Abstract
Familial amyotrophic lateral sclerosis (fALS) caused by mutations in copper–zinc superoxide dismutase (SOD1) is characterized by the presence of SOD1-rich inclusions in spinal cords. Similar inclusions observed in fALS transgenic mice have a fibrillar appearance suggestive of amyloid structure. Metal-free apo-SOD1 is a relatively stable protein and has been shown to form amyloid fibers in vitro only when it has been subjected to severely destabilizing conditions, such as low pH or reduction of its disulfide bonds. Here, by contrast, we show that a small amount of disulfide-reduced apo-SOD1 can rapidly initiate fibrillation of this exceptionally stable and highly structured protein under mild, physiologically accessible conditions, thus providing an unusual demonstration of a specific, physiologically relevant form of a protein acting as an initiating agent for the fibrillation of another form of the same protein. We also show that, once initiated, elongation can proceed via recruitment of either apo- or partially metallated disulfide-intact SOD1 and that the presence of copper, but not zinc, ions inhibits fibrillation. Our findings provide a rare glimpse into the specific changes in a protein that can lead to nucleation and into the ability of amyloid nuclei to recruit diverse forms of the same protein into fibrils.
Keywords: amyloid, amyotrophic lateral sclerosis, neurodegeneration, protein aggregation
The antioxidant metalloprotein copper-zinc superoxide dismutase (SOD1) is a 153-residue, β-rich, homodimeric protein that is abundantly present in the cytoplasm. Each subunit of the mature form contains a copper ion, a zinc ion, and a disulfide bond (1). In vitro studies have shown that the presence of the metal cofactors, copper and zinc, protect the disulfide bond from reduction, suggesting a possible explanation for the persistence of the disulfide bond in the reducing environment of the cytoplasm (2). More than 100 mutations in SOD1 have been linked to the familial form of amyotrophic lateral sclerosis (fALS), a fatal neurodegenerative disease caused by selective death of motor neurons. Neuronal death is attributed to a toxic gain of function by mutant SOD1, but the exact mechanism of toxicity is unknown. Mutations in SOD1 that have been identified in fALS patients occur in 74 positions that are well dispersed over the length of this 153-residue polypeptide (3).
Proteinaceous aggregates have been found in the spinal cords of ALS patients, and immunomicroscopy has confirmed that the protein inclusions present in the spinal cords of SOD1-fALS patients are rich in SOD1 (4, 5). Transgenic mice expressing human SOD1 mutants that cause fALS share many symptoms with their human counterparts, including progressive motor neuron degeneration and the presence of detergent-resistant, SOD1-rich aggregates in their spinal cords (6, 7). These aggregates have recently been shown to consist primarily of full-length, unmodified SOD1 (6). The visible proteinaceous inclusions have a fibrillar appearance and bind thioflavin S, suggesting an amyloid-like structure (7–9). Taken together, these results suggest that the ability to misfold into an amyloid form under physiologically accessible conditions represents a fundamental property of this protein and that this property is related to the toxicity of the ALS-SOD1 mutant proteins.
Bound copper and zinc, the intrasubunit disulfide bond, and the dimeric nature of the protein all contribute to the unusually high stability of mature SOD1, and the absence of one or more of these posttranslational modifications may destabilize the structure of SOD1 sufficiently to allow misfolding into an amyloid form. Previous studies have demonstrated that apo (metal free) SOD1 can be induced to form insoluble, fibrillar structures, but only under destabilizing conditions such as low pH (10), elevated temperature, the presence of trifluoroethanol (11), or removal of the intrasubunit disulfide bond by mutation (12).
In the current study, we report that small amounts of disulfide-reduced, metal-free, monomeric SOD1 can rapidly initiate fibril formation in disulfide-intact forms of SOD1 under mild, physiologically relevant conditions. Once initiated, SOD1 amyloid fibrils elongate by recruiting apo or partially metallated, dimeric, disulfide-intact SOD1. We also show that the presence of a small amount of disulfide-reduced mutant SOD1 can initiate fibrillation in WT SOD1, suggesting that WT SOD1 can participate in the etiology of ALS. Our studies show that even when fibrillation is initiated by the completely immature, disulfide-reduced, apo form of SOD1, more mature and structurally stable forms of SOD1 can participate in the elongation process. These findings have important implications in understanding the role of SOD1 maturation in fALS etiology and demonstrate the ability of WT SOD1 to contribute to ALS pathogenesis. These results also constitute a clear demonstration of an amyloid-forming process in which initiation of fibrillation can be separated mechanistically from elongation.
Results
Apo SOD1, normally dimeric, can be dissociated into monomers by treatment with a chaotrope such as guanidinium hydrochloride (Gdm·HCl) or by reduction of its intrasubunit disulfide bond (13–15), and monomerization has been implicated in SOD1 aggregation pathways (16). To determine the effect of partial monomerization, apo human WT SOD1 was incubated with agitation at 37°C, pH 7.4, in the presence of either 1 M Gdm·HCl or 50 mM DTT, both conditions known to induce partial dimer dissociation into monomers. An exponential rise in thioflavin T (TfT) fluorescence was observed after a lag time of ≈38 (± 4.2) h in the case of Gdm·HCl and 2.1 (± 0.3) h in the case of DTT. (Fig. 1A). Turbidity developed under both conditions, and electron microscopy confirmed the presence of amyloid fibrils (Fig. 1B). Other reducing agents, specifically tris(2-carboxyethyl)phosphine (TCEP) and glutathione, were also effective in promoting rapid SOD1 fibrillation (Fig. 1C). The presence of dioxygen in these reactions did not alter the kinetics, because samples prepared under anaerobic conditions showed similar lag phases [supporting information (SI) Fig. S1]. The ability of glutathione to promote fibrillation under anaerobic conditions suggests that reducing conditions in vivo such as those found in the cytoplasm are capable of promoting fibrillation.
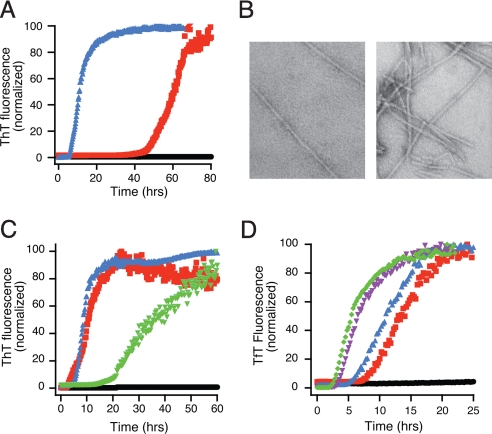
Fig. 1.
Fibrillation of SOD1 can occur in mild conditions. Apo- SOD1 (50 μM) in 10 mM potassium phosphate buffer, pH 7.4 with additions as indicated was incubated with constant agitation at 37 °C in 96-well plates. TfT fluorescence was monitored as an indicator of fibril formation. (A) Fibrillation of SOD1 is stimulated by 1 M Gdm·HCl (red) or 50 mM DTT (blue). No addition is indicated by black circles. (B) Electron micrographs of SOD1 fibrils generated in the presence of 1 M Gdm·HCl (Left) and 5 mM DTT (Right). (Scale: 1 cm = 200 nm). (C) Multiple reducing agents catalyze fibril formation. Apo-SOD1 alone (black circles), with 10 mM DTT (red squares), 10 mM TCEP (blue triangles), or 10 mM glutathione (green triangles). (D) DTT concentration dependence of fibril formation for apo-SOD1. DTT concentrations are: 50 μM (black circles), 250 μM (red squares), 1 mM (blue triangles), 5 mM (purple inverted triangles), and 25 mM (green diamonds).
Gdm·HCl treatment leaves the intrasubunit disulfide bond of SOD1 intact, but 50 mM DTT treatment, in our experience, generates a mixture of disulfide-intact SOD1 (SOD1S-S) and disulfide-reduced SOD1 (SOD12SH). To determine the minimum concentration of DTT required for fibrillation, apo-SOD1 was incubated with 50 μM to 25 mM DTT (Fig. 1D). Surprisingly, as low as 250 μM DTT was capable of promoting the formation of amyloid fibrils. At this concentration of reducing agent, we expect only a small fraction of the protein to be reduced, suggesting that a small amount of disulfide-reduced SOD1 is sufficient to initiate fibrillation.
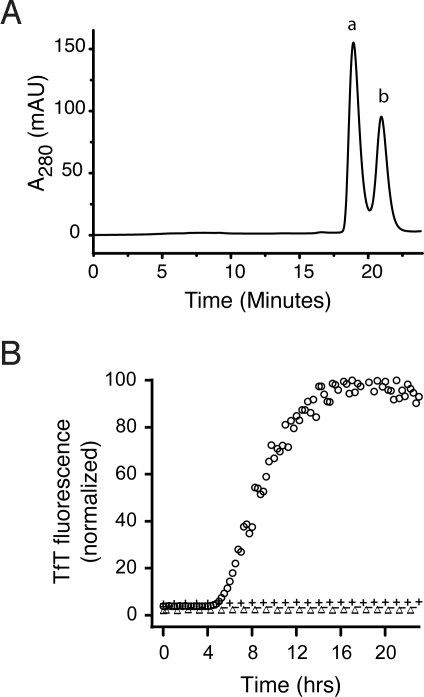
SOD1S-S is a dimer, whereas SOD12SH is a monomer and, thus, size-exclusion chromatography (SEC) can be used to separate them. SEC on an aliquot collected during lag phase from a fibrillation reaction containing 5 mM DTT (Fig. 2A) showed the presence of dimeric SOD1S-S eluting at 19.8 min (peak a) and monomeric SOD12SH eluting at 21.2 min (peak b). Treatment of SOD1 from these 2 peaks with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), a thiol modifying agent, followed by SDS/PAGE demonstrated that 2 extra AMS units per subunit had been added to the SOD1 in peak b, confirming the assignments of peak a as SOD1S-S and peak b as SOD12SH (Fig. S2).
Fig. 2.
Disulfide-reduced SOD1 with free thiol groups initiates fibrillation in the absence of reducing agent. (A) Separation of SOD1SH-SH (peak b) and SOD1S-S (peak a) generated during the lag phase of a fibrillation reaction in 5 mM DTT performed in the manual format by SEC. (B) These peaks were used as isolated or treated with NEM to block free thiols and added to fibrillation reactions without reducing agent. Apo-SOD1 (47.5 μM) in phosphate buffer was incubated with 2.5 μM SOD1 in the following forms: SOD12SH (peak b), ○; SOD1S-S (peak 2), △; SOD1–4NEM (peak b, NEM treated), −; SOD1S-S-2NEM (peak a, NEM treated), +.
To test the hypothesis that the presence of only a small amount of SOD12SH is sufficient to initiate fibrillation of SOD1S-S, we studied the effect of replacing the reducing agent with SEC-purified SOD12SH or SOD1S-S (peaks a and b, respectively, in Fig. 2A). Because agitation alone has been shown to promote misfolding and amyloid formation in many proteins, because of repeated transient exposures to the air–water interface, we also used SOD1S-S that had been isolated from the same reaction as the SOD12SH, and thus had been similarly agitated, as a control. We found that a very small amount of SOD12SH (5% of the overall reaction) was sufficient to induce fibrillation, whereas SEC-purified SOD1S-S had no effect (Fig. 2B). Amyloid fibril formation is proposed to occur by the formation of 1 or more amyloid nuclei during the lag phase, which then recruit soluble protein during the elongation phase to generate mature fibrils. Our results suggest that SOD12SH itself can initiate amyloid formation, even in the absence of reducing agents, by forming nuclei, which then recruit SOD1S-S to form mature fibrils. This mechanism is likely to be relevant to fALS pathogenesis in vivo because disulfide-reduced SOD1 is present in the spinal cords of transgenic ALS mice that eventually develop SOD1-rich aggregates in their terminal stages (17).
In addition to Cys-57 and Cys-146, which form the disulfide bond, each subunit of SOD1 contains 2 reduced cysteine residues, Cys-6 and Cys-111. Previous cell culture and in vitro aggregation studies suggested that these 2 nondisulfide cysteine residues might play an important role in the formation of disulfide-cross-linked SOD1 oligomers and aggregates (18–20). To understand and distinguish the roles of Cys-6 and Cys-111 from those played by the disulfide-forming residues, Cys-57 and Cys-146, we investigated the effect of mutations in these positions on the ability to initiate fibrillation in our present system (Fig. 3A). Preparations of disulfide-reduced WT SOD1 and C6A,C111S SOD1 (AS-SOD1) were generated by overnight treatment of the apoproteins with 10 mM TCEP, followed by SEC-HPLC purification. The same proteins with cysteine thiolates blocked by reaction with N-ethylmaleimide (NEM) were also tested, as were single-cysteine mutants C57S SOD1 and C146S SOD1. In the absence of reducing agents, only disulfide-reduced WT SOD1 and AS-SOD1 were capable of nucleating the conversion of soluble SOD1 into amyloid fibrils. The absolute requirement that both C57 and C146 be present and unalkylated suggests that these residues participate in the formation of amyloid nuclei by intermolecular disulfide cross-linking.
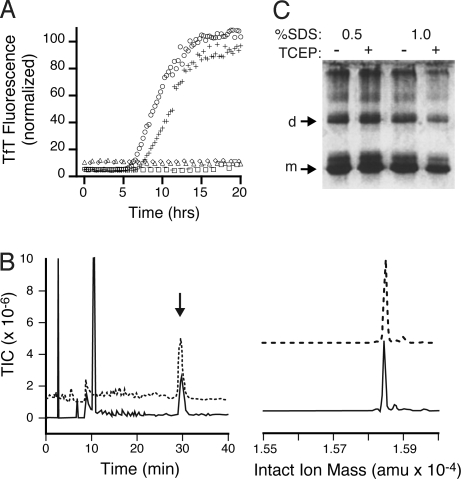
Fig. 3.
Both Cys-57 and Cys-146, but not Cys-6 or Cys-111, are necessary to initiate amyloid formation. (A) 2.5 μM SOD12SH (○), AS2SH (+), SOD1–4NEM (△), AS-2NEM (−), C57S (◇), and C146S (□) were added to 47.5 μM apo-SOD1 in the absence of reducing agents. (B) The amyloid nuclei elongate via noncovalent interactions to form mature fibrils. SOD1 fibrils grown in the presence of 5 mM DTT were denatured in 6.5 M Gdm·HCl with or without 20 mM TCEP and analyzed by HPLC-MS. (Left) Total ion count of Gdm·HCl-denatured (solid line) or Gdm·HCl-and-TCEP-denatured (dashed line) SOD1 fibrils. (Right) Reconstructed mass spectrum of peak at 30 min for LCMS of Gdm·HCl denaturation (solid line) and Gdm·HCl-TCEP denaturation (dashed line) reactions, showing monomeric SOD at 15,845 Da. (C) PAGE of amyloid fibrils grown in 5 mM DTT and denatured in SDS with or without 20 mM TCEP show similar amounts of monomers, dimers, and higher oligomers, suggesting fibril elongation does not involve intermolecular disulfide cross-linking.
In addition to initiation, intermolecular disulfide cross-linking might be involved in fibril elongation. For example, apo-SOD1 was found to form soluble, high molecular weight, disulfide-linked structures that bind thioflavin T (TfT) when incubated for extended periods at 37 °C and pH 7.4, with no agitation (18). To probe for the presence of intermolecular disulfide cross-linking under our conditions, SOD1 fibrils grown in 5 mM DTT were denatured in Gdm·HCl with or without TCEP, and the products were analyzed by HPLC-MS (Fig. 3B). Similar amounts of monomeric SOD1 were observed in both cases (peak at 30 min), suggesting that most of the SOD in the fibrils was not cross-linked by disulfide bonds. No dimers, trimers or higher-order oligomers were detected by HPLC. Gel electrophoresis on fibrils denatured in SDS with or without the addition of TCEP confirmed the presence of similar amounts of monomeric SOD under both conditions. Low amounts of dimers and higher-order oligomers were also detected; their presence may be caused by incomplete denaturation of fibrillar SOD1 in 0.5–1% SDS (Fig. 3C). As before, the presence of TCEP had no effect on the yield of monomeric or oligomeric SOD1. HPLC-MS of AS-SOD1 fibrils formed in the presence of 5 mM DTT and denatured in Gdm·HCl, in the absence of TCEP, yielded similar amounts of monomeric SOD in both cases, showing that the presence of free thiolates at Cys-6 and Cys-111 had no effect on the outcome of the reaction (Fig. S3). These results suggest that the nuclei, whose formation likely involved intermolecular disulfide cross-linking, developed into mature fibrils by recruiting disulfide-intact SOD1 through noncovalent interactions.
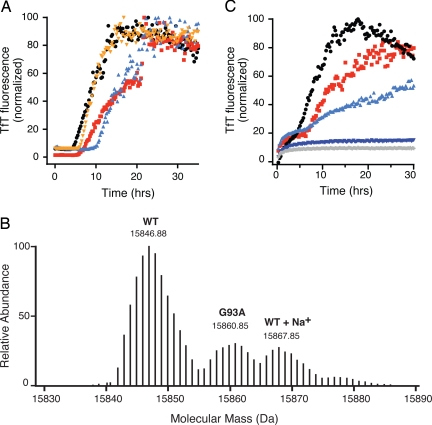
In vitro studies have shown that the intrasubunit disulfide bond in FALS-SOD1 mutant proteins is more susceptible to reduction than that in WT SOD1 and thus is more likely to be present under the reducing conditions prevalent in vivo (2). We therefore tested the ability of reduced ALS-SOD1 mutant proteins to induce fibrillation of WT SOD1. Apo-SOD1S-S was incubated with small amounts of apo-G93A2SH, apo-G37R2SH, and apo-D101N2SH (Fig. 4A) in the absence of reducing agents. In each case, the reduced mutant protein, present at 5% of the overall protein concentration, initiated fibrillation with a lag time comparable with that of WT SOD12SH. The presence of mutant SOD1 in the fibrils was verified by Fourier transform mass spectrometry (FTMS) on the denatured, TCEP-reduced fibrils (Fig. 4B). Small amounts of reduced ALS-SOD1 mutant proteins were also found to initiate fibrillation in 1:1 mixtures of WT and mutant-SOD1 proteins (Fig. S4).
Fig. 4.
Disulfide-reduced mutants can initiate fibrillation of SOD1, while copper, but not zinc, can inhibit fibril elongation. (A) Small amounts of disulfide-reduced fALS-SOD1 mutants can initiate fibril formation of apo-SOD1; 2.5 μM SOD12SH (red squares), G93A2SH (orange circles), G37R2SH (aqua triangles), and D101N2SH (black triangles) were incubated with 47.5 μM apo-SOD1 in the absence of reducing agents. Fibril formation was confirmed in all cases by electron microscopy (data not shown). (B) High-resolution mass spectrometry in an LTQ-FTMS instrument shows that WT SOD1 fibrils grown by the addition of G93A2SH and denatured in the presence of 6.5 M Gdm·HCl and 20 mM TCEP contain both WT SOD1 and G93A. Shown is the isotopically-resolved reconstructed intact-ion mass spectrum from the z = 10 peak. (C) Cu and Zn binding to apo-SOD1 can attenuate and, in some cases, abolish fibril formation initiated by SOD12SH; 2.5 μM SOD12SH was added to 47.5 μM apo-SOD1 (red squares), E2,EZn-SOD1 (black circles), as-isolated SOD1 (light blue triangles), Cu2E2-SOD1 (dark blue triangles), and Cu2Zn2-SOD1 (gray diamonds). As-isolated SOD1 refers to SOD1 purified from yeast. It contains 0.6 eq. copper and 2.5 eq. zinc ions per dimer.
In the native, holo enzyme, each subunit of the SOD1 dimer binds 1 copper and 1 zinc ion, and their presence has a profound impact on the stability of the protein. Fully metallated WT SOD1 melts at a very high 92 °C. The absence of bound copper lowers the melting point to 78 °C, and the successive removal of zinc ions lowers the melting point further to 62 °C (1 zinc per dimer) and 52 °C (apo-SOD1) (1). We examined the effect of adding small amounts of SOD12SH to the different metal-bound forms of SOD1. As shown in Fig. 4C, SOD1 with 1 zinc ion per dimer fibrillated with kinetics very similar to those for apo-SOD1, whereas isolated SOD1 containing zinc ions in both subunits showed a much slower rate of elongation. The absence of an effect caused by the binding of 1 zinc ion per apo-SOD1 dimer is surprising, because 1 zinc per dimer confers considerable structural stabilization in other assays (21). In contrast, the presence of 2 copper ions per dimer, either alone or in conjunction with bound zinc, completely inhibited the formation of amyloid fibrils, showing that copper-bound, disulfide-intact forms of SOD1 are not easily recruited by SOD1 amyloid nuclei.
Discussion
The ability of a specific form of a protein to act as an initiating agent for the fibrillation of another form has been demonstrated infrequently, presumably because of experimental difficulties associated with pinpointing initiation events. The few known instances include defining the minimum peptide sequence required to initiate fibrillation in Ure2p (22) and Aβ (23) and determining the oligomeric status of nuclei and the conformation changes required for their formation in polyglutamine peptides (24) and islet amyloid polypeptide (25). Here, we have been able to demonstrate the requirements for fibril initiation and elongation in SOD1 individually. Our studies show that a specific intermediate in SOD1 maturation pathway, disulfide-reduced, monomeric apo-SOD1, can initiate the rapid conversion of more mature forms of soluble SOD1 into amyloid fibrils. These results may be especially relevant to SOD1 fibrillation mechanisms in vivo because SOD1 monomers, including disulfide-reduced SOD1, have been detected in spinal cord extracts of transgenic mice expressing fALS SOD1 mutants well before they reach terminal stages of the disease (26, 27).
We have also shown that nucleation by disulfide-reduced (monomeric) SOD1 requires the presence of cysteine thiolates at both positions 57 and 146, suggesting that the nucleus is formed by intermolecular disulfide cross-linking using these cysteines. Similar species can form in vivo despite the reducing environment of the cytoplasm as shown by the presence of disulfide-cross-linked SOD1 oligomers in spinal cord extracts from ALS SOD1 transgenic mice. Cys-146 was one of the cysteine residues involved in their formation (28). Once SOD1 nuclei form, elongation proceeds via recruitment of apo- or partially-metallated disulfide-intact SOD1 through noncovalent interactions. Although the binding of zinc offers SOD1 limited protection from being assembled into fibrils, copper alone or in conjunction with zinc prevents SOD1 from being incorporated into amyloid fibrils. Because SOD1 aggregates isolated from the spinal cords of transgenic ALS mice are copper-deficient (17), copper binding may be an essential mechanism for protection from fibrillation in vivo.
It is important to note, however, that our studies do not show that disulfide reduction is absolutely required for initiation. In the presence of Gdm·HCl, disulfide-intact, apo-SOD1 can be induced to form amyloid fibrils, albeit with a significantly longer lag phase, possibly via a mechanism involving disulfide-intact monomers.
We found that small amounts of disulfide-reduced mutants can initiate fibrillation of WT SOD1. This result supports the idea that WT SOD1 participates in disease progression and may help explain the mechanism. Transgenic mice coexpressing WT and mutant SOD1 often show an earlier onset and accelerated progression of ALS-like neurodegenerative symptoms (9, 29, 30). Because the S S bond in ALS mutant proteins is more easily reduced than that in WT SOD1 (2), the reduced forms are more likely to be present in higher quantities in vivo, allowing for earlier nucleus formation, which could be followed by efficient incorporation of the more abundant WT SOD1. This process may be particularly important for unstable mutants such as L126Z or A4V that have low steady-state concentrations in vivo (31), and might require WT protein to sustain fibril growth, but it could also occur with more stable mutants in which WT and mutant proteins are present in approximately equal amounts. Supporting this idea, we found that in 1:1 mixtures of WT and mutant proteins the fibrils incorporated both proteins.
S bond in ALS mutant proteins is more easily reduced than that in WT SOD1 (2), the reduced forms are more likely to be present in higher quantities in vivo, allowing for earlier nucleus formation, which could be followed by efficient incorporation of the more abundant WT SOD1. This process may be particularly important for unstable mutants such as L126Z or A4V that have low steady-state concentrations in vivo (31), and might require WT protein to sustain fibril growth, but it could also occur with more stable mutants in which WT and mutant proteins are present in approximately equal amounts. Supporting this idea, we found that in 1:1 mixtures of WT and mutant proteins the fibrils incorporated both proteins.
In conclusion, our studies have allowed us to separate the processes of initiation and elongation in SOD1 fibril formation and have demonstrated that reduction of the intrasubunit disulfide bond dramatically accelerates the initiation process. Our observation of the initiation of the fibrillation of WT SOD1 by the addition of small amounts of disulfide-reduced ALS SOD1 mutant proteins provides a feasible model for initial stages of the disease process in SOD1-linked fALS.
Materials and Methods
SOD1 Expression and Purification.
WT and fALS SOD1 mutants were expressed in Saccharomyces cerevisiae and purified according to published procedures (14). C6A and C111S SOD1 (AS-SOD1) was expressed in Escherichia coli and purified by using a similar procedure. Purified SOD1 was demetallated by dialysis in a Slide-a-lyzer (Pierce; 10, 000 Da) against 50 mM EDTA, 50 mM NaCl, pH 3.8 as described (14) except that, in the last step, 20 mM potassium phosphate, pH 7.0 was used as the dialysis buffer. Apo-SOD1 was flash-frozen in liquid nitrogen and stored at −20 °C before use. Metal content of apo-SOD1 and metallated SOD1 was determined by inductively coupled plasma (ICP)-MS. Typically, apo-SOD1 contained ≤0.07 equivalents of Cu and Zn per dimer.
Metal Titrations.
Metal titrations were carried out according to the procedures outlined by Goto et al. (32). Briefly, apo-SOD1 was dialyzed against 100 mM sodium acetate, pH 5.5 and concentrated to ≈300 μM with a YM-10 microconcentrator (Millipore/Fisher). One equivalent of copper sulfate or zinc sulfate was added from an aqueous 10 mM stock solution in 2–4 equal aliquots at 1-h intervals with slow stirring at 4 °C. When necessary, the second equivalent of copper or zinc was added in a similar fashion and stirred overnight at 4 °C. To remove unbound metal, the reaction was transferred to a YM-10 microconcentrator, concentrated to 200 μL, and overlaid with an equal volume of 20 mM potassium phosphate, pH 7.4. The solution was concentrated, and the wash step was repeated 2–3 additional times. Cu2Zn2SOD1 was prepared by adding the necessary equivalents of zinc, followed by copper, to 300 μM as-isolated SOD1 in 100 mM sodium acetate, pH 5.5 whose copper and zinc content had been determined by ICP-MS. Zinc binding was allowed to proceed overnight at 4 °C, and then copper was added followed by overnight incubation. The sample was washed in the manner outlined above. Metal incorporation was verified by ICP-MS and UV-visible spectroscopy (32).
Amyloid Formation in Vitro.
All solutions were filtered through a 0.22-μm syringe filter before use. Amyloid conversion was carried out by using 2 formats: automated and manual. Reactions contained 50 μM apo-SOD1 in 10 mM potassium phosphate, pH 7.4 with or without with various concentrations of reducing agents (DTT, TCEP, or glutathione) or 1 M Gdm·HCl, as indicated. In the automated format, 200 μL of solution was pipetted into a chamber in a 96-well plate followed by a Teflon ball (1/8-in diameter) and 2 μL of TfT from a 4 mM aqueous stock. The plate was agitated at 300 rpm (3-mm rotation diameter) in a Fluoroskan plate-reader (Thermo) at 37 °C. Fluorescence measurements were recorded every 15 min by using λex = 444 nm, λem = 485 nm, with an integration time of 200 μs. All reactions were performed in replicates of 3 or more. Reactions carried out anaerobically were assembled in a glove box by using reagent solutions that were purged with argon and left open inside the anaerobic chamber for 1 h before use.
In the manual format, reactions of the same composition as above were performed at a final volume of 1 mL in 2-mL glass vials containing a Teflon ball (1/8-in diameter) and agitated on an orbital shaker at 250 rpm (19-mm rotation diameter). The vials were sealed with plastic caps containing a PTFE septum and purged with nitrogen. At intervals, aliquots were withdrawn by using a Hamilton gas-tight syringe through the septum. TfT fluorescence was measured by diluting 10 μL of the reaction mix into 150 μL of 40 μM TfT stock solution made in 5 mM potassium phosphate, pH 7, and fluorescence emission at 480 nm was recorded on a spectrophotometer using λex = 444 nm (excitation and emission slit widths = 2 mm). Lag times were calculated by fitting TfT fluorescence data to the formula:
where lag phase was calculated as tm − 2/k (33).
SEC-HPLC.
Disulfide-oxidized and -reduced apo-SOD1, which are dimeric and monomeric, respectively, were separated on a 7.5-mm × 30-cm TSK G2000 SW column (Toyosoda) equipped with a guard column on an Agilent 1200 HPLC using a mobile phase containing 50 mM NaCl, 50 mM potassium phosphate (pH 6.7), 5 mM DTT. Apoprotein (0.5–1 mg) reacted overnight with 10 mM TCEP was loaded at a concentration of 3–5 mg/ml. SOD1S-S and SOD12SH collected from HPLC were concentrated by using YM-10 microconcentrators to a final concentration of 3–5 mM, so that the concentration of accompanying DTT never exceeded 50 μM when HPLC-purified SOD1 was added to apo-SOD1 during initiation reactions. This procedure was done to ensure that initiation, where it occurred, was caused by the presence of disulfide-reduced apo-SOD1 and not DTT. As shown in Fig. 1D, apo-SOD1 fibrillation did not occur in DTT concentrations of 50 μM or less.
NEM and AMS Labeling.
Disulfide-intact or disulfide-reduced apo-SOD1 at 3–5 mg/ml was treated with 5 mM NEM at 37 °C for 1 h. Excess NEM was removed by 3 rounds of concentration and redilution in a YM-10 microconcentrator, in 20 mM potassium phosphate, pH 7.4. The presence of NEM bound to cysteines was checked by ESI-MS. AMS labeling was carried out before gel electrophoresis similar to published procedures (34).
Amyloid Denaturation Monitored by Gel Electrophoresis.
Fibrils from 1 well of the 96-well plate were transferred to a 1.5-mL microcentrifuge tube and centrifuged at 16,000 × g. The supernatant was decanted, and the pelleted fibrils were resuspended in deionized water (Nanopure) and centrifuged again to remove all soluble SOD1. The resulting pellet was resuspended in 100 μL of 20 mM potassium phosphate, pH 7.4. Ten microliters of the fibril suspension was incubated with 0.5 or 1% SDS with or without 20 mM TCEP in a total volume of 20 μL for 1 h at 50 °C. Twenty microliters of nondenaturing loading dye (without SDS or β-mercaptopethanol) was added to the reactions, and 20 μL of the resulting solution was loaded on a 5% stacking, 15% resolving SDS polyacrylamide gel and electrophoresed at 150 V until the samples stacked and then at 200 V. The gels were Coomassie-stained to visualize SOD1.
Liquid Chromatography/MS.
SOD1 fibrils from a well from reactions in the 96-well plate were collected by centrifugation at 16,000 × g for 15 min, resuspended in 200 μL of distilled water, and centrifuged again to remove all reducing agent, chaotrope, and soluble SOD1. The fibrils were denatured by incubation in 6.5 M Gdm·HCl with or without 20 mM TCEP at 50°C for 1 h. The reaction was then diluted 1:1 with 5% acetonitrile containing 1% formic acid and loaded on a reverse-phase PL-RPS column connected to an API Sciex mass spectrometer. SOD1 was eluted off the column by using a water/acetonitrile gradient.
FTMS.
High-resolution mass spectrometry was performed on an LTQ-FT instrument from Thermo Fisher, operated in nanospray mode. The sample was denatured according to the above procedure and desalted by using a C18 Zip-Tip (Millipore) according to the manufacturer's protocol, with final elution in 50% acetonitrile containing 0.1% formic acid. The sample (5 μL) was loaded into a pulled-glass nanospray emitter (Proxeon), and a stable spray was established at 1,800 V relative to the ion transfer tube. The instrument was operated in FTMS mode at 100,000 resolution at m/z = 400. After recording the full mass spectrum, an ion isolation experiment was performed on the +10 species centered at 1,587.0 ± 3 Da. The zero-charge spectrum was obtained by using Xtract software (Thermo Fisher). A collision-activated dissociation (CAD) experiment was performed in the ion trap (energy = 12), and the fragments were analyzed by FTMS. The complex MS/MS spectrum was matched to SOD1 sequences by using Prosight PC software (Thermo Fisher), confirming the presence of both WT SOD1 and G93A in the CAD spectrum.
Electron Microscopy.
Fibril samples from a single well in the 96-well plate were spun down at 16,000 × g, washed with water, and resuspended in 100 μL of deionized water. Five microliters of the fibril suspension was deposited on a formavar-coated copper grid (Ted Pella, Inc.). The sample was allowed to adsorb for 5 min, blotted, washed for 30 s with distilled water, blotted and then stained with freshly filtered 2% uranyl acetate (5 μL) for 3 min and blotted again. The grids were air-dried for 30 min before insertion into a JEOL 1200 electron microscope operated at 80 keV. Fibrils were typically visualized at a magnification of 75,000×.
Supplementary Material
Acknowledgments.
We thank Sean Lehman and Emmanuel Koli for protein purification of AS-WT SOD1 and Dr. Amir Liba for metal analysis of SOD1 by ICP-MS. This work was supported by National Institute of Neurological Disorders and Stroke Grant NS-049134.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 18649.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807058105/DCSupplemental.
References
- 1.Valentine JS, Doucette PA, Potter SZ. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 2.Tiwari A, Hayward LJ. Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J Biol Chem. 2003;278:5984–5992. doi: 10.1074/jbc.M210419200. [DOI] [PubMed] [Google Scholar]
- 3.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 4.Kato S, et al. Pathological characterization of astrocytic hyaline inclusions in familial amyotrophic lateral sclerosis. Am J Pathol. 1997;151:611–620. [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata N, et al. Cu/Zn superoxide dismutase-like immunoreactivity in Lewy body-like inclusions of sporadic amyotrophic lateral sclerosis. Neurosci Lett. 1994;179:149–152. doi: 10.1016/0304-3940(94)90956-3. [DOI] [PubMed] [Google Scholar]
- 6.Shaw BF, et al. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J Biol Chem. 2008;283:8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J. Copper-binding-site-null SOD 1 causes ALS in transgenic mice: Aggregates of non-native SOD 1 delineate a common feature. Hum Mol Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 8.Basso M, et al. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J Biol Chem. 2006;281:33325–33335. doi: 10.1074/jbc.M603489200. [DOI] [PubMed] [Google Scholar]
- 9.Jaarsma D, Teuling E, Haasdijk ED, De Zeeuw CI, Hoogenraad CC. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J Neurosci. 2008;28:2075–2088. doi: 10.1523/JNEUROSCI.5258-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiDonato M, et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J Mol Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 11.Stathopulos PB, et al. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc Natl Acad Sci USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa Y, Kaneko K, Yamanaka K, O'Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in familial form of ALS. J Biol Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnesano F, et al. The unusually stable quaternary structure of human Cu, Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 14.Doucette PA, et al. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant: Insights into the molecular basis for dimer stability. J Biol Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- 15.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakhit R, et al. Monomeric Cu/Zn superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial ALS. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson PA, et al. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 18.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niwa J, et al. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J Biol Chem. 2007;282:28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- 20.Karch CM, Borchelt DR. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potter SZ, et al. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 22.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 23.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. On the nucleation of amyloid β-protein monomer folding. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wetzel R. Kinetics and thermodynamics of amyloid fibrils assembly. Acc Chem Res. 2008;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 25.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 26.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 27.Zetterstrom P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa Y, Fu R, Deng HX, Siddique T, O'Halloran TV. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc Natl Acad Sci USA. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng HX, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaarsma D, et al. Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman EK, Wilcox HM, Scott RW, Siman R. Proteasome inhibition enhances the stability of mouse Cu/Zn superoxide dismutase with mutations linked to familial amyotrophic lateral sclerosis. J Neurol Sci. 1996;139:15–20. [PubMed] [Google Scholar]
- 32.Goto JJ, et al. Loss of in vitro metal ion binding specificity in mutant copper-zinc superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2000;275:1007–1014. doi: 10.1074/jbc.275.2.1007. [DOI] [PubMed] [Google Scholar]
- 33.Cohlberg JA, Li J, Uversky VN, Fink AL. Heparin and other glycosaminoglycans stimulate the formation of amyloid fibrils from-synuclein in vitro. Biochemistry. 2002;41:1502–1511. doi: 10.1021/bi011711s. [DOI] [PubMed] [Google Scholar]
- 34.Furukawa Y, Torres AS, O'Halloran TV. Oxygen-induced maturation of SOD1: A key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.