Abstract
Newly synthesized proteins must form their native structures in the crowded environment of the cell, while avoiding non-native conformations that can lead to aggregation. Yet remarkably little is known about the progressive folding of polypeptide chains during chain synthesis by the ribosome, or of the influence of this folding environment on productive folding in vivo. P22 tailspike is a homotrimeric protein that is prone to aggregation via misfolding of its central β-helix domain in vitro. We have produced stalled ribosome:tailspike nascent chain complexes of four fixed lengths in vivo, in order to assess co-translational folding of newly synthesized tailspike chains as a function of chain length. Partially synthesized, ribosome-bound nascent tailspike chains populate stable conformations with some native-state structural features even prior to the appearance of the entire β-helix domain, regardless of the presence of the chaperone trigger factor, yet these conformations are distinct from the conformations of released, refolded tailspike truncations. These results suggest that organization of the aggregation-prone β-helix domain occurs co-translationally, prior to chain release, to a conformation that is distinct from the accessible energy minimum conformation for the truncated free chain in solution.
As a protein is synthesized by the ribosome, it begins to fold into a three-dimensional shape, and must simultaneously avoid aggregation with other proteins in the crowded cell. In this seemingly hostile environment, where total protein concentrations can exceed 200–300 mg/mL, de novo protein folding during and after translation is, for many proteins, more efficient than in vitro refolding1. In other words, many proteins that can fold productively in the cell will aggregate severely under in vitro refolding reactions, presumably due to differences between the dominant folding pathway used. For example, experiments with both bacterial and firefly luciferase have demonstrated that these nascent chains adopt conformations co-translationally that are not populated during refolding from denaturant2,3, and these co-translational conformations fold to the native state much more efficiently than the conformations populated during refolding from denaturant. However, there continues to be debate as to what extent large, multi-domain proteins can fold co-translationally in prokaryotes4–7.
While molecular chaperones certainly play a role in efficient protein folding in the cell, less than 20% of E. coli cytoplasmic proteins require an interaction with one of the three major chaperone systems (trigger factor (TF), DnaK/DnaJ, or GroEL/ES) in order to fold correctly under normal growth conditions8. And remarkably, both chaperone systems responsible for the recognition of newly synthesized polypeptide chains in the E. coli cytoplasm (TF and DnaK/J) can be deleted simultaneously without abolishing cell viability9–11. Hence for many proteins, co-translational folding alone may be sufficient to partition newly synthesized polypeptide chains away from aggregation pathways and towards a productive folding pathway12.
How might protein folding be affected by the vectorial appearance of the polypeptide chain? Studies of purified, C-terminal truncations of single-domain proteins suggest that the formation of stable, folded, native-like structure requires the presence of an entire domain13,14; for many proteins, deleting only a few C-terminal residues is sufficient to globally destabilize domain structure. Yet some native structural topologies will presumably be more affected by co-translational folding than others. For example, the formation of β-sheet structure can require contacts between residues that are distant in the primary sequence, and hence is inherently more complex than the formation of α-helical structure. While recent reports have described the potential for α-helices to form co-translationally, even within the ribosomal exit tunnel15,16, it remains to be determined how the residues comprising a single β-strand or other incomplete portions of a β-sheet avoid non-native intra- or intermolecular contacts while waiting for their native partner residues to appear outside the ribosome17. Similar issues arise when considering vectorial folding of multimeric proteins.
The homotrimeric tailspike protein from Salmonella phage P22 is comprised almost entirely of β-sheet structure; its largest structural motif is a thirteen-rung parallel β-helix (Fig. 1). In vitro, tailspike folds via first the organization of the β-helix domain, followed by chain trimerization, and finally additional folding steps that include the interdigitation of the C-terminal domain, which imparts high thermostability18–20. The proper formation of the β-helix domain must occur prior to trimerization, and tailspike temperature sensitive for folding (tsf) and suppressor (su) mutations are found exclusively in the β-helix domain21–23. In vivo, tailspike is expressed primarily as a soluble protein at 30°–37°C, although some aggregation occurs even under physiological conditions18. In vitro, tailspike refolding is far less efficient, with significant aggregation occurring at 20°C24. Neither GroEL nor DnaK has been shown to participate in productive tailspike folding in vivo 25,26; the role of TF is undetermined. Previous qualitative studies from our laboratory27 and others28,29 have demonstrated that full length tailspike nascent chains have significant native-like organization of the β-helix domain prior to release of the nascent chain from the ribosome. Trimerization, however, does not occur co-translationally30. Here, we show that tailspike β-sheet formation occurs in a processive manner, during translation of successive rungs; i.e., prior to the presence of the entire β-helix domain. In addition, we show that co-translational folding traps the nascent chain in a conformation that is distinct from the conformations accessible to the free polypeptide chain.
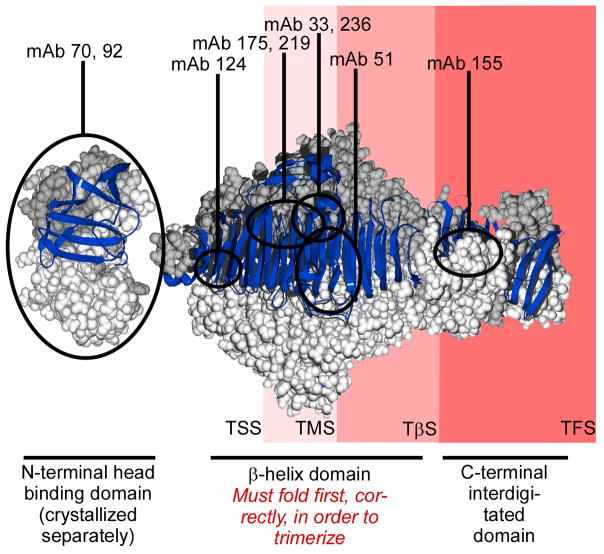
Figure 1. Tailspike structure and truncation constructs.
Tailspike trimer crystal structure42; one monomer chain is depicted as a blue ribbon and the other two chains are depicted as light and dark grey space filling models. mAb epitope boundaries32 are indicated by black circles. The portion of the tailspike polypeptide chain exposed outside the ribosome for each tailspike nascent chain construct is indicated by the red shading behind the crystal structure.
Results
Selection of tailspike nascent chain lengths
We used the 17-residue “stall sequence” from the E. coli SecM protein to efficiently stall tailspike translation in vivo27 at selected locations. Yet in any study of nascent chain conformations on stalled ribosomes, there is a risk that the equilibrium conformation of a stalled nascent chain might not accurately reflect the conformation of that nascent chain populated during unstalled translation. In order to minimize this possible discrepancy, we selected nascent chain lengths that correspond to the locations of statistically significant clusters of rare codons in the endogenous tailspike mRNA sequence, including detected translation pause sites (Clarke & Clark, submitted). Due to translational pausing, nascent chains of these lengths have more time to reach an equilibrium conformation, even during unstalled translation. The tailspike nascent chains described here therefore may more closely resemble biologically relevant translation intermediates than nascent chains of other lengths. Moreover, nascent chains of these lengths expose structurally interesting portions of the tailspike polypeptide chain outside the ribosome exit tunnel (Fig. 1): TSS (residues 1–229) exposes the N-terminal domain and the first three rungs of the β-helix domain; TMS (residues 1–406) exposes the N-terminal domain and the first half (8 rungs) of the β-helix; TβS (residues 1–556) exposes the N-terminal domain and the entire β-helix; and TFS (residues 1–666) exposes the entire tailspike chain, with the exception of the extreme C-terminal residues that are masked by the ribosome exit tunnel.
Many conformation-sensitive mAbs bind tailspike nascent chains as they emerge from the ribosome
A panel of 9 anti-tailspike monoclonal antibodies (mAbs) developed in the Goldberg laboratory bind to a range of tailspike structural features, with the majority recognizing epitopes in the β-helix domain31,32 (Fig. 1). Previous studies have shown that, in most cases, mAb binding is sensitive to the folding status of tailspike29,31,33, including a broad range of recognition profiles for tailspike in vitro refolding intermediates32. The anti-tailspike mAbs, therefore, provide a selective probe with which to assay tailspike conformational states in complicated mixtures such as cell lysates. Overall, antibody binding as a function of refolding time suggests a model for in vitro tailspike refolding (and particularly the organization of the β-helix domain) that begins with the folding of the extreme N- and C-termini of the β-helix32. The following work demonstrates how the co-translational appearance of the β-helix affects this organizational pattern.
Previous, qualitative results have shown that full-length tailspike nascent chains form some native-like structure while associated with the ribosome, based on the binding of three anti-tailspike monoclonal antibodies27,28. We have extended this observation, quantitatively characterizing the binding of 9 mAbs to four tailspike nascent chain lengths, using the competition ELISA method34 (Fig. 2; see also Supplementary Fig. S1). Of course, for each nascent chain length, only the subset of the antibodies whose epitopes have been synthesized can be used to provide information on the tailspike nascent chain conformation31.
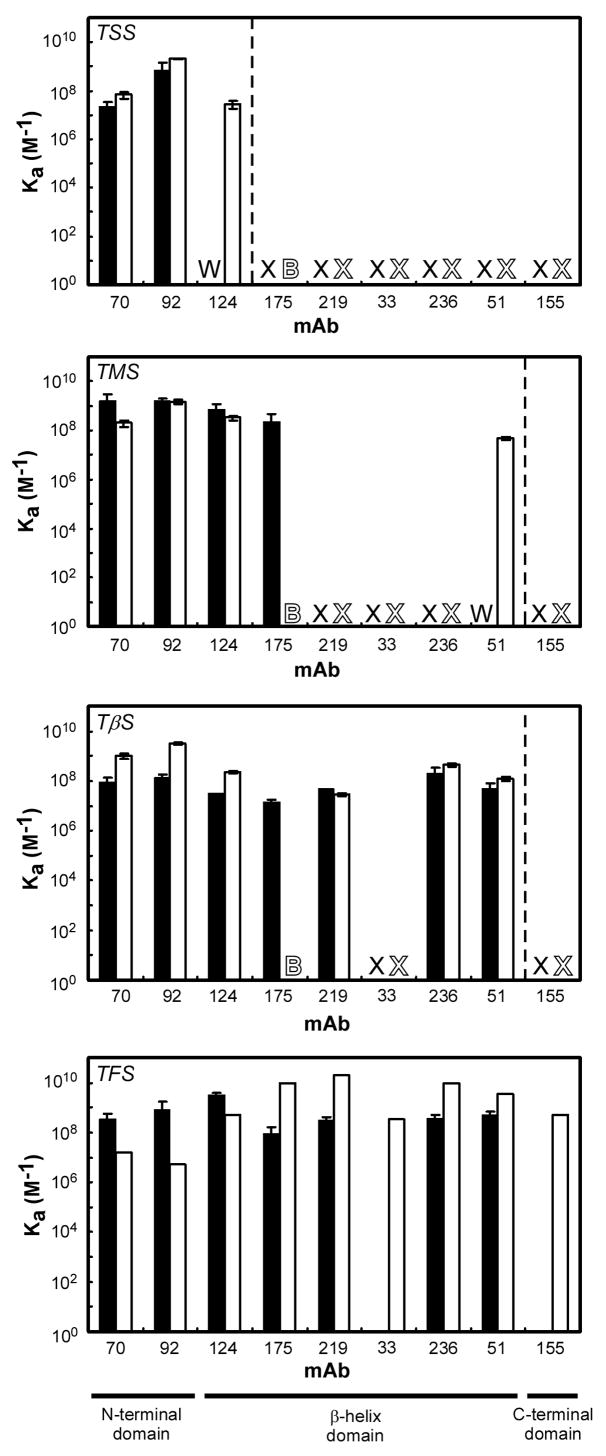
Figure 2. Binding of anti-tailspike mAbs to ribosome bound tailspike nascent chains and corresponding released chains.
mAb binding affinity to ribosome bound tailspike nascent chains (solid bars), and mAb binding affinity to the either the corresponding released tailspike chain (for TSS, TMS and TβS), or native tailspike (for TFS) (open bars). ‘X’ indicates no measurable binding affinity was detected; ‘W’ indicates a weak affinity, too low to be quantified (Ka < 106 M−1); ‘B’ signifies high background binding of mAb 175 to other components in the cell lysate supernatant. Error bars represent the standard deviation between experiments performed in triplicate. mAbs to the right of the vertical dashed lines have epitopes not yet synthesized at these nascent chain lengths. Results for mAb 236 binding to TFS, TβS and TSS nascent chains were previously reported27.
For the shortest tailspike nascent chain construct (TSS), two of the three mAbs with epitopes encoded within this nascent chain bound tightly. Indeed, mAbs 70 and 92 bind with a higher affinity to TSS than to native trimeric tailspike. In addition, mAb 124 binding can be detected qualitatively, but the affinity constant is too low to be measured accurately (<106 M−1). For TMS, which truncates tailspike in the middle of the β-helix domain, binding was detected for four of the expected seven mAbs. The most N-terminal of the anti-native mAbs, 175, binds TMS nascent chains tightly (Ka = 2 × 108 M−1), though not at tightly as to native trimer (Ka = 1010 M−1). mAb 51 binding to TMS nascent chains can be detected qualitatively, but the affinity constant is too low to be measured. For TβS, seven of the expected eight mAbs (i.e., all except mAb 33) bound to the nascent chain. Finally, for TFS, all mAbs except 155, which recognizes the most C-terminal interdigitated portion of the tailspike polypeptide chain, and mAb 33, in the center of the β-helix, showed binding affinities comparable to those measured for native tailspike32. In all cases, mAbs that bound to TFS nascent chains bound very tightly (≥107 M−1), and in some cases more tightly than to native trimeric tailspike. Interestingly, mAbs 175, 219, and 236, whose epitopes are all located in the β-helix and show low to no affinity for early in vitro tailspike refolding intermediates32, all bound tightly to ribosome-bound tailspike nascent chains of several lengths, including (for mAb 175) TMS, which contains an incomplete β-helix domain.
Limited proteolysis reveals a stable fragment present in longer nascent chains
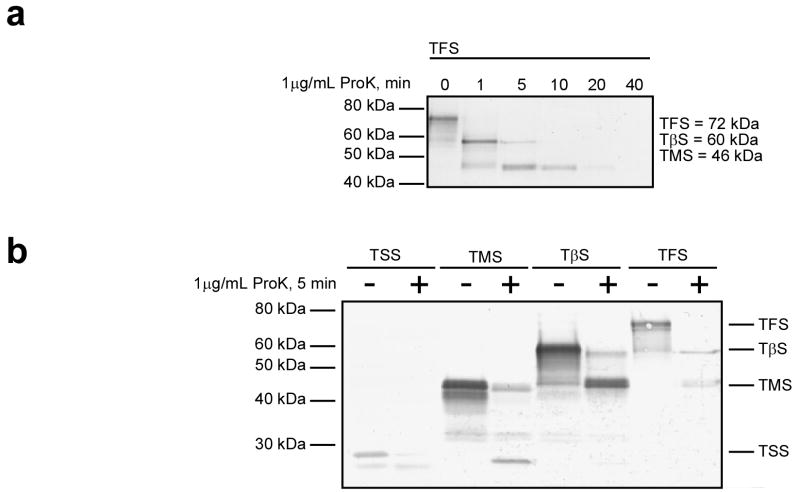
As an alternative means of measuring structure formation, we subjected stalled ribosome:nascent chain complexes (RNCs) to limited proteolytic digestion with proteinase K, a nonspecific protease. We treated RNCs with proteinase K for varying times at 4°C (Fig. 3). We performed control experiments to insure that protease digestion did not affect the structure of the ribosome itself (data not shown). TSS was rapidly digested with no detectable intermediates or protected fragments. TMS showed more resistance to protease digestion: after 5 minutes, detectable amounts of full length TMS remained, although a significant amount was digested to a protease-resistant fragment of approximately 25 kDa. Both TβS and TFS were rapidly digested to a relatively protease-resistant 47 kDa fragment, indistinguishable in size from the intact TMS nascent chain. Interestingly, at intermediate times, TFS was digested to a meta-stable 60 kDa fragment as well, corresponding to the size of TβS (Fig. 3a). Identical results were obtained for western blotting performed using the two anti-N-terminal antibodies or a mixture of all anti-tailspike mAbs (not shown).
Figure 3. Protease digestion of ribosome-bound tailspike nascent chains.
(a) Ribosomes bearing TFS nascent chains were treated with 1 μg/mL proteinase K for the indicated times at 4°C. The resulting fragments were separated on 5–14% denaturing polyacrylamide gels, transferred to a PVDF membrane, and immunoblotted using mAbs 70 and 92. (b) Ribosomes bearing TFS, TβS, TMS or TSS nascent chains were incubated for 5 minutes at 4°C with or without 1 μg/mL proteinase K. Digestion products were detected as in (a). Similar results were observed for room-temperature reactions, although the reactions proceeded more rapidly (data not shown).
Released, refolded truncated chains populate conformations distinct from the conformations of ribosome-bound nascent chains
Tailspike C-terminal truncations bearing the SecM stall sequence are eventually released from the ribosome. The majority aggregate (not shown), but a small fraction remains soluble, and can be separated from RNCs by sucrose gradient ultracentrifugation27. We measured the binding of the anti-tailspike mAbs to these released, truncated tailspike chains (Fig. 2). For released TSS, the two N-terminal, less conformationally-sensitive mAbs, 70 and 92, still bind with high affinity. For TMS, the four mAbs that bind the nascent chain attached to the ribosome still bind to the released chain, with one exception: we observed high background binding in the empty vector control samples for mAb 175 binding to released chains, making accurate quantification impossible. In addition, both released TSS and released TMS are recognized by an additional mAb: mAb 124 to released TSS and mAb 51 to released TMS. In both cases, the epitope for this additional mAb is located very close to the C-terminus of the construct, and ribosome release may increase antibody access to the nascent chain. For TβS, all seven mAbs that bind to TβS nascent chains bind with similar affinities to TβS released chains, again with the caveat that background binding for mAb 175 to released chains was unusually high.
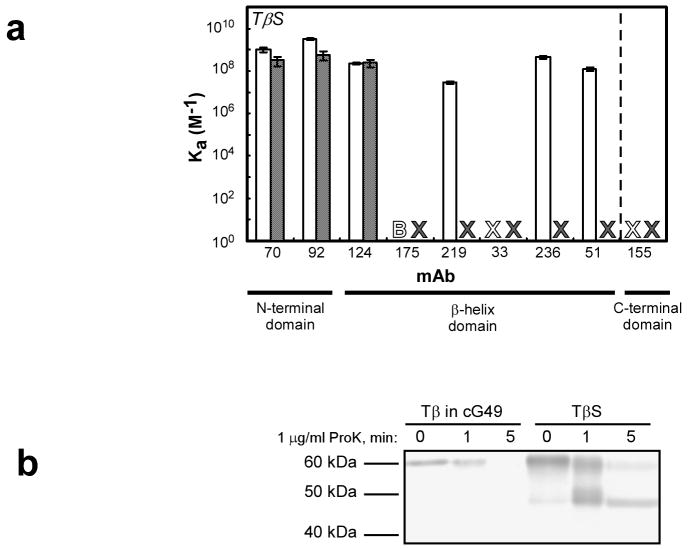
To test whether these tailspike truncations can also adopt ordered, native-like structure independent of the ribosome, we expressed and purified the tailspike truncation constructs as free chains (without the SecM stall sequence; designated TS, TM, and Tβ). As with released chains bearing the SecM stall sequence, all three chain lengths aggregated severely when expressed intracellularly, even at 30°C (data not shown). After purification under denaturing conditions, we attempted to refold each truncated chain length. However, only the Tβ construct produced detectable levels of soluble protein. We measured the binding of the anti-tailspike mAbs to this refolded Tβ. The three mAbs with the most N-terminal epitopes (70, 92 and 124) bound refolded Tβ with high affinity. In contrast, the other mAbs that bind to the TβS nascent chain, all of which have epitopes located further into the β-helix domain, showed essentially no binding affinity to Tβ (Fig. 4a).
Figure 4. Conformational differences between refolded Tβ and the corresponding nascent, or released, polypeptide chain.
(a) mAb binding affinity to TβS released chains (open bars), and Tβ refolded, truncated protein (shaded bars). An ‘X’ indicates that no measurable binding affinity could be detected, and a ‘B’ signifies high background binding (see Figure 2). Error is reported as in Figure 2. (b) Proteolysis of refolded Tβ. Refolded Tβ was diluted to a concentration similar to that of ribosome-bound TβS in a solution containing ribosomes expressing an unrelated stalled nascent chain (cG49). Digestion of ribosome-bound TβS is shown for comparison. The proteolysis reactions were performed at 4°C and were visualized by western blotting as described in Figure 3.
To further investigate the relationship between the conformations of refolded truncated chains and ribosome-bound nascent chains, we subjected truncated, refolded Tβ to limited proteolysis with proteinase K under similar conditions to those used for digesting ribosome-bound tailspike nascent chains. To account for the possibility that the protease resistance of ribosome-bound TβS was due to ribosomal proteins competing with TβS as substrate for proteinase K, we digested Tβ at in the presence of ribosomes expressing an unrelated ribosome-bound stalled nascent chain. Under these conditions, refolded Tβ remains resistant to proteolytic digestion for approximately the same length of time as ribosome-bound TβS, indicating the refolded, truncated construct has adopted a well-ordered conformation. However, proteinase K treatment of refolded Tβ did not produce a protease-resistant 47 kDa fragment, unlike digestion of ribosome-bound TβS (Fig. 4b).
The lack of mAb binding to the β-helix and the change in protease digestion pattern of refolded Tβ was surprising, as previous studies on the isolated β-helix domain of tailspike (Bhx) demonstrated this domain refolds under low salt conditions to an active, native-like conformation35,36. When we refolded Tβ using the low salt buffer used for Bhx refolding, we obtained far-UV CD and tryptophan fluorescence emission spectra largely similar to those obtained for Bhx35,36, despite the additional N-terminal domain present in the Tβ construct. Yet Tβ refolded under low salt conditions also did not produce the characteristic 47 kDa protease-resistant fragment (Supplementary Fig. S2). To control for the influence of other cellular components (for example, molecular chaperones) on the conformation of soluble Tβ, we also refolded Tβ in the presence of a cleared E. coli cell lysate. However, the presence of cellular factors, including molecular chaperones and ribosomes, had no effect on the yield of refolded Tβ, the appearance of the 47 kDa protease-resistant fragment (Supplementary Fig. S3), or the mAb binding profile (not shown). Taken together with the mAb binding results above, these results indicate that the β-helix domain, upon dilution from denaturant, does not populate the same conformations as the corresponding nascent chain when associated with the ribosome, or even after ribosome release.
Molecular chaperones are not required for co-translational folding of tailspike nascent chains
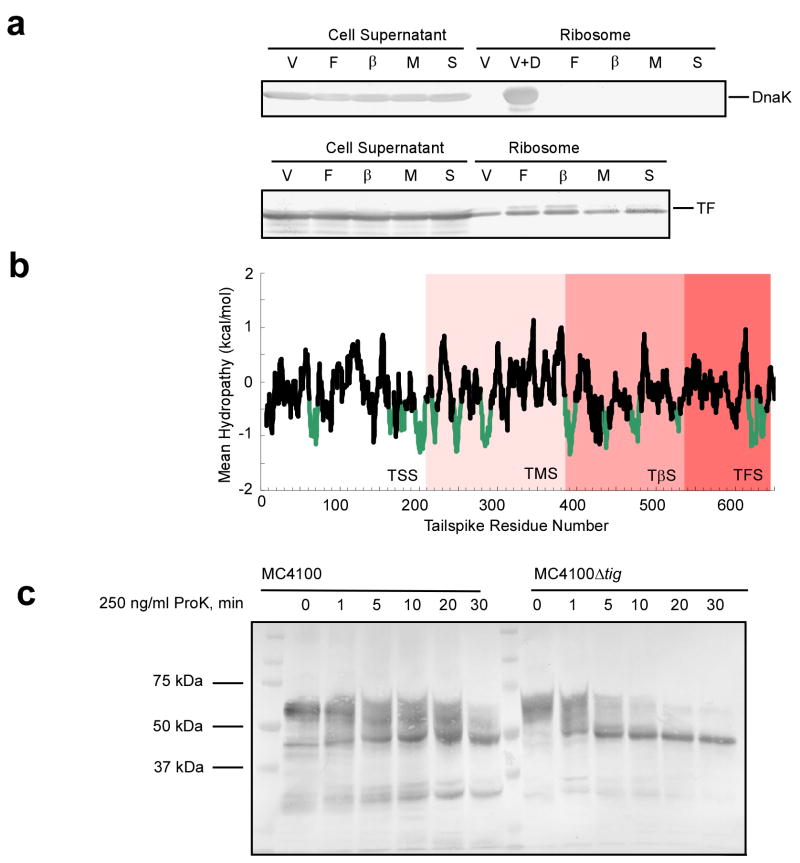
Molecular chaperones are known to play an important role in the co-translational folding of several proteins. In order to account for the effects of chaperones on tailspike folding, we first measured recruitment of the ribosome-associated chaperones DnaK and TF. While we were unable to demonstrate recruitment of DnaK to ribosomes bearing tailspike nascent chains, we were able to detect some recruitment of TF to tailspike RNCs (Fig. 5a). TF has been shown to bind hydrophobic stretches of nascent polypeptides37. When we analyzed the hydropathy of the tailspike amino acid sequence, we found several stretches of hydrophobic amino acids, which may provide binding sites for TF (Fig. 5b). To test whether TF plays a role in tailspike co-translational folding, we expressed TβS nascent chains in an E. coli strain lacking TF (MC4100Δtig), and a parent strain with TF (MC4100). Time-resolved proteinase K digestion of these TβS RNCs revealed that the 47 kDa stable fragment appears regardless of the presence or absence of TF, and is the major digestion product of TβS from either cell line (Fig. 5c). MC4100 is not a protease deficient strain, and the presence of many other bands besides the major digestion products might be due to endogenous E. coli proteases. Moreover, the digestion of TβS is slightly faster in the TF deletion strain, which could be indicative of TF restricting access of the protease to the ribosome-bound nascent chain. Regardless, these experiments show that tailspike nascent chains are able to form native-like structures in a co-translational manner, without a requirement for TF chaperone activity.
Figure 5. Chaperone association with ribosome-bound tailspike nascent chains.
(a) Cell lysates were centrifuged through sucrose cushions. The unpelleted lysate supernatant was removed, and the ribosomes were resuspended. Aliquots of both the unpelleted lysate and the ribosomes were examined by western blotting using anti-DnaK (top) or anti TF (bottom) antibodies. V, empty vector control; F, TFS; β, TβS; M, TMS; S, TSS. For DnaK, a vector control sample spiked with DnaK (1:1 DnaK:ribosome) is included as a positive control (V+D). (b) Hydropathy plot of tailspike. Mean hydropathy of the tailspike sequence was calculated according to published procedures37. Putative TF binding sites, as indicated by a mean hydropathy of less than −0.5 kcal/mol, are shown in green. (c) Proteinase K digestion of TβS RNCs from MC4100 or MC4100Δtig cells. Proteolysis reactions were preformed at 4°C and visualized by western blotting as described in Figure 3.
Discussion
The mAb binding and protease protection studies reported here demonstrate that the tailspike β-helix domain can fold to a native-like, compact conformation prior to release from the ribosome. Moreover, formation of this compact, native-like conformation begins prior to the complete synthesis of the β-helix domain, as demonstrated by the proteolytic protection and high affinity mAb binding to the TMS nascent chain. Some threshold length of the β-helix appears to be required for the formation of stable structure, however, as the TSS nascent chain, with only three rungs of the β-helix exposed from the ribosome, is quickly digested.
Two mAbs, 33 and 155, did not recognize any of the four tailspike nascent chain lengths studied here. Under in vitro refolding conditions, both recognize only native trimer29,32. Moreover, the epitope for mAb 155 lies in the C-terminal interdigitated domain. It is likely that native-like structure in the interdigitated C-terminus forms only after post-translational trimerization, i.e., even the TFS nascent chains are unlikely to adopt a native-like structure that requires interactions between the three monomer chains. The conformational epitope for mAb 33, like mAb 155, might require trimerization in order to form, as suggested by the in vitro refolding experiments. Surprisingly, however, the mAb 33 epitope is located in the center of the β-helix, near the mAb 236 epitope, and mAb 236 binds tightly to TβS and TFS nascent chains. Nevertheless, subtle conformational differences between these epitopes might exist; alternatively, the mAb 33 epitope may be masked by an interaction between the nascent chain and the surface of the ribosome.
It is an interesting to note that the size of the fragment produced upon proteinase K digestion of TβS and TFS is indistinguishable from the size of the TMS species (47 kDa); i.e., this 47 kDa fragment corresponds to the chain length produced during an endogenous rare-codon-derived translation pause point (Clarke & Clark, submitted). While the exact identity of the TFS and TβS 47 kDa fragment is unknown, it is recognized by mAbs 70 and 92, both of which recognize epitopes located in the tailspike N-terminal domain. Because the size of the fragment is much larger than the N-terminal domain itself (16 kDa), a fragment of 47 kDa could include the entire N-terminal domain and still accommodate a sizeable portion (7 rungs) of the β-helix domain. These results indicate that proteinase K digestion occurs primarily at the C-terminus of the nascent chain, suggesting this more recently synthesized portion is less stably folded than the N-terminus of the nascent chain (Fig. 6).
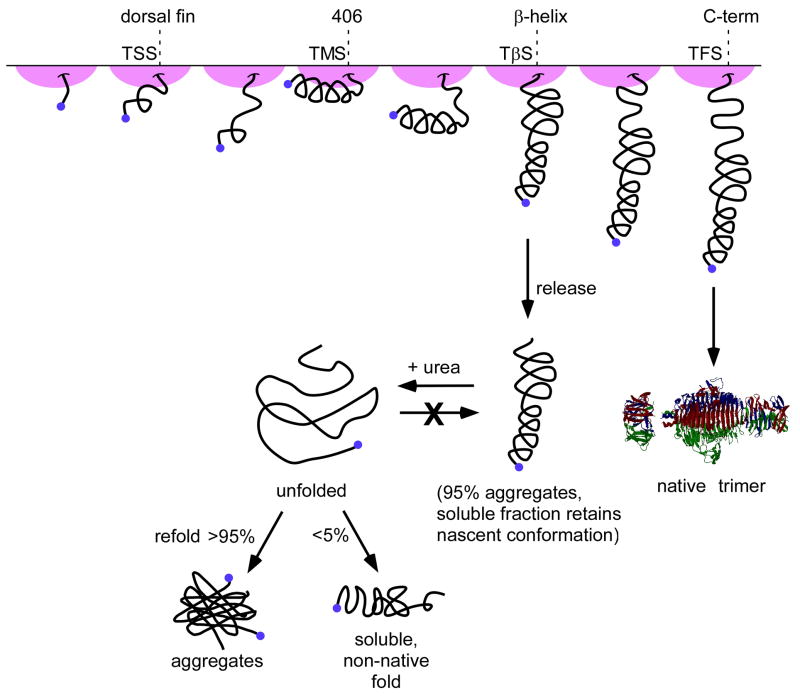
Figure 6. Model for co-translational folding of tailspike nascent chains.
When the first rungs of the β-helix emerge from the ribosome exit tunnel (TSS), little native-like or compact structure can form. As more of the β-helix is translated, a relatively stable compact structure containing native-like interactions forms throughout the first seven rungs of the β-helix domain (TMS). Once the entire β-helix domain is exposed from the ribosome (TβS), native-like compact structure forms throughout a significant portion of the chain, including the mAb 51 epitope (around residue 406). Based on the results of the protease protection experiments, the tailspike C-terminal interdigitated domain may be in a less compact conformation than the C-terminus of the β-helix domain. Free from the ribosome, truncated chains tend to aggregate, but the fraction remaining in solution maintains the conformation adopted while ribosome-bound. Chemical denaturation and refolding yields primarily insoluble aggregates, but the small amount of Tβ that refolds to a soluble, compact structure adopts a conformation that is distinct from the conformation of TβS while it is ribosome-bound.
When tailspike nascent chains are released prematurely from the ribosome, the truncated polypeptide chains primarily aggregate, although a small fraction remains soluble and maintain conformations similar to those observed for the corresponding ribosome-bound nascent chain. A small fraction of the aggregated released TβS chains, after solublization in chemical denaturant and dilution into buffer, are able to refold into a soluble, “well-behaved” conformation. Yet surprisingly, this conformation is distinct from the conformation of nascent TβS chains on the ribosome, or released chains (Fig. 6). This suggests that the refolding of denatured, truncated tailspike chains results in a fundamentally different final conformation (or ensemble of conformations) than the folding of newly synthesized, ribosome-bound tailspike chains. More generally, it appears that co-translational folding ushers the ribosome-bound tailspike polypeptide chain into an energy minimum that is inaccessible to free truncated polypeptide chains with untethered termini.
The tendency of released (and refolded) C-terminal tailspike truncations to aggregate is not surprising. For example, in vitro, the TM and TS truncations are expected to be aggregation-prone via intermolecular interactions involving their uncapped C-terminal ends of the β-helix domain, as has been predicted for uncapped β-structure in general38. Tethering the C-terminus of the nascent chain to the ribosome would shield the partially folded β-helix from such associations. Alternatively, the ribosome may play a more active role, functioning as a scaffold or template for incompletely synthesized tailspike chains. While it is unclear what portions of the ribosome surface might serve as a scaffold, interactions between full-length tailspike chains and 30S ribosome subunits have been reported previously28. The exit tunnel and the immediate surroundings likely play a role in protecting the nascent chains, as mAb 51, whose epitope is located near the C-terminus of TMS, can bind released, but not ribosome-associated, TMS. The same is likely true for mAb 124 binding to released (but not nascent) TSS.
Some have speculated that interactions with TF delay co-translational folding in prokaryotes37. TF might therefore explain inefficiencies reported for the folding of multi-domain proteins in prokaryotic translation systems4,7. Yet while protease digestion of nascent TβS on ribosomes lacking TF is slightly accelerated, we did not observe an effect of TF on tailspike nascent chain conformations. Hence while TF is bound to ribosomes translating tailspike nascent chains, it is not required for the formation of protease-resistant structure. Tailspike nascent chains fold to a native-like structure co-translationally, on prokaryotic ribosomes, and this process is neither aided nor retarded by the presence of TF.
Co-translational folding may represent a general strategy for proteins—and in particular large, multi-domain proteins—to fold productively and avoid aggregation during biogenesis. For tailspike folding in vivo, we propose that co-translational folding of the β-helix domain allows tailspike to preferentially partition into the productive folding pathway and away from the aggregation-prone conformations that dominate refolding in vitro (Fig. 6). Previous studies have demonstrated that early, proper folding of the β-helix domain is crucial to the assembly of the native trimer39,40. More generally, co-translational folding may represent a strategy for efficient folding of other β-sheet topologies. The ribosome, perhaps in concert with TF, can act as a temporary cap on the C-terminal strands as they emerge from the ribosome. As seen for free, truncated tailspike polypeptides, exposure of uncapped β-strands provides an excellent nucleus for aggregation that can be shielded during co-translational folding on the ribosome.
Methods
Plasmids
Construction of plasmids pET21b/TFS, /TβS, and /TSS was described previously27. pET21b/TMS was constructed similarly, using the primers 5′-GGGAATCCATATGACAGACATCACTGCAAACG-3′ and 5′-CCCGAGCTCCTCCGGATTCATGTCAGTGTC-3′. Plasmids pET21b/Tβ, /TM and /TS, lacking the C-terminal SecM stall sequence, were constructed by amplifying the tailspike coding regions from the corresponding stall plasmids using the same 5′ primer, and a 3′ primer incorporating an XhoI restriction site. PCR products were cloned into the pET21b vector (Novagen) between the NdeI and XhoI sites, placing the tailspike coding sequence in frame with the C-terminal His tag.
Stalled RNC Preparation
Ribosomes bearing nascent chains stalled at the SecM stall sequence were produced and purified as described previously27.
Preparation of Truncated, Released Tailspike Polypeptide Chains
Cleared lysates from cells expressing the tailspike truncations with the SecM stall sequence were centrifuged though a sucrose cushion as described previously for purifying stalled RNCs27. Released, truncated tailspike chains were collected from the top of the sucrose cushion following ultracentrifugation.
Purification of Truncated, Refolded Tailspike Polypeptide Chains
E. coli strain BL21(DE3)pLysS transformed with pET21b/Tβ, /TM or /TS, were grown in LB supplemented with 100 μg/mL ampicillin at 37°C to an OD600 of ~0.5. Expression was induced by adding IPTG to 500 μM; growth continued for 4 hours. Cells were collected by centrifugation and the pellet was frozen overnight at −20°C. The pellet was thawed on ice and resuspended in CR buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8). Lysozyme was added to 1 mg/mL and the mixture was incubated on ice for 30 minutes, followed by sonication (twelve 30 sec bursts separated by 30 sec rests) on ice. The resulting partially lysed cells were frozen for 90 minutes at −80°C, thawed, and sonicated as before. The lysate was then centrifuged at 10,000 × g for 25 minutes. The pellet was resuspended in PR buffer (50 mM NaH2PO4, 300 mM NaCl, 6 M GdnHCl, pH 8) and filter sterilized.
His-tagged proteins were purified by passage through Ni-NTA superflow resin (Qiagen). The column was washed with 50 mM NaH2PO4, 300 mM NaCl, 6 M GdnHCl, 20 mM imidazole, pH 8 (wash buffer), followed by an imidazole gradient (20–250 mM) to elute the His-tagged protein.
Fractions containing truncated tailspike proteins were concentrated using an Amicon Ultra 10,000 MWCO filter. Imidazole was removed by washing with PR buffer. Concentrated protein was refolded by rapid 50-fold dilution into refolding buffer (50 mM NaH2PO4, 300 mM NaCl, 2 mM EDTA, 3 mM β-mercaptoethanol, pH 8), followed by incubation at 0°C for at least 48 h. Low salt refolding was preformed as described previously36. Purified protein was concentrated using Amicon Ultra 10,000 MWCO filters.
ELISA/Kd Measurements
Competition ELISAs were used to measure the binding of anti-tailspike monoclonal antibodies to stalled ribosome nascent chain complexes and free truncated tailspike proteins, performed as previously described27,32. Dissociation constants were converted to association constants to facilitate graphical comparison of binding results (Fig. 2). Due to experimental limitations, it was not possible to measure affinity constants < 106 M−1. The concentration of free truncated chains was determined by absorbance at 280 nm using the molar extinction coefficient determined from the following equation41:
Proteolysis
Limited proteolysis of RNCs bearing TFS, TβS, TMS or TSS nascent chains was initiated by mixing an aliquot of each ribosome sample with a solution of proteinase K. The final protease concentration was 1 μg/mL. Longer time points were initiated first, and reactions were terminated at the end of the experiment by addition of a one-fifth volume of 100% trichloroacetic acid (w/v). Samples were incubated 30 minutes at −20°C to completely precipitate proteins. The precipitates were centrifuged and washed with acetone:hydrochloric acid (19:1). Washed pellets then prepared for SDS-PAGE and western blotting as described below. Refolded Tβ was first diluted to 200 nM in R buffer (50 mM Tris, 10 mM MgCl2, 150 mM KCl, pH 7.5) containing ribosomes bearing an unrelated stalled nascent chain (cG49) prior to addition of proteinase K.
Supplementary Material
Acknowledgments
We are grateful to Mary C. Finn and Daniel Lano for help with the preparation of truncated polypeptide chains, Krastyu Ugrinov for ribosome-bound cG49 nascent chains, Bernd Bukau for providing the anti-TF antibody, and Jose Barral and Ulrich Hartl for providing the MC4100-based E. coli strains. This work was supported by an award from the NIH (GM 74807).
Abbreviations used
- CD
circular dichroism
- ELISA
enzyme linked immunosorbent assay
- GdnHCl
guanidinium hydrochloride
- IPTG
isopropylgalactopyranoside
- mAb
monoclonal antibody
- PCR
polymerase chain reaction
- RNC
ribosome-nascent chain complex
- TCA
trichloroacetic acid
- TF
trigger factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ellis RJ. Macromolecular crowding: Obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 2.Frydman J, Erdjument-Bromage H, Tempst P, Hartl FU. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat Struct Biol. 1999;6:697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 3.Fedorov AN, Baldwin TO. Process of biosynthetic protein folding determines the rapid formation of native structure. J Mol Biol. 1999;294:579–586. doi: 10.1006/jmbi.1999.3281. [DOI] [PubMed] [Google Scholar]
- 4.Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 5.Nicola AV, Chen W, Helenius A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nat Cell Biol. 1999;1:341–345. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- 6.Kolb VA, Makeyev EV, Spirin AS. Co-translational folding of an eukaryotic multidomain protein in a prokaryotic translation system. J Biol Chem. 2000;275:16597–16601. doi: 10.1074/jbc.M002030200. [DOI] [PubMed] [Google Scholar]
- 7.Hirano N, Sawasaki T, Tozawa Y, Endo Y, Takai K. Tolerance for random recombination of domains in prokaryotic and eukaryotic translation systems: Limited interdomain misfolding in a eukaryotic translation system. Proteins. 2006;64:343–354. doi: 10.1002/prot.21008. [DOI] [PubMed] [Google Scholar]
- 8.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 9.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 10.Deuerling E, Patzelt H, Vorderwulbecke S, Rauch T, Kramer G, Schaffitzel E, Mogk A, Schulze-Specking A, Langen H, Bukau B. Trigger factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol. 2003;47:1317–28. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 11.Genevaux P, Keppel F, Schwager F, Langendijk-Genevaux PS, Hartl FU, Georgopoulos C. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 2004;5:195–200. doi: 10.1038/sj.embor.7400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark PL. Protein folding in the cell: Reshaping the folding funnel. Trends Biochem Sci. 2004;29:527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Neira JL, Fersht AR. Exploring the folding funnel of a polypeptide chain by biophysical studies on protein fragments. J Mol Biol. 1999;285:1309–1333. doi: 10.1006/jmbi.1998.2249. [DOI] [PubMed] [Google Scholar]
- 14.Chow CC, Chow C, Raghunathan V, Huppert TJ, Kimball EB, Cavagnero S. Chain length dependence of apomyoglobin folding: Structural evolution from misfolded sheets to native helices. Biochemistry. 2003;42:7090–7099. doi: 10.1021/bi0273056. [DOI] [PubMed] [Google Scholar]
- 15.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 17.Evans MS, Clarke TFI, Clark PL. Conformations of co-translational folding intermediates. Protein Pept Lett. 2005;12:189–195. doi: 10.2174/0929866053005908. [DOI] [PubMed] [Google Scholar]
- 18.Goldenberg D, King J. Trimeric intermediate in the in vivo folding and subunit assembly pathways of the tailspike endorhamnosidase of bacteriophage P22. Proc Natl Acad Sci USA. 1982;79:3403–3407. doi: 10.1073/pnas.79.11.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King J, Haase-Pettingell C, Robinson AS, Speed M, Mitraki A. Thermolabile folding intermediates: Inclusion body precursors and chaperonin substrates. FASEB J. 1996;10:57–66. doi: 10.1096/fasebj.10.1.8566549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benton CB, King J, Clark PL. Characterization of the protrimer intermediate in the folding pathway of the interdigitated β-helix tailspike protein. Biochemistry. 2002;41:5093–5103. doi: 10.1021/bi0115582. [DOI] [PubMed] [Google Scholar]
- 21.Mitraki A, Fane B, Haase-Pettingell C, Sturtevant J, King J. Global suppression of protein folding defects and inclusion body formation. Science. 1991;253:54–58. doi: 10.1126/science.1648264. [DOI] [PubMed] [Google Scholar]
- 22.Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal structure of P22 tailspike protein: Interdigitated subunits in a thermostable trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- 23.Haase-Pettingell C, King J. Prevalence of temperature sensitive folding mutations in the parallel β-coil domain of phage P22 tailspike endorhamnosidase. J Mol Biol. 1997;267:88–102. doi: 10.1006/jmbi.1996.0841. [DOI] [PubMed] [Google Scholar]
- 24.Betts SD, King J. Cold rescue of the single-chain tailspike intermediate at the junction between productive folding and off-pathway aggregation. Protein Sci. 1998;7:1516–1523. doi: 10.1002/pro.5560070704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sather SK, King J. Intracellular trapping of a cytoplasmic folding intermediate of the phage P22 tailspike using iodoacetamide. J Biol Chem. 1994;269:25268–25276. [PubMed] [Google Scholar]
- 26.Gordon CL, Sather SK, Casjens S, King J. Selective in vivo rescue by GroEL/ES of thermolabile folding intermediates to phage P22 structural proteins. J Biol Chem. 1994;269:27941–27951. [PubMed] [Google Scholar]
- 27.Evans MS, Ugrinov KG, Frese MA, Clark PL. Homogeneous stalled ribosome nascent chain complexes produced in vivo or in vitro. Nature Meth. 2005;2:757–762. doi: 10.1038/nmeth790. [DOI] [PubMed] [Google Scholar]
- 28.Clark PL, King J. A newly synthesized, ribosome-bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates. J Biol Chem. 2001;276:25411–25420. doi: 10.1074/jbc.M008490200. [DOI] [PubMed] [Google Scholar]
- 29.Friguet B, Djavadi-Ohaniance L, King J, Goldberg ME. In vitro and ribosome-bound folding intermediates of P22 tailspike protein detected with monoclonal antibodies. J Biol Chem. 1994;269:15945–15949. [PubMed] [Google Scholar]
- 30.Goldenberg D, Berget P, King J. Maturation of the tail spike endorhamnosidase of Salmonella phage P22. J Biol Chem. 1982;257:7864–7871. [PubMed] [Google Scholar]
- 31.Friguet B, Djavadi-Ohaniance L, Haase-Pettingell CA, King J, Goldberg ME. Properties of monoclonal antibodies selected for probing the conformation of wild type and mutant forms of the P22 tailspike endorhamnosidase. J Biol Chem. 1990;265:10347–10351. [PubMed] [Google Scholar]
- 32.Jain M, Evans MS, King J, Clark PL. Monoclonal antibody epitope mapping describes tailspike β-helix folding and aggregation intermediates. J Biol Chem. 2005;280:23032–23040. doi: 10.1074/jbc.M501963200. [DOI] [PubMed] [Google Scholar]
- 33.Speed MA, Morshead T, Wang DI, King J. Conformation of P22 tailspike folding and aggregation intermediates probed by monoclonal antibodies. Protein Sci. 1997;6:99–108. doi: 10.1002/pro.5560060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 35.Danner M, Fuchs A, Miller S, Seckler R. Folding and assembly of phage P22 tailspike endorhamnosidase lacking the N-terminal, head binding domain. Eur J Biochem. 1993;215:653–661. doi: 10.1111/j.1432-1033.1993.tb18076.x. [DOI] [PubMed] [Google Scholar]
- 36.Miller S, Schuler B, Seckler R. A reversibly unfolding fragment of P22 tailspike protein with native structure: The isolated β-helix domain. Biochemistry. 1998;37:9160–9168. doi: 10.1021/bi980190e. [DOI] [PubMed] [Google Scholar]
- 37.Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- 38.Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchs A, Seiderer C, Seckler R. In vitro folding pathway of phage P22 tailspike protein. Biochemistry. 1991;30:6598–6604. doi: 10.1021/bi00240a032. [DOI] [PubMed] [Google Scholar]
- 40.Simkovsky R, King J. An elongated spine of buried core residues necessary for in vivo folding of the parallel β-helix of P22 tailspike adhesin. Proc Natl Acad Sci USA. 2006;103:3575–3580. doi: 10.1073/pnas.0509087103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: Crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.