Abstract
The C-terminal domains of Dnmt3a and Dnmt3L form elongated heterotetramers (3L-3a-3a-3L). Analytical ultracentrifugation confirmed the Dnmt3a-C/3L-C complex exists as a 2:2 heterotetramer in solution. The 3a–3a interface is the DNA-binding site, while both interfaces are essential for AdoMet binding and catalytic activity. Hairpin bisulfite analysis shows correlated methylation of two CG sites in a distance of ∼8-10 bp in the opposite DNA strands, which corresponds to the geometry of the two active sites in one Dnmt3a-C/3L-C tetramer. Correlated methylation was also observed for two CG sites at similar distances in the same DNA strand, which can be attributed to the binding of two tetramers next to each other. DNA-binding experiments show that Dnmt3a-C/3L-C complexes multimerize on the DNA. Scanning force microscopy demonstrates filament formation rather than binding of single tetramers and shows that protein–DNA filament formation leads to a 1.5-fold shortening of the DNA length.
In mammals and other vertebrates, DNA methylation occurs at the C5 position of cytosine in CG dinucleotides (1,2). DNA methylation, together with histone modifications, plays an important role in modulating chromatin structure, thus controlling gene expression and many other chromatin-dependent processes including parental imprinting, maintenance of the genome integrity, X-chromosome inactivation as well as protection against endogenous retroviruses and transposons (2,3). Errors in DNA methylation contribute to both the initiation and the progression of various cancers (4,5). All active DNA methyltransferases (MTases) identified in mammals are indispensable for the normal embryonic development in mice (6,7).
The Dnmt3 family of mammalian DNA MTases comprises two active enzymes, Dnmt3a and 3b, which establish genomic methylation patterns, and one regulatory factor, Dnmt3-like protein (Dnmt3L). Dnmt3a and 3b show expression in several fetal and adult tissues (8), and knockout of either of them causes a strong developmental phenotype (7). Dnmt3a and 3b both contain a variable region at the N-terminus, followed by a PWWP domain, a PHD-like domain, and a C-terminal catalytic domain that is active in an isolated form. The smaller Dnmt3L contains only the N-terminal PHD-like domain, that interacts with H3 histone tails unmethylated at lysine 4 (H3K4) (9), and the C-terminal MTase-like domain, that interacts with Dnmt3a (and 3b) (10–12). Dnmt3L co-localizes and co-immunoprecipitates with both Dnmt3a and 3b (13), and enhances de novo methylation by both MTases in vitro (11,12,14,15) and in cells (16). These data suggest that Dnmt3L is a probe for H3K4 methylation, and if the methylation is absent from chromatin then Dnmt3L induces de novo DNA methylation by activating Dnmt3a (or 3b).
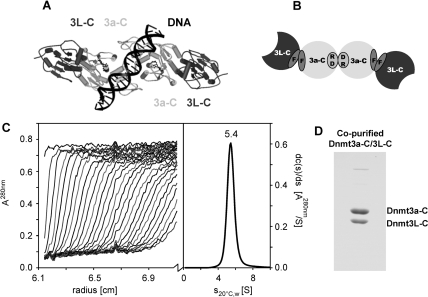
The minimal regions required for activity of Dnmt3a and 3b (17) and for the interaction between Dnmt3L and Dnmt3a (or 3b) are in the C-terminal domains of the proteins (10–12,18). In the crystal structure, the Dnmt3a-C/3L-C complex is a 2:2 heterotetramer with two 3L–3a interfaces and one 3a–3a interface (3L–3a–3a–3L) (19). The 3L–3a interface is mainly hydrophobic, represented by two pairs of phenylalanine, F728 and F768 of mouse Dnmt3a and F297 and F337 of mouse Dnmt3L; we termed it the double-F (FF) interface (Figure 1A and B). Dimerization via the 3a–3a interface occurs mainly by hydrophilic interactions, in particular by two reciprocal salt bridges between R881 and D872; we termed it the RD interface. Both interfaces are large with more than 900 Å2 per protomer (19). Molecular modeling of DNA into the Dnmt3a-C dimer showed that the two active sites are located in the major groove about 40 Å apart (Figure 1A), suggesting that dimeric Dnmt3a could methylate two CG sites separated by about one DNA helical turn (∼10 bp) in one binding event. Known biological targets of the Dnmt3a/3L enzymes were shown to contain their CG target sites in a periodic arrangement with a period of ∼10 bp, which fits to the distance of the two active sites in the complex (19). More recently, a 10-bp correlation of non-CpG DNA methylation by Arabidosis thaliana DRM2 (which is related to Dnmt3a) has been observed (20).
Figure 1.
Multimeric state of Dnmt3a-C/3L-C. (A) Model of the Dnmt3a-C/3L-C tetramer, colored in dark and light grey for Dnmt3L-C and Dnmt3a-C, respectively. Modeling of the DNA suggests that the two active sites could methylate two CG sites spaced by ∼10 bp in the opposite DNA strands. (B) Schematic picture of the Dnmt3a-C/3L-C tetramer, the 3a-3L (FF) and 3a–3a (RD) interfaces are indicated. (C) Analytical ultracentrifugation of the Dnmt3a-C/3L-C complex, using concentration corresponding to A280nm of 0.72. Scans were taken every 11.3 min (left panel) and were used to calculate the differential sedimentation coefficient distribution (right panel). (D) SDS-gel showing co-purification of Dnmt3a-C and Dnmt3L-C.
Here, we determined the multimeric state of Dnmt3a-C/3L-C in solution and the importance of tetramer formation for AdoMet binding, DNA binding and methylation by the complex. Using scanning force microscopy (SFM) imaging we observed a nucleoprotein filament formation by Dnmt3a-C/3L-C and investigated its consequences for the pattern of DNA methylation introduced by Dnmt3a-C/3L-C.
EXPERIMENTAL PROCEDURES
Analytical ultracentrifugation
Analytical ultracentrifugation of the co-purified Dnmt3a-C/3L-C complex was done with an An50-Ti 8-place rotor in a Beckman-Coulter model XL-A centrifuge equipped with UV absorption optics. Sedimentation velocity experiments were run at speeds of 50 000 r.p.m. at 4°C in 20 mM Tris–HCl pH 8.0, 0.3 M KCl, 0.67 M glycerol, 16 mM β-mercaptoethanol (for details see Supplementary Text 2).
Mutagenesis, protein expression and protein purification
The sequence encoding C-terminal domains of the mouse Dnmt3a (residues 608–908) and Dnmt3L (residues 208–421) were subcloned as N-terminal 6-His fusion proteins into pET-28a vector (Novagen) (3,11). Selected Dnmt3a-C and Dnmt3L-C variants were generated using the megaprimer site-directed mutagenesis method as described (21). Introduction of the mutations was confirmed by restriction marker site analysis and DNA sequencing. Protein expression and purification was carried out as described (11,19) (for details see Supplementary Text 2).
Methylation kinetics
Time courses of DNA methylation were measured by the incorporation of titrated methyl groups from labeled S-[methyl-3H] adenosyl-methionine (Perkin Elmer) into biotinylated oligonucleotides (Bt-GAG AAG CTG GGA CTT CCG GGA GGA GAG TGC/GCA CTC TCC TCC CGG AAG TCC CAG CTT CTC) using the avidin-biotin methylation kinetic assay as described (22). The reactions were carried out in the methylation buffer [20 mM HEPES pH 7.2, 1 mM EDTA, 50 mM KCl, 25 μg/ml bovine serum albumin (BSA)] at 37°C, using 1 μM substrate DNA, 0.76 μM AdoMet and 2.5 μM Dnmt3a-C wild-type (wt) or mutant enzyme, 2.5 μM Dnmt3L-C wt or mutant protein or 1.25 μM co-purified tetramer. To determine relative catalytic activities of the wt and mutants, equal concentrations of enzymes were used in the reactions. To determine the initial slope, the data were fitted by linear regression of the initial part of the reaction progress curves. All kinetic analyses were carried our at least in triplicate.
AdoMet binding
The cofactor binding was analyzed by UV-crosslinking experiments basically as described (11) using 6 μM wt Dnmt3L-C, 3 μM Dnmt3a-C (wt or mutant) and 1.4 μM [methyl-3H]AdoMet in methylation buffer supplemented with 10% glycerol and 1% β-mercaptoethanol. An excess of Dnmt3L-C was used to overcome the potentially weakened binding of Dnmt3L-C to the Dnmt3a-C variants, because it was the aim of this experiment to determine the effect of the Dnmt3L-C-induced conformational change of Dnmt3a-C on AdoMet binding (for details see Supplementary Text 2).
Electrophoretic mobility shift assay (EMSA)
DNA binding by Dnmt3a-C wt and mutants (1–10 μM) after preincubation with equimolar amounts of Dnmt3L-C was investigated by EMSA using a 146-bp DNA (30 nM) in reaction buffer (20 nM HEPES pH 7.5, 1 mM EDTA, 100 mM KCl, 0.2 mM sinefungin, 0.5 mg/ml BSA) basically as described (19). Sinefungin is an analogon of AdoMet that does not allow DNA methylation but supports DNA binding of Dnmt3a-C (data not shown). The gel was then scanned with a phosphoimager system (Fuji). For quantitative analysis the intensities of the gel regions corresponding to free DNA, fully occupied DNA and binding intermediates were determined at different protein concentrations and fitted to noncooperative and to cooperative binding models (for details see Supplementary Text 2).
Active site footprinting by hairpin bisulfite methylation analysis
DNA methylation by the Dnmt3a-C/3L-C co-purified complex was investigated by a hairpin bisulfite approach (23). Two sets of oligonucleotides were used as substrate containing 10 or 13 GAC repeats which form 9 or 12 CG sites in identical sequence context, respectively. 9xCG substrate: d(AAT TGA CGA CGA CGA CGA CGA CGA CGA CGA CGAC)/d(GAT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGTC); 12xCG substrate d(AAT TGA CGA CGA CGA CGA CGA CGA CGA CGA CGA CGA CGA CGA C)/d(GAT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGTC. The annealed oligonucleotides (100 nM) were incubated with 2.5 μM Dnmt3a-C/3L-C complex for 2 h in methylation buffer supplemented by 0.32 mM AdoMet. The methylation reaction was stopped by freezing the sample in liquid nitrogen, followed by proteinase K digestion (New England Biolabs). Afterwards, the hairpin loop and the adaptors were ligated to the methylated substrates, DNA was subjected to bisulfite conversion essentially as described (24,25), cloned into Topo-TA vector (Invitrogen) and individual clones were sequenced.
SFM
SFM experiments were carried out using preincubated Dnmt3a-C/3L-C complex and a 509-bp DNA fragment, which can be visualized easily in SFM. The DNA was derived from the CG island upstream of the human SUHW1 gene and contains 58 CG sites roughly equally distributed over the entire DNA length. DNA–protein filaments were generated in a total volume of 30 μl by combining 12 nM DNA with 100 nM Dnmt3a-C/3L-C tetramer in 50 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA and 100 μM of sinefungin. The two protein components (200 nM each) where preincubated in the binding buffer for 30 min at room temperature. After addition of DNA, samples were incubated for additional 30 min at room temperature to allow for DNA binding. The complex solution (1 μl) was mixed with 9 μl of 10 mM MgCl2 solution and deposited on freshly cleaved mica (Plano, Wetzlar), allowed to adhere for 40 s and then washed with 1 ml of bi-distilled water. The sample was then dried using compressed air. Protein–DNA filaments where observed by tapping mode in air using a Multimode SFM with a Nanoscope III controller (Digital instruments, Santa Barbara, CA). We used NSTNCHF silicon cantilevers (Nascatec, Stuttgart) with a nominal spring constant of 50 N/m and a resonance frequency of ∼150 kHz. All images were obtained with a scanning speed of 0.75–1 Hz and a resolution of 512 × 512 pixels. To remove background slope, raw images were flattened using the Nanoscope software. DNA–protein complexes were regarded as a filament if the height exceeded 150% of the height observed for DNA molecule alone and were at least 20 nm wide. Filaments were evaluated using the section tool of the Nanoscope V6r12 software.
Compaction of the DNA in nucleoprotein filaments was calculated using Equation (1):
| 1 |
With C, compaction factor, L, length of the filament; L0, length of the free DNA (170 nm); f: fraction of the molecule covered by protein.
RESULTS
Oligomeric state of Dnmt3a-C/3L-C
We investigated the oligomeric state of the Dnmt3a-C/3L-C complex in solution by analytical ultracentrifugation. The co-purified Dnmt3a-C/3L-C complex sedimented in a single boundary with s20°C,water = 5.4 S (Figure 1C). SDS gel electrophoresis showed the complex to contain both proteins in a 1:1 stoichiometric ratio (Figure 1D). The frictional ratios calculated from the measured sedimentation rate constant would be 1.5 for a 2:2 and 0.93 for a 1:1 Dnmt3a-C/3L-C hydrated complex. Since a ratio < 1.0 is not possible, the measured sedimentation rate is too fast for a 1:1 heterodimer and is only compatible with a 2:2 heterotetramer.
Catalytic activity of Dnmt3a-C/3L-C and its interface mutants
In order to study the functional importance of Dnmt3a–3a interaction at the RD interface, we mutated R881 to alanine (R881A) in Dnmt3a-C. At the Dnmt3a–3L interface (the FF interface), we exchanged F728 in Dnmt3a-C to alanine (F728A) and to aspartic acid (F728D), assuming that the F to D exchange, which introduces a charged residue instead of a hydrophobic one, would result in a stronger phenotype than the F to A mutation. We also made the corresponding mutations in Dnmt3L-C at the FF interface (F297A and F297D).
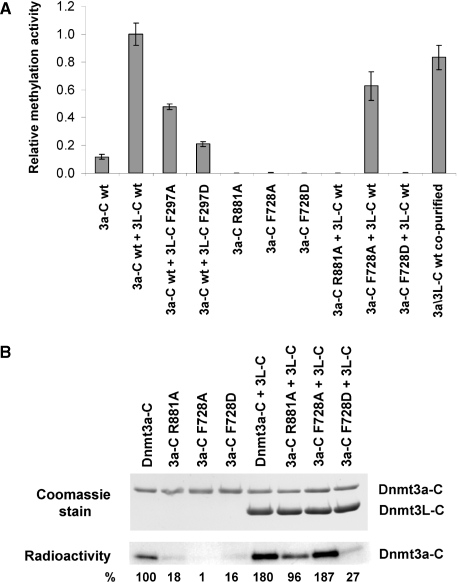
We compared the catalytic activity of wt Dnmt3a-C and its variants using roughly equal concentrations of the enzymes and an unmethylated oligonucleotide substrate with one centrally placed CG site. All the Dnmt3a-C variants turned out to be catalytically inactive (Figure 2A and Supplementary Figure 1). To ensure that the lack of activity was not caused by protein misfolding, we determined the near UV circular dichroism (CD) spectra of the Dnmt3a-C wt and its mutants. As shown in Supplementary Figure 2, the wt enzyme and R881A mutant CD spectra superimpose, indicating that the mutant protein is folded as the wt protein. The CD spectra of the F728A and F728D mutants also demonstrate that the proteins are folded. However, both showed a small, but reproducible shift in the shorter wavelength region indicating a change in the secondary structure composition.
Figure 2.
Effects of interface mutations on catalytic activity and cofactor binding. (A) Catalytic activity of the Dnmt3a-C wt and its interface variants analyzed in the absence and presence of Dnmt3L-C by transfer of radioactive methyl groups to a 30-bp oligonucleotide containing one centrally placed CG site. Time courses of DNA methylation were determined over 20 min and used to determine initial slopes. All experiments were carried out at least in triplicate, error bar indicate the standard deviation of the initial slopes. Examples of individual DNA methylation reactions are shown in Supplementary Figure 1. (B) AdoMet binding of Dnmt3a-C wt and its mutants in the absence and presence of Dnmt3L-C, as determined by UV-crosslinking of the cofactor to the proteins. After UV crosslink, the proteins were separated on two identical SDS–polyacrylamide gels. The upper panel shows the gel stained with Coomassie, the lower panel shows an autoradiogram indicating the amount of AdoMet bound to the proteins. Densitometric quantification of the signals is shown below the gels in % of the activity observed with wt Dnmt3a-C.
Next, we compared the activity of the wt Dnmt3a-C/3L-C complex and its variants. Since the co-purified Dnmt3a-C/3L-C complex showed similar activity to the complex formed by preincubation of equimolar amounts of purified Dnmt3a-C and Dnmt3L-C for 30 min (Figure 2A), in all subsequent experiments the complexes formed by preincubation of both proteins were used. After incubation with Dnmt3L-C, the activity of F728A reached 70% of the level observed for the wt complex, but F728D showed almost no activity (Figure 2A). The R881A variant was completely inactive (Figure 2A) even after incubation with excess of Dnmt3L-C (data not shown). Mutations at the FF interface in Dnmt3L-C (F297A and F297D) reduced the activity of Dnmt3a-C/3L-C by 50% and 75%, respectively (Figure 2A). In addition, the activity of Dnmt3a-C F728A variant was nearly abolished when incubated with the F297A mutants of Dnmt3L-C (Supplementary Figure 1B), because two exchanges at the FF interface (one in Dnmt3a-C and one in Dnmt3L-C) disturb the interface more strongly or prevent complex formation. Hence all the three interface residues (R881 and F728 of Dnmt3a and F297 of Dnmt3L)—though located distant from the catalytic center of the enzyme—are important for the catalytic function of Dnmt3a-C and Dnmt3a-C/3L-C complexes.
AdoMet binding by Dnmt3a-C and its interface mutants
AdoMet binding to Dnmt3a-C can be detected in a UV crosslinking assay (11), whereas Dnmt3L-C does not bind to AdoMet (11,15,19). In the absence of Dnmt3L-C, Dnmt3a-C has reduced AdoMet binding (Figure 2B) (11,15), and its three mutants showed a strong reduction in AdoMet binding in comparison to the wt protein. When preincubated with Dnmt3L-C, the AdoMet binding of the Dnmt3a-C F728A mutant was efficiently rescued, but little rescue was seen for F728D (Figure 2B). This result indicates that binding of Dnmt3L-C to the Dnmt3a-C F728A variant can recover its AdoMet-binding ability, presumably by stabilizing the FF interface, but the interaction is weak or lost between Dnmt3L-C and F728D variant. Addition of Dnmt3L-C only partially rescued the AdoMet binding by the Dnmt3a-C R881A, indicating that the mutational defect in the RD interface cannot be overcome by binding of Dnmt3L at the FF interface. Reduction of AdoMet binding by disruption of either of the two interfaces can be explained structurally by interaction networks which connect residues important for AdoMet binding with interface structural elements [Supplementary Figure 3 and (19)].
DNA binding by Dnmt3a-C/3L-C and its interface mutants
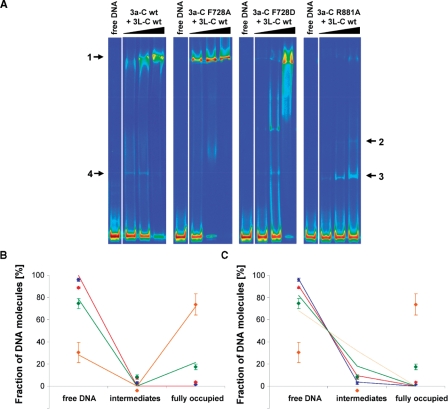
Using EMSAs, we have previously shown that Dnmt3a-C and Dnmt3a-C/3L-C complexes multimerize on the DNA, and that Dnmt3L-C binds very weakly to DNA (19). The binding process shows the co-existence of DNA fully occupied by protein (band 1 in Figure 3A) and free DNA with low amounts of incompletely occupied complexes over a range of protein concentrations (Figure 3A). We determined the amount of the free DNA, as well as DNA partially and fully occupied by protein by densitometry. Fitting of the binding profile was not possible with a non-cooperative binding model suggesting a cooperative binding mechanism (Figure 3B and C).
Figure 3.
DNA binding of Dnmt3a-C/3L-C and effect of interface mutations on DNA binding. (A) DNA binding of Dnmt3a-C/3L-C wt and mutants determined by EMSA experiment after incubating a 146-bp fluorescently labeled DNA with increasing amounts of protein. It has been shown previously that band 4 observed with wt Dnmt3a-C/3L-C is due to Dnmt3L-C binding to the DNA (19). With Dnmt3a-C F728D and R881A a ladder of bands is also observed in absence of Dnmt3L-C (data not shown). (B and C) Densitometric analysis of the complexes formed by Dnmt3a-C/3L-C at four different enzyme concentrations: 1 µM (blue), 2.5 µM (red), 5 µM (green) and 10 µM (orange). Data are averages from two independent gels, fitted to a fully cooperative (B) and a noncooperative (C) binding model. Error bars represent the deviations between the gels.
To study the role of the RD and FF interfaces in DNA binding, we performed gel retardation experiments with the interface variants as well. After preincubation with Dnmt3L-C, the F728A variant bound DNA slightly better than the wt complex (Figure 3A), whereas Dnmt3a-C F728D/3L-C show increased amount of smear and incompletely occupied complexes (Figure 3A). In contrast, the R881A mutant only weakly bound DNA and formed smaller complexes, as shown by an appearance of a ladder of bands, corresponding to one, two or even more protein complexes bound to the DNA (Figure 3A, bands 2 and 3). These results demonstrate that the RD interface is required for DNA binding and the FF interface is supportive but not essential.
Protein–DNA filament formation by Dnmt3a-C/3L-C complex
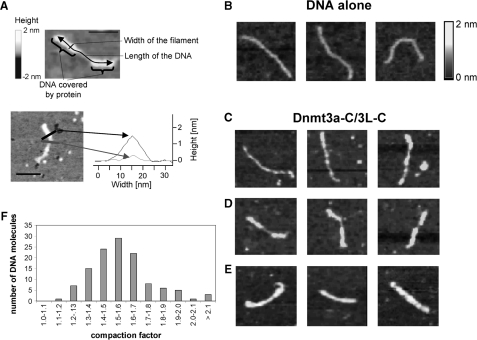
Dnmt3a-C/3L-C protein–DNA complexes were studied by SFM experiments using a CG rich substrate. DNA bound by protein could be distinguished from free DNA by the change in height and width (Figure 4A). The results showed that Dnmt3a-C/3L-C complexes polymerize on the DNA forming a nucleoprotein filament (Figure 4B–E). We observed many complexes in which the DNA was fully occupied by protein, although the protein was used in substoichiometric amounts with respect to binding sites (100 nM tetramer, 12-nM DNA corresponding to ∼600 nM binding sites, when assuming that one tetramer binds to ∼10 bp, as suggested by the hairpin bisulfite experiments described below). Similarly as in the gel retardation assay, we observed the co-existence of free DNA and DNA covered completely with Dnmt3a-C/3L-C. Furthermore, multimeric binding of complexes was predominantly observed rather than binding of single tetramers. This is in contrast to results obtained with the EcoRV MTase where mostly single complexes were found using the same technique (26). These observations suggest cooperative binding of Dnmt3a-C/3L-C to DNA.
Figure 4.
Scanning force microscopy of Dnmt3a-C/3L-C DNA filaments. (A) Dimensions of SFM complexes. In the upper part, a partially occupied DNA molecule is shown. Regions covered with protein are indicated and the length and width of the filaments defined. The height of the filament is defined by the Z-axis and encoded in the grayscale of the image. In the lower part, height profiles of sections through a protein/DNA filament (black) and an unoccupied DNA (gray) are shown. The scale bars are 50 nm. (B–E) Examples of SFM complexes: (B) Images showing free DNA. (C–E). Images showing examples of Dnmt3a-C/3L-C DNA filaments with low (C), heavy (D) or full (E) occupancy with protein. In each panel in B–E the frame size is 150 nm. (F) Distribution of DNA length compaction factors of 116 filaments. The distribution of the lengths’ observed for free DNA and fully occupied DNA are shown in Supplementary Figure 4.
For each DNA molecule the length of the nucleoprotein complex region and the length of the unbound region were determined and the factor of shortening of the DNA length calculated (Figure 4F). Naked DNA (194 molecules) had a length of 171 ± 18 nm (standard deviation) (Supplementary Figure 4). The filaments fully occupied with protein (33 examples) were 117 ± 11 nm in length, which corresponds to a shortening of the DNA length by ∼1.5-fold (average 1.47). For 116 examples, combining all partially occupied filaments with more than 20% protein coverage and all fully occupied filaments, the shortening factor shows a distribution that peaks at 1.5 as well (Figure 4F).
Product pattern of methylated DNA generated by Dnmt3a-C/3L-C
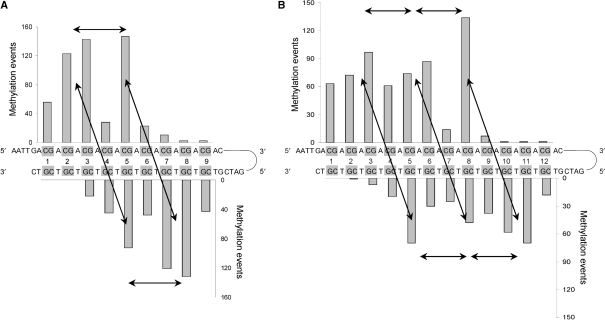
To determine the binding of several Dnmt3a-C/3L-C tetramers to the DNA and map the positions of the individual active sites with respect to the DNA, we used two substrates having either 9 or 12 CG sites next to each other. Methylation was investigated by the hairpin bisulfite approach that allows analyzing the methylation of both DNA strands in a single substrate molecule. The distribution of the methylation marks in the 9xCG and 12xCG substrates (with 5′ overhangs of four nucleotides) was investigated after sequencing of 260 and 169 clones, respectively. The results showed a pronounced, nonrandom distribution of methylation events (Figure 5). To assess the statistical significance of the preferential methylation at particular sites, P-values were calculated assuming a binomial distribution of the methylation events (Supplementary Text 1). For the 9xCG substrate we observed statistically significant preferential methylation at CG2 and CG3 in the upper strand, CG5 in both strands and CG7 and CG8 in the lower strand (Figure 5A). Similarly, the 12xCG substrate was preferentially methylated to a significant degree at CG2 and CG3, CG5 and CG6 and CG8 in the upper strand and CG5, CG8, CG10 and CG11 in the lower strand (Figure 5B). Hence, the multimeric complex positions its active sites such that it methylates the DNA in distances of 8–10 bp in both strands of the DNA. Both substrates show 3–4 CG sites at the 3′ parts of each strand that were hardly methylated. This suggests binding of the tetramer to DNA in an orientation such that two CG sites are methylated in a distance of 8–10 bp with both strands being methylated towards the 5′ end.
Figure 5.
Methylation pattern generated by Dnmt3a-C/3L-C. DNA methylation analysis by hairpin bisulfite experiments. Double-stranded oligonucleotide substrates containing either 9 (A) or 12 (B) equally spaced CG sites were methylated by Dnmt3a-C/3L-C complex and the methylation pattern was analyzed by the hairpin bisulfite method. 260 and 169 clones were analyzed for the 9xCG and 12xCG substrates, respectively. The P-values for the enrichment of methylation at the particular sites are given in Supplementary Text 1. Arrows connect peaks of methylation separated by 8–10 bp in the same or opposite DNA strand.
DISCUSSION
The catalytic domain of Dnmt3a, Dnmt3a-C, has two equally sized surface areas involved in protein–protein interactions, the RD and FF interface. Both interfaces are required for the catalytic activity of a Dnmt3a-C/3L-C heterotetramer, which needs to bind the cofactor AdoMet, bind the substrate DNA (mainly via the RD interface), and transfer the methyl groups. Gel retardation experiments suggest a cooperative multimerization of Dnmt3a-C/3L-C on DNA. In a previous study Dnmt3a-C DNA binding and Dnmt3L-C interaction were investigated by various techniques showing that Dnmt3L-C stimulates AdoMet binding, DNA binding and DNA methylation by Dnmt3a-C (11). In addition, Surface Plasmon Resonance was used to study formation of the protein–DNA complex. In this experimental system under the conditions of no or short preincubation and the covalent attachment of one of the partner to the surface, formation of binary Dnmt3a-C/DNA but not ternary Dnmt3a-C/3L-C/DNA complexes was observed. Therefore, the interaction of Dnmt3a and Dnmt3L was interpreted as being transient, and Dnmt3L-C was proposed to act as a DNA exchange factor for Dnmt3a-C. Here, we have confirmed that preincubated Dnmt3a-C and Dnmt3L-C are present in the protein–DNA complexes (Supplementary Figure 5) indicating that that this model is no longer valid.
SFM imaging shows that the Dnmt3a-C/3L-C complex forms protein/nucleic acid filaments. The apparent width of the filamentous protein–DNA complexes when measured in air was ∼20 nm (Figure 4A). To approximate the real dimensions of the fiber, the correction for tip geometry of the SFM cantilever could be estimated to 2–5 nm, because the apparent width of naked DNA was 4–7 nm while its true diameter is 2 nm. These numbers suggest that the physical diameter of the filaments is about 15–18 nm which is similar to the longest dimension of Dnmt3a-C/3L-C tetramer in the crystal structure (16 nm) (19). During filament formation, the apparent length of the DNA shortens about 1.5-fold. This relative small degree of shortening suggests that the DNA component in the nucleoprotein complex travels in a relatively straight wiggly path.
In the modeled tetramer–DNA complex, the two active sites of the two central Dnmt3a-C molecules would methylate opposite DNA strands at a distance of about 10 bp (19), which fits to the result of the hairpin bisulfite experiments. However, in these experiments we observed a coupled methylation also at sites separated by 8–10 bp on the same DNA strands, similarly as observed previously on longer DNA (19). This result suggests a regular arrangement of more than one tetramer on the DNA. The finding that CG sites can be methylated in both strands indicates that two Dnmt3a-C molecules from adjacent tetramers can approach each other such that one would interact with the CG site in the upper strand of the DNA and the second in the lower strand (Supplementary Figure 6).
In extrapolation, we suggest that the nucleoprotein filaments observed in EMSA and SFM studies consist of many Dnmt3a-C/3L-C tetramers bound next to each other on the DNA. Such nucleoprotein filaments could efficiently methylate targets presenting CG sites in a periodic pattern of 8–10 bp. Such periodic distribution of CG sites has been observed in the differentially methylated DNA sequences associated with maternally imprinted genes (19), methylation of which is lost in the female germ cells of Dnmt3L (27) and germline Dnmt3a knock out mice (28). Further experiments will be required to elucidate Dnmt3 binding to differently modified chromatin structures in the cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The BMBF BioFuture program; the DFG (JE 252/4 and JE 252/6); and US NIH GM49245 (to D.J. and X.C). Funding for open access charges: DFG.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 6.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 7.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 8.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margot JB, Ehrenhofer-Murray AE, Leonhardt H. Interactions within the mammalian DNA methyltransferase family. BMC Mol. Biol. 2003;4:7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 12.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 13.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 14.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 15.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 16.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 18.Dhayalan A, Jurkowski TP, Laser H, Reinhardt R, Jia D, Cheng X, Jeltsch A. Mapping of protein-protein interaction sites by the ‘absence of interference’ approach. J. Mol. Biol. 2008;376:1091–1099. doi: 10.1016/j.jmb.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeltsch A, Lanio T. Site-directed mutagenesis by polymerase chain reaction. Methods Mol. Biol. 2002;182:85–94. doi: 10.1385/1-59259-194-9:085. [DOI] [PubMed] [Google Scholar]
- 22.Roth M, Jeltsch A. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem. 2000;381:269–272. doi: 10.1515/BC.2000.035. [DOI] [PubMed] [Google Scholar]
- 23.Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KM, Manley NC, Vary J.C., Jr, Morgan T, Hansen RS, Stoger R. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl Acad. Sci. USA. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurkowski TP, Anspach N, Kulishova L, Nellen W, Jeltsch A. The M.EcoRV DNA-(adenine N6)-methyltransferase uses DNA bending for recognition of an expanded EcoDam recognition site. J. Biol. Chem. 2007;282:36942–36952. doi: 10.1074/jbc.M706933200. [DOI] [PubMed] [Google Scholar]
- 27.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 28.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.