Over-represented KEGG pathways were identified after multiple testing adjustments (adjusted P-value <5 × 10−2) and are listed by viral protein. For each pathway, number of HHCV is given, with the relative contribution of IMAP Y2H dataset in brackets. Shaded entries denote discussed pathways.
IJT network (insulin–Jak/STAT–TGFβ network)
Chronic infection by HCV is associated with an increased risk for metabolic disorders with the development of steatosis. Insulin resistance is a common feature of this process. It also contributes to liver fibrosis and is a predictor of a poor response to interferon-α (IFN-α) anti-viral therapy (D'Souza et al, 2005; Romero-Gomez et al, 2005). Conversely, IFN-α can prevent fibrosis progression (Poynard et al, 2002). TGFβ has a crucial function in maintaining cell growth and differentiation in the liver. It is a strong profibrogenic cytokine whose production is frequently enhanced during infection. Impaired TGFβ response is also observed during HCV infection (Schuppan et al, 2003). Although insulin, TGFβ and Jak/STAT pathways have been suspected to be involved in these clinical features (Romero-Gomez, 2006), their closely related perturbation during HCV infection remains largely unexplained. We thus used a network approach to identify cellular proteins targeted by HCV and localized at the interface of these pathways. The resulting interaction map was constructed to form the IJT network (insulin–Jak/STAT–TGFβ network, Figure 4A; Supplementary methods). Sixty-six HHCV proteins are connecting two pathways, whereas 30 HHCV proteins are connecting the three pathways. Interaction of these proteins with HCV proteins may thus induce functional perturbations that could expand to adjacent pathways. One of these proteins is PLSCR1 (Scramblase 1), connecting insulin and Jak/STAT pathways. Known to be involved in the redistribution of plasma membrane phospholipids (Sahu et al, 2007), this protein is also a potential activator of genes in response to interferon, and its knockdown with siRNA favours viral replication (Dong et al, 2004). Interestingly, PLSCR1−/− mice also exhibit an onset of insulin resistance (Wiedmer et al, 2004). Although not annotated in the insulin or Jak/STAT pathways, PLSCR1 thus appears essential for the functionality of these pathways. By interacting with PLSCR1, CORE could therefore interfere with both Jak/STAT and insulin pathways. Another example is the nuclear factor Yin Yang 1 (YY1), which exhibits a more central position in the IJT network as it connects the three pathways. HCV CORE interaction with YY1 has been previously shown to be functional relieving NPM1 expression. This observation could be extrapolated to PPARδ expression and SMADs transcriptional activity in support of insulin and TGFβ pathway modulation (Kurisaki et al, 2003; Mai et al, 2006; He et al, 2008). Interestingly, BCL6, targeted by NS5A, is another transcriptional repressor at the interface of the three pathways that inhibit Smad signalling (Wang et al, 2008). It also exerts an effect as a corepressor of PPARδ (Lee et al, 2003) and it regulates the expression of a subset of Jak/STAT pathway target genes (Arbouzova et al, 2006). Thus, perturbation of these pathways can reasonably be expected as a consequence of BCL6 or YY1 targeting by HCV. Also central in the IJT network, NOTCH1 has been reported to interfere functionally with the three pathways. Literature analysis revealed that many of the proteins at the interface are actually known to have an important function in the regulation of one, two or the three pathways without being annotated in the KEGG database. These are only illustrative examples of cellular targets most likely to be involved in HCV-induced phenotypes. Although this molecular approach of the pathology is applicable to basal element of a system (proteins in this work) some of the clinical phenotypes observed in chronic HCV infection are most likely to result from the integrative effect of protein interactions depicted in the IJT network. In addition, the robustness property of a network can confer its ability to remain functional in face of different perturbations despite the deregulation of a single protein.
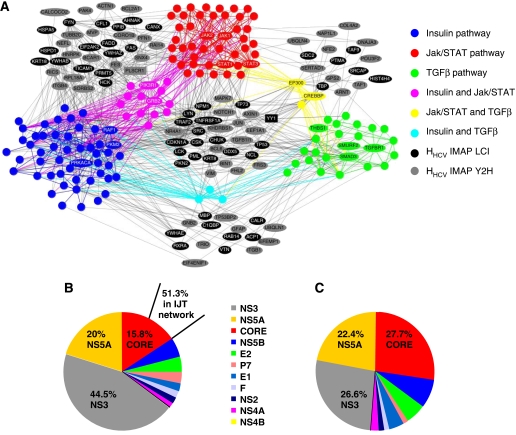
Figure 4.
IJT network. (A) Graphical representation of IJT network. Protein (nodes) members of insulin (blue), Jak/STAT (red) and TGFβ (green) pathways according to KEGG annotation, and their interactions (edges) are shown (proteins interacting with HCV proteins are named). Proteins shared by two pathways are shown in secondary colours (pink, yellow and cyan). Grey and black nodes are neighbours that connect the KEGG pathways and that interact with HCV proteins (grey: protein from the IMAP Y2H data set; black: protein from the IMAP LCI data set). Neighbours interacting with HCV but not connecting the KEGG pathways are not represented. Discussed protein examples PLSCR1 and YY1 are in box. References to visualization tools are provided in supplementary files (network visualization). IJT network construction in Supplementary methods. (B) Relative contribution of each viral protein in V–HHCV. Percentage interactions for the three most interacting viral proteins, relative to the total number of interactions as listed in Supplementary Table SI, are shown. A total of 51.3% of CORE interactions are concentrated in the IJT network. (C) Relative contribution of each viral protein in IJT network. Percentage interactions for the three most interacting viral proteins, relative to the total number of viral protein interactions with proteins of IJT network, are shown.
Another issue that became apparent in the IJT network is that CORE protein mediates proportionally more interactions than the other HCV proteins (Figure 4B and C). Indeed, preferential interaction with IJT network was observed only with CORE (51.3%, Supplementary Table SVI). As a consequence, CORE makes 27.7% of the interactions in the IJT network, corresponding to a significant enrichment (exact Fisher test P-value <10−4). More precisely, this CORE's interactors are over-represented in Jak-STAT and TGFβ pathways (exact Fisher test P-value <0.05) and in HHCV connecting insulin–Jak/STAT and insulin–TGFβ pathways (exact Fisher test P-value <0.05, Supplementary Table SVI). CORE thus appears as a major perturbator of the IJT network. Interestingly, transgenic mice expressing CORE develop insulin resistance (Shintani et al, 2004; Pazienza et al, 2007). A proposed mechanism was that CORE-induced SOCS3 promotes proteasomal degradation of IRS1 and IRS2 through ubiquitination (Kawaguchi et al, 2004). As SOCS3 is also a negative regulator of Jak/STAT pathway, this could explain the occurrence of IFN-α resistance. Clearly, the IJT network indicates that the action of CORE is most likely to be much more complex that previously thought. Although the IJT network cannot yet be analysed dynamically, it remains that it provides a unique way of deciphering some of the complex disorders associated with chronicity. It is also worth considering that the IJT network may identify a series of genes involved in diseases, such as steatosis and fibrogenesis, in the absence of viral infection.
Focal adhesion
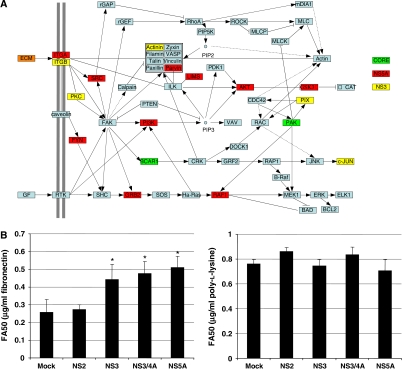
Focal adhesion was over-represented as a new function targeted by NS3 and NS5A proteins, with a major contribution of data generated by IMAP Y2H screens (Table II). Integrin-linked focal adhesion complexes control cell adhesion to extracellular matrix (ECM) and association of these complexes with actin-cytoskeleton has an important function in cell migration. Upon binding to the ECM, both α and β integrin subunits recruit proteins establishing a physical link between the actin-cytoskeleton and signal transduction pathways. When deregulated, this functional process can lead to perturbation of cell mobility, detachment from the ECM and tumour initiation and progression. Figure 5A shows KEGG focal adhesion pathway with proteins targeted by HCV, mainly NS3 and NS5A proteins. Impact of single expression of NS3, NS3/4A or NS5A on focal adhesion functionality was assessed using a cellular adhesion assay on fibronectin and poly-L-lysine. These viral proteins significantly inhibited cell adhesion to fibronectin compared with NS2-expressing cells (an HCV protein with no interactor in the focal adhesion pathway) or mock-transfected cells, with 40% increase of FA50 (matrix concentration for half maximum adhesion) (Figure 5B left, Student's t-test P-value <0.05). By contrast, adhesion to poly-L-lysine, which does not engage integrins, was not affected (Figure 5B right). The same inhibition level was observed for NS3/4A and NS3, suggesting that the enzyme activity of this protease does not have a major effect on focal adhesion perturbation. In addition to initiation and progression of cancer, the engagement of focal adhesion by HCV could have consequences on viral spreading. Interference with several steps of the actin-cytoskeleton remodelling has been described for retroviruses, which can exploit this process to surf along cellular protrusions of target cells to reach the entry site (Lehmann et al, 2005). It is conceivable that a related process, involving binding of the viral envelop to integrins, could be exploited by HCV to favour its transmission. This intriguing hypothesis will, however, be difficult to test until an efficient infection system of polarized cell is available.
Figure 5.
Interaction of HCV with focal adhesion. (A) Schematic representation of focal adhesion adapted from KEGG (ID: Hs04510). Proteins targeted by CORE, NS5A and NS3 HCV proteins are shown in yellow, red and green, respectively. ECM is a generic term for proteins of the extracellular matrix, some of which are targeted by HCV proteins (orange). (B) Functional validation of focal adhesion perturbation by NS3 and NS5A. Ninety-six-well plates were coated with fibronectin (left) or poly-L-lysine (right) at various concentrations. The 293T cells expressing NS2, NS3, NS3/4A or NS5A were plated on the matrix for 30 min. Adherent cells were stained with crystal violet. FA50 is the matrix concentration for half maximum adhesion. Values represent means of triplicate with standard deviation. *Student's t-test P-value <0.05.
Conclusion
In a network approach of HCV infection, the interaction map identifies all connections potentially needed for the virus to replicate and escape host defence. Whether all interactions really occur and have functional consequences is the open question of all interactome studies. The answer to this question necessitates the integration of system-level data sets of different origins that will set the stage for complex systems analysis of the infection. In a complex biological system, function cannot be predicted without understanding the component parts and their interactions and will result from the combination of theoretical knowledge of the cellular network with biological measurement of the interactions. Biological measurement, however, is still in the realm of low-throughput biology and needs major experimental improvement before prediction becomes the rule rather than the exception. Another fascinating challenge of this approach is to identify molecular signatures common to several viruses at the protein network level to develop original large-spectrum anti-viral molecules. A major step towards this goal is the high-throughput screening of a large variety of viruses, which is the aim of I-MAP.
Materials and methods
Construction of the HCV ORFeome
All HCV protein sequences were cloned in full length and domains except NS4B, for which no domain has been designed, using the euHCVdb facilities (http://euhcvdb.ibcp.fr; Combet et al, 2007) (Supplementary Figure S1). NS4A–NS3 fusion protein, as well as NS4A–NS3 protease domain were constructed (Kim et al, 1996; Taremi et al, 1998). All 27 ORFs from the HCV genotype 1b, isolate con1 (AJ238799) (Lohmann et al, 1999), were cloned in a Gateway recombinational cloning system (Walhout et al, 2000). Each ORF was PCR-amplified (with KOD polymerase, Novagen) using attB1.1 and attB2.1 recombination sites fused to forward and reverse primers, then cloned into pDONR223 (Rual et al, 2004). All entry clones were sequence-verified.
Yeast Two-hybrid (Y2H) screens
HCV ORFs were transferred from pDONR223 into bait vector (pPC97) to be expressed as Gal4–DB fusions in yeast. Two different screening methods were used (IMAP1 and IMAP2). For IMAP1, bait vectors were introduced in MAV203 yeast strain, and both human spleen and fetal brain AD-cDNA libraries (Invitrogen) were screened by transformation as described (Li et al, 2004). All primary positive clones (selected on SD−W−L−H+3−AT) were tested by further phenotypic assay using two additional reporter genes: LacZ (X-Gal colorimetric assay) and URA3 (growth assay on 5-FOA supplemented medium). Positive clones that displayed at least two out of three positive phenotypes were retested in fresh yeasts: bait vectors were retransformed into MAV203 and each prey cDNA (obtained by colony PCR, see below) were transformed in combination with linearized prey vector (gap repair; Walhout and Vidal, 2001). Clones that did not retest were discarded. AD-cDNA were PCR-amplified and inserts were sequenced to identify interactors. IMAP2 screens were performed by yeast mating, using AH109 and Y187 yeast strains (Clontech; Albers et al, 2005). Bait vectors were transformed into AH109 (bait strain), and human spleen and fetal brain AD-cDNA libraries (Invitrogen) were transformed into Y187 (prey strain). Single bait strains were mated with prey strain, then diploids were plated on SD−W−L−H+3−AT medium. Positive clones were maintained onto this selective medium for 15 days to eliminate any contaminant AD-cDNA plasmid (Vidalain et al, 2004). AD-cDNAs were PCR-amplified and inserts were sequenced.
Text-mining of interactions between HCV and human proteins
Literature-curated interactions (LCI), describing binary interactions between cellular and HCV proteins, were extracted from BIND database and PubMed (publications before August 2007) by using an automatic text-mining pipeline completed by expert curation process. For the text-mining approach, all abstracts related to ‘HCV' and ‘protein interactions' keywords were retrieved, subjected to a sentencizer (sentence partition) and a part-of-speech tagger for gene name (based on NCBI gene name and aliases) and interaction verbs (Rebholz-Schuhmann et al, 2008) (interact, bind, attach and so on). Sentences presenting co-occurrences of at least one human gene name, one viral gene name and one interaction term were prioritized to curation by human expert.
Validation by co-affinity purification
Cellular ORFs (interacting domains found in Y2H screens) were cloned by recombinational cloning from a pool of human cDNA library or the MGC cDNA plasmids using KOD polymerase (Toyobo) into pDONR207 (Invitrogen). After validation by sequencing, these ORFs were transferred into pCi-neo-3 × FLAG gateway-converted. HCV ORFs were transferred into pDEST27 (GST fusion in N-term). A total of 4 × 105 HEK-293T cells were then co-transfected (6 μl JetPei, Polyplus) with 1.5 μg of each pair of plasmid. Controls are GST-alone against 3 × FLAG-tagged prey. Two days after transfection, cells were harvested and lysed (0.5% NP-40, 20 mM Tris–HCl (pH 8.0), 180 mM NaCl, 1 mM EDTA and Roche complete protease inhibitor cocktail). Cell lysates were cleared by centrifugation for 20 min at 13 000 r.p.m. at 4°C and soluble protein complexes were purified by incubating 300 μg of cleared cell lysate with 40 μl glutathione sepharose 4B beads (GE Healthcare). Beads were then washed extensively with lysis buffer and proteins were separated on SDS–PAGE and transferred to nitrocellulose membrane. A total of 50 μg of cleared cell lysate was analysed by western blot to check the amount of 3 × FLAG-tagged cell protein. GST-tagged viral proteins and 3 × FLAG-tagged cellular proteins were detected using standard immunoblotting techniques using anti-GST (Covance) and anti-FLAG M2 (Sigma) monoclonal antibodies.
Integrated human interactome network (H–H network)
Only physical and direct binary protein-protein interactions were retrieved from BIND (Bader et al, 2003), BioGRID (Stark et al, 2006), DIP (Xenarios et al, 2002), GeneRIF (Lu et al, 2007), HPRD (Peri et al, 2004), IntAct (Kerrien et al, 2007), MINT (Chatr-aryamontri et al, 2007) and Reactome (Vastrik et al, 2007). NCBI official gene names were used to unify protein ACC, protein ID, gene name, symbol or alias defined in different genome reference databases (i.e ENSEMBL, UNIPROT, NCBI, INTACT, HPRD and so on) and to eliminate interaction redundancy due to the existence of different protein isoforms for a single gene. Thus, the gene name was used in the text to identify the proteins. Finally, only non-redundant protein–protein interactions were retained for building the human interactome data set.
Topological analysis
The R (http://www.r-project.org/) statistical environment was used to perform statistical analysis and the igraph R package (http://cneurocvs.rmki.kfki.hu/igraph/) to compute network connected components, centrality (degree, betweenness) and shortest path measures.
The Wilcoxon–Mann–Whitney rank sum test (the U-test) was chosen to statistically challenge observed differences. The U-test is a non-parametric alternative to the paired Student's t-test for the case of two related samples or repeated measurements on a single sample. The generalized linear model and ANOVA analysis was used to respectively model and test the separate and additive effects of degree and betweenness on the probability that HCV proteins interact with human proteins.
Functional analysis using KEGG annotations
Cellular pathway data were retrieved from KEGG (Aoki-Kinoshita and Kanehisa, 2007) and the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) and were used to annotate NCBI gene functions. For each viral–host protein interactors, the enrichment of specific KEGG pathway was tested by using an exact Fisher test (P-value <5 10−2) followed by the Benjamini and Hochberg multiple test correction (Benjamini et al, 2001) to control false discovery rate.
Cell-adhesion assay
Serial dilutions (from 20 to 0.04 μg/ml) of fibronectin or poly-L-lysine in PBS were coated on 96-well microplates overnight at 4°C. Non-specific binding sites were saturated at room temperature with PBS 1% BSA for 1 h. HEK 293T cells were transfected with pCi-neo-3 × FLAG NS2, NS3, NS3/4A or NS5A (JetPei, Polyplus), collected 2 days later with 2 mM EDTA in PBS, spread in triplicate at 1 × 105 cell per well in serum-free medium with 0.1% BSA and incubated for 30 min at 37°C. Non-adherent cells were washed away and adherent cells were fixed with 3.7% paraformaldehyde. Cells were stained with 0.5% crystal violet in 20% methanol for 20 min at room temperature and washed five times in H2O. Staining was extracted from 50% ethanol in 50 mM sodium citrate, pH 4.5, and the absorbance was read at 590 nm on an ELISA reader (MRX microplate reader, Dynatech Laboratories). Values were normalized to 100% adhesion at 10 μg/ml. The percentage of adhesion was determined for each cell type at each matrix concentration. 50% of maximum adhesions (FA50) were calculated from the curves (Supplementary Figure S3) (adapted from Miao et al, 2000).
Supplementary Material
Supplementary Information Supplementary methods, Figure S1-S3, Table legend
Supplementary Table SI HHCV listing
Supplementary Table SII Characteristics of IMAP1 and IMAP2 screens
Supplementary Table SIII Listing of human proteins interacting with more than one viral protein
Supplementary Table SIV List of human protein- protein interactions
Supplementary Table SV Topological analyis of the HCV-human network
Supplementary Table SVI HCV Protein distribution and enrichment in IJT network
Acknowledgments
This work was funded by ANRS, INSERM and the French Ministry of Industry. We acknowledge L Meyniel for critical reading of the manuscript. We also acknowledge Lyon Biopöle. V Navratil is supported by a grant from INRA.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albers M, Kranz H, Kober I, Kaiser C, Klink M, Suckow J, Kern R, Koegl M (2005) Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics 4: 205–213 [DOI] [PubMed] [Google Scholar]
- Aoki-Kinoshita KF, Kanehisa M (2007) Gene annotation and pathway mapping in KEGG. Methods Mol Biol 396: 71–92 [DOI] [PubMed] [Google Scholar]
- Appel N, Schaller T, Penin F, Bartenschlager R (2006) From structure to function: new insights into hepatitis C virus RNA replication. J Biol Chem 281: 9833–9836 [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Bach EA, Zeidler MP (2006) Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol 16: 80–88 [DOI] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res 31: 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284 [DOI] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E (2007) Epstein–Barr virus and virus human protein interaction maps. Proc Natl Acad Sci USA 104: 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res 35: D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Le Mercier P, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F, Deleage G (2007) euHCVdb: the European hepatitis C virus database. Nucleic Acids Res 35: D363–D366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza R, Sabin CA, Foster GR (2005) Insulin resistance plays a significant role in liver fibrosis in chronic hepatitis C and in the response to antiviral therapy. Am J Gastroenterol 100: 1509–1515 [DOI] [PubMed] [Google Scholar]
- Dong B, Zhou Q, Zhao J, Zhou A, Harty RN, Bose S, Banerjee A, Slee R, Guenther J, Williams BR, Wiedmer T, Sims PJ, Silverman RH (2004) Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J Virol 78: 8983–8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MD, Murali TM, Sobral BW (2008) The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog 4: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman D, Light S, Bjorklund AK, Elofsson A (2006) What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol 7: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS et al. (2006) Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet 38: 285–293 [DOI] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL (2007) The human disease network. Proc Natl Acad Sci USA 104: 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Oh E, Jeong H, Kahng B, Kim D (2002) Classification of scale-free networks. Proc Natl Acad Sci USA 99: 12583–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CQ, Ding NZ, Fan W (2008) YY1 repressing peroxisome proliferator-activated receptor delta promoter. Mol Cell Biochem 308: 247–252 [DOI] [PubMed] [Google Scholar]
- Hernandez P, Huerta-Cepas J, Montaner D, Al-Shahrour F, Valls J, Gomez L, Capella G, Dopazo J, Pujana MA (2007) Evidence for systems-level molecular mechanisms of tumorigenesis. BMC Genomics 8: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Jedynak BM, Bader JS (2007) Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput Biol 3: e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Joy MP, Brock A, Ingber DE, Huang S (2005) High-betweenness proteins in the yeast protein interaction network. J Biomed Biotechnol 2005: 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Yoshida T, Harada M, Hisamoto T, Nagao Y, Ide T, Taniguchi E, Kumemura H, Hanada S, Maeyama M, Baba S, Koga H, Kumashiro R, Ueno T, Ogata H, Yoshimura A, Sata M (2004) Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3. Am J Pathol 165: 1499–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Rome LH (1986) Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol 103: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B (2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O'Malley ET, Harbeson SL, Rice CM, Murcko MA, Caron PR, Thomson JA (1996) Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87: 343–355 [DOI] [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, Ten Dijke P, Heldin CH, Ericsson J, Moustakas A (2003) Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol Cell Biol 23: 4494–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Chawla A, Urbiztondo N, Liao D, Boisvert WA, Evans RM, Curtiss LK (2003) Transcriptional repression of atherogenic inflammation: modulation by PPARdelta. Science 302: 453–457 [DOI] [PubMed] [Google Scholar]
- Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W (2005) Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol 170: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY (2006) A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125: 801–814 [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285: 110–113 [DOI] [PubMed] [Google Scholar]
- Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP (2004) Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology 126: 586–597 [DOI] [PubMed] [Google Scholar]
- Lozach PY, Lortat-Jacob H, de Lacroix de Lavalette A, Staropoli I, Foung S, Amara A, Houles C, Fieschi F, Schwartz O, Virelizier JL, Arenzana-Seisdedos F, Altmeyer R (2003) DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem 278: 20358–20366 [DOI] [PubMed] [Google Scholar]
- Lu Z, Cohen KB, Hunter L (2007) GeneRIF quality assurance as summary revision. Pac Symp Biocomput 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai RT, Yeh TS, Kao CF, Sun SK, Huang HH, Wu Lee YH (2006) Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene 25: 448–462 [DOI] [PubMed] [Google Scholar]
- Miao H, Burnett E, Kinch M, Simon E, Wang B (2000) Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat Cell Biol 2: 62–69 [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM (2007) Replication of hepatitis C virus. Nat Rev Microbiol 5: 453–463 [DOI] [PubMed] [Google Scholar]
- Negro F (2006) Insulin resistance and HCV: will new knowledge modify clinical management? J Hepatol 45: 514–519 [DOI] [PubMed] [Google Scholar]
- Pazienza V, Clement S, Pugnale P, Conzelman S, Foti M, Mangia A, Negro F (2007) The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology 45: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, Rashmi BP, Shanker K, Padma N, Niranjan V, Harsha HC, Talreja N, Vrushabendra BM, Ramya MA, Yatish AJ, Joy M (2004) Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res 32: D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynard T, McHutchison J, Manns M, Trepo C, Lindsay K, Goodman Z, Ling MH, Albrecht J (2002) Impact of pegylated interferon alfa-2b and ribavirin on liver fibrosis in patients with chronic hepatitis C. Gastroenterology 122: 1303–1313 [DOI] [PubMed] [Google Scholar]
- Ramirez F, Schlicker A, Assenov Y, Lengauer T, Albrecht M (2007) Computational analysis of human protein interaction networks. Proteomics 7: 2541–2552 [DOI] [PubMed] [Google Scholar]
- Rebholz-Schuhmann D, Arregui M, Gaudan S, Kirsch H, Jimeno A (2008) Text processing through Web services: calling whatizit. Bioinformatics 24: 296–298 [DOI] [PubMed] [Google Scholar]
- Romero-Gomez M (2006) Insulin resistance and hepatitis C. World J Gastroenterol 12: 7075–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Gomez M, Del Mar Viloria M, Andrade RJ, Salmeron J, Diago M, Fernandez-Rodriguez CM, Corpas R, Cruz M, Grande L, Vazquez L, Munoz-De-Rueda P, Lopez-Serrano P, Gila A, Gutierrez ML, Perez C, Ruiz-Extremera A, Suarez E, Castillo J (2005) Insulin resistance impairs sustained response rate to peginterferon plus ribavirin in chronic hepatitis C patients. Gastroenterology 128: 636–641 [DOI] [PubMed] [Google Scholar]
- Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, Clingingsmith TR, Hartley JL, Esposito D, Cheo D, Moore T, Simmons B, Sequerra R, Bosak S, Doucette-Stamm L, Le Peuch C (2004) Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res 14: 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sahu SK, Gummadi SN, Manoj N, Aradhyam GK (2007) Phospholipid scramblases: an overview. Arch Biochem Biophys 462: 103–114 [DOI] [PubMed] [Google Scholar]
- Schuppan D, Krebs A, Bauer M, Hahn EG (2003) Hepatitis C and liver fibrosis. Cell Death Differ 10(Suppl 1): S59–S67 [DOI] [PubMed] [Google Scholar]
- Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Tsukamoto K, Kimura S, Moriya K, Koike K (2004) Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology 126: 840–848 [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Strader DB, Wright T, Thomas DL, Seeff LB (2004) Diagnosis, management, and treatment of hepatitis C. Hepatology 39: 1147–1171 [DOI] [PubMed] [Google Scholar]
- Tan SL, Ganji G, Paeper B, Proll S, Katze MG (2007) Systems biology and the host response to viral infection. Nat Biotechnol 25: 1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taremi SS, Beyer B, Maher M, Yao N, Prosise W, Weber PC, Malcolm BA (1998) Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci 7: 2143–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311: 239–242 [DOI] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L (2007) Reactome: a knowledge base of biologic pathways and processes. Genome Biol 8: R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidalain PO, Boxem M, Ge H, Li S, Vidal M (2004) Increasing specificity in high-throughput yeast two-hybrid experiments. Methods 32: 363–370 [DOI] [PubMed] [Google Scholar]
- Wachi S, Yoneda K, Wu R (2005) Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics 21: 4205–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 328: 575–592 [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Vidal M (2001) High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306 [DOI] [PubMed] [Google Scholar]
- Wang D, Long J, Dai F, Liang M, Feng XH, Lin X (2008) BCL6 represses Smad signaling in transforming growth factor-beta resistance. Cancer Res 68: 783–789 [DOI] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20: 2138–2139 [DOI] [PubMed] [Google Scholar]
- Wiedmer T, Zhao J, Li L, Zhou Q, Hevener A, Olefsky JM, Curtiss LK, Sims PJ (2004) Adiposity, dyslipidemia, and insulin resistance in mice with targeted deletion of phospholipid scramblase 3 (PLSCR3). Proc Natl Acad Sci USA 101: 13296–13301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30: 303–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Supplementary methods, Figure S1-S3, Table legend
Supplementary Table SI HHCV listing
Supplementary Table SII Characteristics of IMAP1 and IMAP2 screens
Supplementary Table SIII Listing of human proteins interacting with more than one viral protein
Supplementary Table SIV List of human protein- protein interactions
Supplementary Table SV Topological analyis of the HCV-human network
Supplementary Table SVI HCV Protein distribution and enrichment in IJT network