Abstract
The high-risk human papillomaviruses (HPVs) are the causative agents of nearly all cervical cancers and are etiologically linked to additional human cancers, including those of anal, oral, and laryngeal origin. The main transforming genes of the high-risk HPVs are E6 and E7. E6, in addition to its role in p53 degradation, induces hTERT mRNA transcription in genital keratinocytes via interactions with Myc protein, thereby increasing cellular telomerase activity. While the HPV type 16 E6 and E7 genes efficiently immortalize human keratinocytes, they appear to only prolong the life span of human fibroblasts. To examine the molecular basis for this cell-type dependency, we examined the correlation between the ability of E6 to transactivate endogenous and exogenous hTERT promoters and to immortalize genital keratinocytes and fibroblasts. Confirming earlier studies, the E6 and E7 genes were incapable of immortalizing human fibroblasts but did delay senescence. Despite the lack of immortalization, E6 was functional in the fibroblasts, mediating p53 degradation and strongly transactivating an exogenous hTERT promoter. However, E6 failed to transactivate the endogenous hTERT promoter. Coordinately with this failure, we observed that Myc protein was not associated with the endogenous hTERT promoter, most likely due to the extremely low level of Myc expression in these cells and/or to differences in chromatin structure, in contrast with hTERT promoters that we found to be activated by E6 (i.e., the endogenous hTERT promoter in primary keratinoctyes and the exogenous hTERT core promoter in fibroblasts), where Myc is associated with the promoter in either a quiescent or an E6-induced state. These findings are consistent with those of our previous studies on mutagenesis and the knockdown of small interfering RNA, which demonstrated a requirement for Myc in the induction of the hTERT promoter by E6 and suggested that occupancy of the promoter by Myc determines the responsiveness of E6 and the downstream induction of telomerase and cell immortalization.
The high-risk papillomaviruses (HPVs; e.g., HPV type 16 [HPV-16] and HPV-18) are associated with anogenital carcinomas (65, 66), laryngeal carcinomas, and head and neck carcinomas (15). These HPVs carry two oncogenes, E6 and E7, which are retained and expressed in HPV-positive cervical cancers (1, 2, 50) and are required for maintenance of the tumorigenic phenotype (35, 36). The E6 and E7 proteins were first identified as targeting the p53 and Rb tumor suppressor pathways in host cells (8, 9, 35, 36, 47, 48), thereby disrupting cell cycle controls.
Telomerase is a specialized reverse transcriptase that synthesizes repeat DNA sequences at the ends of chromosomes termed telomeres (17). The absence of telomerase activity in most normal human cells results in the progressive shortening of telomeres with each cell division (19, 56, 60), eventuating in growth arrest or replicative senescence (6, 19). In contrast to most human somatic cells, immortalized and cancer cells contain detectable telomerase activity and consequently maintain their telomere length and proliferative potential (18, 25, 52, 63).
Our previous studies and those of other laboratories have shown that E6-mediated hTERT transactivation is independent of p53 degradation and interactions with PDZ proteins (12, 14, 22, 26, 30). However, as demonstrated in studies of small interfering RNA (siRNA) knockdown, hTERT transactivation by E6 requires the cellular ubiquitin ligase E6AP as well as Myc (12, 14, 30, 58). We have also shown that E6 and Myc associate in vivo and bind coordinately with promoter activation to the hTERT promoter in primary human foreskin keratinocytes (HFKs) (58).
The HPV-16 E6 oncoprotein increases cellular telomerase activity (27, 54), predominantly by inducing transcription of the hTERT gene (13, 32, 42, 57). The hTERT protein is the catalytic, rate-limiting subunit of the telomerase enzyme complex and is selectively expressed in a small subset of normal cells (stem cells), tumor tissues, and tumor-derived cell lines (33, 40, 45, 55). Interestingly, overexpression of hTERT protein or an hTERT promoter transactivator (Myc) can substitute for E6 in the immortalization of primary HFKs (26, 28), indicating that telomerase activation constitutes a major immortalizing activity of E6.
To further explore the relationship between E6, Myc, telomerase, and cell immortalization, we transduced primary HFKs and human foreskin fibroblasts (HFFs) with E6, E7, or both E6 and E7. Although the E6 and E7 genes can immortalize human foreskin keratinocytes (20, 37), they (HPV-16 DNA) fail to immortalize HFFs (43). The intent of this study was to determine whether genital keratinoctyes and fibroblasts differ in their regulation of telomerase and their response to E6 expression. We found that E6 and E7 were expressed in both keratinocytes and fibroblasts and induced the degradation of p53 and pRb, respectively. In addition, the E6 protein effectively induced an exogenous hTERT promoter in fibroblasts, and both E6 and Myc associated with this promoter. However, E6 could not induce the endogenous hTERT promoter or increase cellular telomerase activity in fibroblasts. This failure to induce the hTERT promoter correlated with a lack of promoter-bound Myc protein, an observation that is compatible with recent findings showing that overexpression of Myc can induce the fibroblast hTERT promoter, activate telomerase, and facilitate immortalization (4). Overall, our data demonstrate that E6 induces the hTERT promoter and activates telomerase, but only in cells in which Myc resides on the hTERT promoter.
MATERIALS AND METHODS
Plasmids and retroviruses.
We used the following previously described (57, 58) vectors and plasmids: the pJS55 vector, pJS55-16E6, pJS55-16E6-AU1, pJSS55-16E7, the pLXSN vector and pLXSN-16E6, pLXSN-16E7, pLXSN-16E6E7, pGL3-basic (pGL3B), and the pGL3B-hTERT core promoter (previously defined as pGL3B-255). Retrovirus-packaging cells (SD3443 cells) were transfected with a pLXSN vector containing either E6 or E7 or both E6 and E7, as described above, using Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer (54). Culture supernatants containing retroviruses were collected 24 h after transfection.
Cell cultures and generation of stable cell lines.
Primary HFKs and HFFs were cultured from neonatal foreskins as described previously (49) and were maintained in keratinocyte growth media (Invitrogen) supplemented with gentamicin (50 μg/ml) and complete Dulbecco's modified Eagle's medium, respectively. Primary HFKs (passage 2) and HFFs (passage 5) were transduced with amphotropic LXSN retroviruses expressing HPV-16 E6, E7, or both E6 and E7 (see above). Retrovirus-transduced cells were selected in G418 (100 μg/ml) for 5 days. Resistant colonies were pooled and passaged every 3 to 4 days (at a 1:4 ratio for HFKs and a 1:8 ratio for HFFs). HeLa cells were maintained in complete Dulbecco's modified Eagle's medium. All cells were cultured on plastic tissue culture dishes or in flasks.
RT-PCR.
Total cellular RNA was isolated with TRIzol reagent (Invitrogen) and treated with a DNA-free kit (Ambion) according to the manufacturer's instructions. cDNA synthesis and PCR were performed as described previously (28-30). The primers used for reverse transcription-PCR (RT-PCR) were 5′-CAACAAACCGTTGTGTGAT-3′ and 5′-CGTGTTCTTGATGATCTGC-3′ for unspliced E6 (29); 5′-ATGCATGGAGATACACCTAC-3′ and 5′-CATTAACAGGTCTTCCAAAG-3′ for E7; 5′-ATGCCCCTCAACGTTAGCTTC-3′ and 5′-AAGCCGCTCCACATACAGTC-3′ for Myc mRNA; 5′-TGAGCGATAACGATGACATC-3′ and 5′-CATCGAAGGCAGAGATGGTG-3′ for Max; 5′-CGGCGGTTCGGATGAACATC-3′ and 5′-GGTCGATTTGGTGAACGGCT-3′ for Mad1; and 5′-CTCAGACACCATGGGGAAGGTGA-3′ and 5′-ATGATCTTGAGGCTGTTGTCATA-3′ for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA.
Real-time qRT-PCR.
TaqMan real-time quantitative RT-PCR (qRT-PCR) was performed on a Bio-Rad iCycler MyiQ, using previously reported (10, 29, 30) primers and probes for the quantitation of hTERT mRNA (sense primer, 5′-TGACACCTCACCTCACCCAC-3′; anti-sense primer, 5-CACTGTCTTCCGCAAGTTCAC-3′; and TaqMan probe, 5′-ACCCTGGTCCGAGGTGTCCCTGAG-3′) and Myc mRNA (sense primer, 5′-ACCACCAGCAGCGACTCTGA-3′; anti-sense primer, 5′-TCCAGCAGAAGGTGATCCAGACT-3′; and probe, 5′-GAGGCAGGCTCCTGGCAAAAGGTC-3′). To detect Mad and Max, qRT-PCR was performed, using Bio-Rad Sybr green iQ mixture and the primer described above for RT-PCR. GAPDH was used as an internal control, and expression levels were analyzed using iQ5 software with the normalized expression (ΔΔCT) method according to the manufacturer's (Bio-Rad's) guidelines.
Western blots.
Cells from stable cell lines and cells treated with siRNA duplexes were washed once with phosphate-buffered saline and lysed in 2× sodium dodecyl sulfate (SDS) gel electrophoresis sample buffer. Proteins were separated on a 4% to 20% Tris-glycine gradient gel (Invitrogen) and were electrophoretically transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were blocked in 5% dry milk-phosphate-buffered saline-Tween 20 or 5% bovine serum albumin-phosphate-buffered saline-Tween 20 and incubated with one of the following primary antibodies: anti-p53, anti-Myc, anti-Max, anti-Mad (all from Santa Cruz Biotechnology), pRb (Cell Signaling Technology), or β-actin (Sigma). The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Santa Cruz Biotechnology), used at a dilution of 1:10,000. The membranes were visualized by using a Western blotting chemiluminescence luminol reagent (Santa Cruz Biotechnology).
Luciferase assay.
Primary HFKs or HFFs (1 × 105) were seeded onto 24-well plates and grown overnight. Transient transfections were performed, using Lipofectamine 2000 reagent (Invitrogen) according to the protocol provided by the manufacturer. Cotransfections were performed, using 0.5 μg of a core hTERT reporter plasmid (pGL3B-hTERT) and 20 ng of each expression vector (pJS55-16E6, pJS55-16E7, or both), or empty vectors as controls for basal promoter activity. Cells were also cotransfected with 2 ng of plasmid pRL-CMV (Promega), which contains the Renilla reniformis luciferase gene, and used as transfection controls. Firefly and Renilla luciferase activities were measured 24 h after transfection, using the Dual-Luciferase Reporter assay system (Promega).
Real-time q-TRAP.
Human keratinocytes and fibroblasts were lysed and then analyzed by real-time quantitative telomeric repeat amplification protocol (q-TRAP) assays as described previously (10, 29, 30).
ChIP assays.
HFKs, HFFs, or HeLa cells were grown to 80 to 90% confluence in 100-mm dishes. Chromatin immunoprecipitation (ChIP) assays were performed as described previously (58). Normal rabbit IgG (Santa Cruz Biotechnology), rabbit anti-Myc polyclonal antibody (N-262; Santa Cruz Biotechnology), and monoclonal anti-AU1 (Covance) were used for immunoprecipitation assays for HFKs, HFFs, and HeLa cells, respectively. PCR products were separated on a 1.8% agarose gel and visualized by ethidium bromide staining.
RESULTS
E6 and E7 disrupt the p53 and pRb pathways in both HFK and HFF cells.
The HPV-16 E6 and E7 genes are necessary and sufficient to immortalize primary HFKs and ectocervical keratinocytes (20, 38). While E6 directs the ubiquitin-dependent degradation of p53, it also has functions that are p53 independent, including telomerase activation and cell transformation (12, 35, 36). To determine whether the immortalizing activity of E6 and E7 correlated with tumor suppressor inactivation or telomerase induction, we used retroviruses to generate cell strains expressing an empty retroviral vector (pLXSN), E6 (pLXSN-16E6), E7 (pLXSN-16E7), or E6 and E7 (pLXSN-16E6E7), as described in Materials and Methods. The above cell lines were then passed serially in vitro to assay for immortalization. When cells reached 80% confluence, they were split at a 1:4 ratio for HFKs and a 1:8 ratio for HFFs. Therefore, one split would correspond to two or three cell population doublings (PDs) for HFKs and HFFs, respectively.
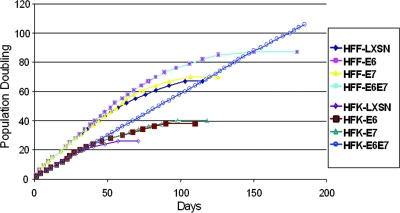
As shown in Fig. 1, HFKs and HFFs transduced with LXSN ceased proliferating at 22 to 24 PDs and 61 to 64 PDs, respectively. E6 alone extended the life span of HFKs to 32 to 34 PDs and HFFs to 82 to 85 PDs, and E7 alone extended cell divisions to 36 to 38 PDs and 67 to 70 PDs, respectively. Neither E6 nor E7 could independently immortalize HFKs or HFFs. As anticipated, the combined activity of E6 and E7 allowed HFKs, but not HFFs, to bypass senescence and become immortalized. We explored the possibility that the E6 and E7 proteins might be differentially targeting the p53 and pRb pathways in these two different human cell types or differentially inducing the hTERT promoter and thereby activating telomerase.
FIG. 1.
HPV E6 and E7 oncoproteins are sufficient to immortalize primary HFKs but not primary HFFs. Primary HFKs and HFFs were transduced with the indicated pLXSN-based retroviruses with E6, E7, E6E7 or an empty vector and selected as previously described. Cultures were passed continuously in vitro as described in the text, and the number of cell doublings was calculated and plotted versus the time in culture. Cultures that did not proliferate and expand in 20 days for HFKs and 30 days for HFFs were considered senescent and were terminated at the indicated times. This experiment was repeated a second time with similar results.
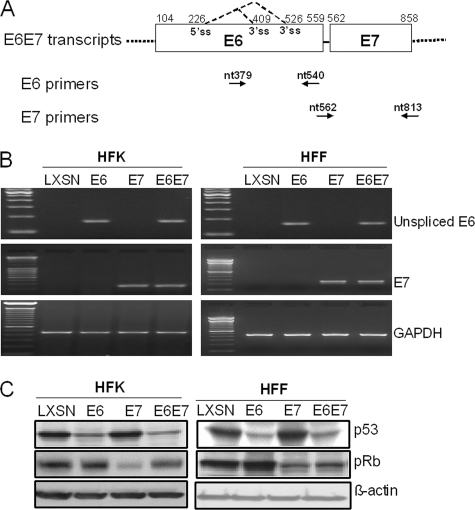
First, we confirmed that E6 and E7 were expressed in both HFK and HFF cells. RT-PCR was performed with E6- or E7-specific primers. As anticipated, E6 mRNA was expressed in HFKs and HFFs transduced with the E6 and E6E7 retroviruses, while E7 mRNA was expressed in the E7- and E6E7-transduced cells. HFKs and HFFs transduced with the pLXSN vector served as the control, and GAPDH was used to normalize gene expression (Fig. 2A). We also performed reactions without reverse transcriptase to confirm that PCR products came from mRNA, not DNA (data not shown).
FIG. 2.
Characterization of HFK and HFF strains expressing HPV E6 and E7 proteins. (A) Diagram of HPV-16 E6 and E7 mRNA expression and locations of primers used in this study. The E6 and E7 open reading frames are shown as open boxes. The dotted lines flanking the open boxes represent vector sequences. The alternative splicing sites in E6 are depicted as dotted lines. The numbers above the E6E7 transcripts are the nucleotide positions of the first nucleotide of the start codon and the last nucleotide of the stop codon of both E6 and E7 or the first nucleotide of the 5′ splicing site (5′ ss) or the last nucleotide of the 3′ splicing sites (3′ ss) in the HPV-16 genome. The primers used in this study are depicted below the transcript lines as arrows and numbers showing the locations (as nucleotide [nt] positions) of primers in the genome. (B) Confirmation of E6 and E7 mRNA expression. Primary HFKs and HFFs were transduced with pLXSN expressing 16E6, E7, E6E7, or an empty vector as previously described. Following antibiotic selection, the cell strains were analyzed for E6 and E7 mRNAs. Total cellular RNA was isolated from the transduced cell strains and treated with a DNA-free kit (Ambion), and RT-PCR was performed with the sets of HPV-16 unspliced E6- and E7-specific primers described in Materials and Methods. GAPDH mRNA was detected as an internal control. PCR products were analyzed on 2% agarose gels. (C) Expression of p53 and pRb proteins. The stable cell lines described above were lysed in electrophoresis sample buffer. The proteins were separated on 4- to 20%-gradient gels, transferred to a PVDF membrane, and reacted with mouse anti-p53 monoclonal antibody or rabbit anti-pRb. Anti-β-actin antibody was used to verify equal loading of samples. Low amounts of p53 protein were observed in E6- and E6E7-expressing cells, and a decreased level of pRb protein was noted in E7-expressing cells.
To determine whether the E6 and E7 proteins were functional in both cell types, we assayed the levels of tumor suppressors p53 and pRb by Western blotting, using β-actin as a loading control (Fig. 2B). Regardless of cell type, the p53 levels were decreased in E6-transduced cells, and the pRb protein levels were reduced in E7-expressing cells, indicating that the HPV oncoproteins were functional in both cell types and also that inactivation of p53 and pRb is insufficient for immortalization of HFFs.
E6 is sufficient to induce the exogenous hTERT promoter in both HFKs and HFFs.
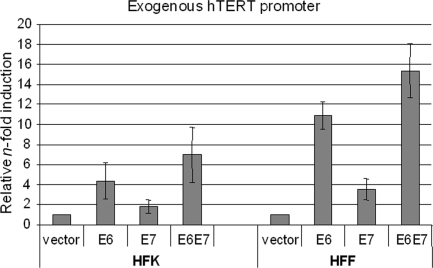
To determine whether the failure of E6E7 to immortalize HFF cells might be due to the inability of E6 to activate the hTERT promoter, we first analyzed the effect of E6 and E7 on an exogenous hTERT promoter luciferase construct. HFKs and HFFs were transfected with the control vector, pJS55-E6, pJS55-E7, or both, at the same time that they were transfected with the hTERT promoter reporter. In HFKs, E6 induced the hTERT promoter three- to fivefold compared to the control vector (Fig. 3, left side), and E7 increased promoter activity twofold. Together, E6 and E7 enhanced the hTERT promoter activity 7- to 10-fold. Surprisingly, we found that the exogenous hTERT promoter was induced to even greater levels in HFF cells, despite the fact that these cells cannot be immortalized by the E6 and E7 genes (Fig. 3, right side). The level of hTERT promoter activity in the HFF cells was proportionately higher, 9- to 12-, 3- to 5-, and 14- to 18-fold, for E6, E7, and E6E7, respectively.
FIG. 3.
E6 and E7 induce an exogenous core hTERT promoter in both HFKs and HFFs. Primary HFKs and HFFs were cotransfected with wild-type hTERT core promoter (pGL3B-hTERT) and either E6, E7, or E6E7. The pRL-CMV R. reniformis reporter plasmid was also transfected into the cells to standardize for transfection efficiency. Luciferase activity was measured 24 h after transfection, using the Dual-Luciferase Reporter assay system (Promega). Relative n-fold activation reflects normalized luciferase activity induced by E6 and E7 compared to the normalized activity of the control vector. The value of pGL3B-hTERT activity with the empty vector was set to 1. Error bars show standard deviations for at least three independent experiments. E6 is sufficient to induce the hTERT promoter in both HFK and HFF cells.
E6 cannot induce endogenous hTERT transcription or telomerase activity in HFFs.
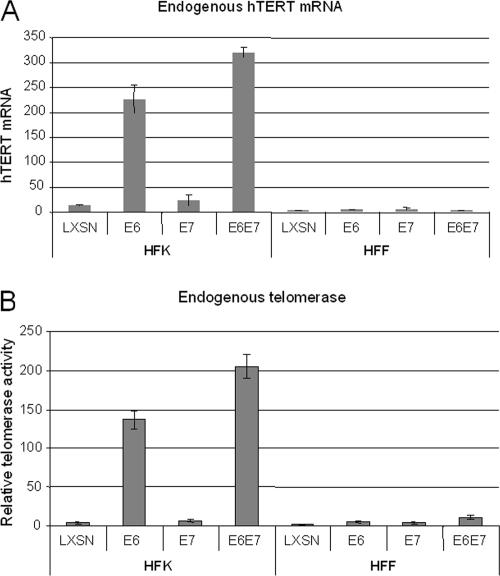
To validate the studies with the exogenous promoter, we performed qRT-PCR with endogenous hTERT mRNA in HFK and HFF cells. Unexpectedly, we found that neither the E6 nor E6E7 genes could induce the hTERT promoter (Fig. 4A). This finding contrasted with the ability of these genes to effectively induce both the endogenous (Fig. 4A) and exogenous hTERT promoters in HFKs (Fig. 3). To further confirm that this mRNA analysis reflected the downstream activation of telomerase, we prepared lysates from the cells and quantified the telomerase activity by using a q-TRAP assay (see Materials and Methods). As anticipated, E6 increased telomerase activity in HFKs (Fig. 4A). However, consistent with the levels of endogenous hTERT mRNA, we did not detect any increase in telomerase activity in HFF cells. These data indicate not only that E6 and E6E7 are insufficient to induce the endogenous hTERT promoter and telomerase in HFF cells but also that the activity of the transfected hTERT promoter constructs does not reliably reflect the activity of the endogenous promoter when compared between two different cell types.
FIG. 4.
E6 is sufficient to induce endogenous hTERT transcription and telomerase activity in primary HFKs but not in HFFs. (A) hTERT mRNA expression in stable keratinocyte and fibroblast cell lines. RNAs were used for detection of hTERT mRNA by qRT-PCR as described in Materials and Methods. A considerable amount of hTERT mRNA was detected in HFKs expressing E6 or both E6 and E7, but there was no detectable mRNA in HFFs expressing either the control vector (LXSN), E6, E7, or both E6 and E7. (B) Telomerase activity. A q-TRAP assay was done as described in Materials and Methods. Telomerase activity was observed in HFKs expressing E6 or both E6 and E7. There was no detectable telomerase activity in HFFs expressing either the control vector, E6, E7, or both E6 and E7.
Myc resides on the endogenous HFK hTERT promoter but not the HFF promoter.
Myc is known to be a direct activator of telomerase in both human keratinocytes and fibroblasts (4, 5, 28, 61), and our previous studies have shown that E6 and Myc physically interact and bind to the hTERT promoter (30, 51, 58). Indeed, the presence of Myc is required for E6-mediated induction of the hTERT promoter. Thus, it was possible that E6 was unable to activate the hTERT promoter because Myc was not present on the hTERT promoter in HFF cells, as it is in noninduced HFK cells.
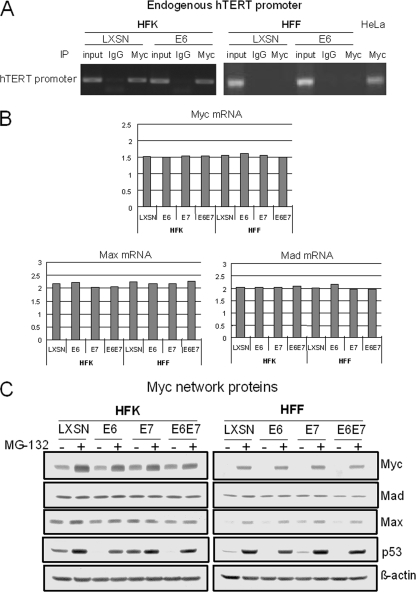
To test this hypothesis, we performed ChIP assays on both cell types with a Myc antibody. In primary HFKs expressing either the vector or E6, Myc was bound to the endogenous hTERT promoter (Fig. 5A), as reported previously (30, 58). However, we did not observe a signal for Myc binding to the endogenous hTERT promoter in HFFs, even when they expressed E6 (Fig. 5A). Myc was bound to the endogenous hTERT promoter in telomerase-positive HeLa cells (used as a positive control). Therefore, these experiments indicate that Myc is present on the hTERT promoter of telomerase-quiescent HFK cells but not on the promoter of telomerase-quiescent HFF cells. The inaccessibility of Myc to the hTERT promoter in HFF cells could derive from either differences in cellular Myc expression or differences in chromatin structure, which prevent Myc from accessing the promoter. We evaluated Myc expression levels, as discussed in the next section.
FIG. 5.
Myc expression and occupancy on the hTERT promoter differ in HFKs and HFFs. (A) Myc binds to endogenous hTERT promoter in primary HFKs but not in primary HFFs. Myc binding to the endogenous hTERT promoter was assayed by ChIP. Myc binding to the hTERT promoter was evaluated in the absence and presence of E6. HeLa cells were used as positive controls for Myc binding to the endogenous hTERT promoter. Myc binds to the endogenous hTERT promoter in HFKs with or without E6, but it does not bind to the promoter in primary HFFs. (B) Quantitation of Myc, Mad, and Max mRNAs. RNAs were subjected to Sybr green-based real-time RT-PCR on a Bio-Rad iQ5 system according to the manufacturer's instructions. GAPDH was used as an internal control, and data were analyzed using the normalized expression (ΔΔCT) method according to the manufacturer's (Bio-Rad's) guidelines. (C) Expression of Myc, Max, Mad, and p53 proteins. HFKs and HFFs expressing either the control vector, E6, E7, or both E6 and E7 were plated into 100-mm dishes in duplicate. After cells reached 80 to 90% confluence, a set of cells was treated with MG-132 for 4 h. All cell extracts were harvested with 2× SDS sample buffer. The same amounts of cell extracts were loaded onto SDS-4 to 20% polyacrylamide gels for electrophoresis, and protein was transferred to a PVDF membrane and blotted with anti-Myc, anti-Max, anti-Mad, and β-actin. Expression of both Myc and Max was lower in HFFs than in HFKs. There was no significant difference in the levels of Mad in HFKs and HFFs.
Myc protein is expressed at very low levels in HFF cells.
Our initial experiments employed RT-PCR to detect Myc mRNA expression in the two cell types, using both qualitative and qRT-PCR (Fig. 5B and C). In addition, we assayed the abundance of mRNA for two other transcription factors that modulate Myc activity, Max and Mad (11, 16, 21, 31, 39, 44, 46). In Fig. 5B and C, it is apparent that Myc is expressed at the mRNA level in both HFK and HFF cells. It is also apparent that there are similar levels of Mad and Max mRNA in these cells. It is also important to note that E6, E7, and E6E7 do not alter the expression of Myc or the Myc-related proteins.
However, the critical question is whether the Myc protein levels were different between HFK and HFF cells, which we explored by using Western blotting with a Myc antibody. As shown in Fig. 5C, Myc protein was detectable in HFKs expressing vector, E6, E7, or E6E7, and the viral oncoproteins did not alter the expression of Myc or its partners, Max and Mad. Unlike the Myc mRNA studies, we found gross differences in the levels of Myc protein between HFK and HFF cells. That is, there were very low levels of Myc protein in HFF cells compared to that in HFK cells, and it was extremely difficult to detect Myc protein in fibroblasts without the use of MG-132 proteasome inhibitor to prevent Myc degradation. p53 protein was blotted as a control, since it is degraded by E6, and as expected, MG-132 led to an increased level of p53 protein in E6-expressing cells. Thus, our data suggest that the low levels of Myc protein in HFFs might be due to rapid protein turnover. Interestingly, there is a coordinately low level of the Myc-associated Max protein in HFFs, and we could visualize Max reproducibly only when using a proteasome inhibitor. Unlike Myc and Max, however, the Mad protein is expressed at similar levels in both HFK and HFF cells, and there is little or no change in the level of this protein with either E6, E7, or E6E7 or in the presence of a proteasome inhibitor. In summary, the low levels of Myc protein detected in HFF cells might be the etiologic basis for its absence on the endogenous hTERT promoter and the nonresponsiveness of this promoter to E6.
Myc protein expression increases the ability of E6 to engage the endogenous hTERT promoter.
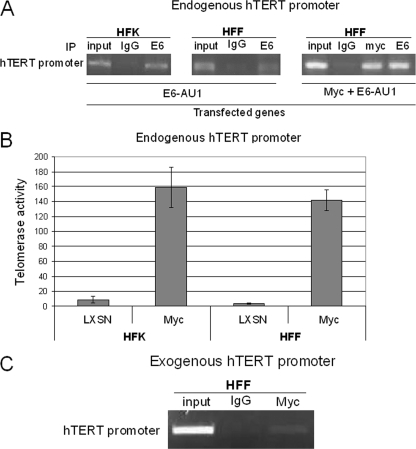
One possibility for the lack of E6's ability to induce the HFF hTERT promoter is that it cannot associate stably with the promoter without Myc binding, which is clearly lacking in HFF cells. To determine if Myc protein might modulate the association of E6 with the hTERT promoter, we transfected both HFF and HFK cells with epitope-tagged E6 (E6-AU1), which is known to retain its ability to induce the hTERT promoter. ChIP experiments with monoclonal anti-AU1 antibody or an IgG control demonstrated that E6-AU1 bound to the hTERT promoter in both types of cells (Fig. 6A), suggesting that E6 might associate with other promoter-associated proteins (e.g., NFX1-91), without the activation of telomerase. However, when the PCR signal was normalized to input, it appeared that E6 binding to the HFK promoter was stronger than that to the HFF promoter (Fig. 6A). This moderate, quantitative difference in E6 binding to the HFK and HFK promoters, however, does not seem to explain the complete absence of hTERT induction by E6 in HFF cells. More likely, it is the absence of Myc on the HFF promoter that is responsible for its lack of responsiveness to E6. Most important, while Myc was sufficient to induce telomerase in both HFKs and HFFs by itself (Fig. 6B), the forced expression of Myc in HFFs significantly increased E6 association with the endogenous hTERT promoter (Fig. 6A, right).
FIG. 6.
Myc and E6 associate with the hTERT promoter in HFFs. (A) Myc-mediated E6 binding to hTERT promoter. HFKs or HFFs transfected with E6-AU1 or both Myc and E6-AU1 were subjected to ChIP assays as described above, using rabbit anti-Myc antibody (N262; Santa Cruz Biotechnology) and monoclonal anti-AU1 antibody (Covance). (B) Myc induces telomerase activity in both HFKs and HFFs. HFKs and HFFs were transduced with pLXSN-Myc retrovirus, and cell lysates were analyzed with q-TRAP assays. Myc alone induces a similar level of telomerase activity in both HFKs and HFFs. (C) Endogenous Myc binds to exogenous hTERT promoter in HFFs. The plasmid pGL3B-hTERT was transfected to HFFs, and a Myc antibody or rabbit IgG was used for IP.
The E6-activated, exogenous hTERT promoter in HFF cells is associated with Myc.
The above data strongly suggest that Myc occupancy on the hTERT promoter correlates with the ability of E6 to induce hTERT transcription. However, as shown in Fig. 3, E6 is sufficient to induce an exogenous hTERT core promoter in HFFs, despite their very low level of Myc protein. This observation provides a unique system to define whether Myc protein levels or the chromatin structure might be the principal determinants of Myc/promoter binding. If Myc is associated with the exogenous hTERT promoter in HFF cells, the “open” chromatin conformation of transfected plasmid DNA may be more accessible to low levels of Myc than the endogenous promoter. If Myc is not associated with the exogenous promoter, it indicates not only that Myc levels regulate hTERT promoter association but also that E6 can induce the hTERT promoter without Myc. To test these possibilities, we transfected the hTERT core promoter and E6 into HFFs and performed a ChIP assay with a Myc antibody. Our data showed that Myc could be detected on the exogenous hTERT core promoter (Fig. 6C), although it was not detectable on the endogenous hTERT promoter (Fig. 5A). Thus, it appears that only hTERT promoters associated with Myc protein are inducible by E6. This conclusion, however, is qualified by the possibility that our ChIP assay conditions may not have been sufficiently sensitive to detect very low levels of Myc on the endogenous promoter.
DISCUSSION
The activation of telomerase in epithelial cells by HPV E6 is believed to be critical for cell immortalization (26). Although this function of E6 is independent of p53 degradation and PDZ binding ability, it appears to require E6AP (3, 12, 22, 24, 30). In contrast to the above studies, a single recent report suggests that E6AP is not required for hTERT promoter induction (51). The etiology of these experimental differences is currently unclear. However, independent studies have shown that the specific knockdown of E6AP interferes with hTERT promoter induction by E6 (3, 12, 22, 24, 30).
We, as well as other researchers, have shown that E6 activates hTERT transcription through Myc binding sites in the hTERT promoter (13, 32, 42, 57). However, E6 neither induces Myc expression (Fig. 5C) (12, 13, 42, 51, 57, 58) nor changes Myc binding to the hTERT promoter (12, 51, 58). Myc protein binds to the promoter in the presence or absence of E6 protein in keratinocytes (12, 51, 58). Interestingly, E6 associates with Myc in vivo and in vitro, and both bind to the core hTERT promoter (58), suggesting that there is a functional, cooperative interaction between these proteins on the hTERT promoter. The importance of Myc/hTERT promoter binding has been verified not only by promoter mutagenesis (13, 32, 42, 57, 59, 61) but also by siRNA knockdown of Myc expression and the expression of Mad and Mnt (Myc antagonists), all of which significantly inhibit E6-induced telomerase activity (29, 58; our unpublished data). E6 also increases the acetylation of histones resident on the promoter (12, 22; our unpublished data), suggesting that it might alter local chromatin structure and enhance promoter activity.
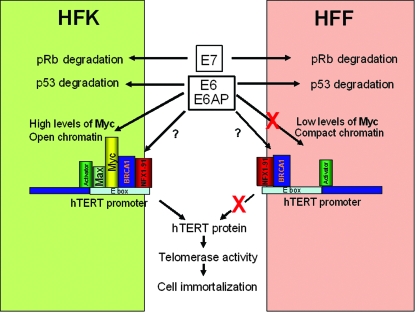
In this study, we examined the correlation between the ability of E6 to induce the hTERT promoter and to immortalize two very different cell types, HFKs and HFFs. It is well documented that the efficient immortalization of HFK cells requires the E6 and E7 genes and that these same two genes are incapable of immortalizing HFF cells (7, 26). Our current study provides a potential model to explain this differential cellular immortalization (Fig. 7).
FIG. 7.
A proposed model for E6 regulation of the hTERT promoter and cell immortalization. Using information from this and previous studies (12, 14, 30, 57, 58, 62, 64), we propose a model to explain how HPV E6 might regulate the hTERT promoter in a cell-type-specific manner. The ability of E6 and E7 to target p53 and Rb in HFF and HFK cells does not differ. However, the low levels of Myc protein in fibroblasts (HFF) relative to keratinoctyes (HFK) is probably a major determinant for the lack of Myc on the endogenous HFF hTERT promoter. Contributions of chromatin structure, however, might also contribute to the limited access of Myc and other regulatory proteins to the HFF hTERT promoter. Since E6 can bind to the HFF hTERT promoter in the absence of detectable Myc, it appears that other proteins resident on this site might mediate its binding. In separate studies, for example, it has been documented that E6 interacts with Myc, BRCA1, and NFX-1.
First, our data indicate that the E6 and E7 genes are expressed in both HFK and HFF cells and that two tumor-suppressor proteins, p53 and pRb, are similarly inactivated. When considered in the context of previous publications, our study suggests that p53 inactivation is neither necessary nor sufficient for cell immortalization. In this study, p53 inactivation (along with E7 expression) was not sufficient to induce the immortalization of HFF cells, and in previous studies of HFK cells, E6-mediated degradation of p53 was not required for cell immortalization (26, 28). The critical conclusion, however, is that the differential abilities of E6 and E7 to immortalize HFK and HFF cells do not reside in the differential expression levels/activities of these genes in the corresponding cells.
Second, the difference between HFK and HFF cells actually appears to derive from variations in the association of Myc protein with the hTERT promoter. HFK cells have Myc associated with the endogenous hTERT promoter, whereas HFF cells do not. The etiology of this difference might derive from the low level of Myc protein that we observed in the HFF cells (Fig. 5C) and that others have observed in normal human WI-38 fibroblasts (34). Compatible with the proposal that Myc protein levels might regulate hTERT responsiveness is the observation by several laboratories that Myc overexpression is sufficient to induce telomerase in these cells (Fig. 6B) (4, 5). More importantly, transduction of Myc into such fibroblasts is sufficient for cell immortalization (4, 23, 34, 41).
Interestingly, our study indicates that the low level of Myc protein in foreskin fibroblasts derives from posttranscriptional controls. The Myc gene is expressed similarly at the mRNA level in HFF and HFK cells, but the level of Myc protein is dramatically different. The ability to restore higher Myc protein levels in HFF cells with proteasome inhibitors suggests that proteolysis might contribute to the very low levels of Myc protein in HFF cells.
However, the level of Myc protein cannot be the complete explanation for the difference between HFF and HFK responses to E6. While HFF cells have nearly undetectable Myc protein levels, they provide a very suitable environment for the transactivation of an exogenous hTERT promoter by E6. Indeed, the exogenous hTERT promoter is activated to an even greater extent in HFF cells than in HFK cells, which suggests that there might be an additional mechanism regulating Myc access to the endogenous hTERT promoter, such as chromatin structure. In general, transfected plasmids are not highly decorated with chromatin proteins and exhibit a more “open” conformation than endogenous genes (53). It is therefore possible that the “open” nature of the exogenous hTERT promoter permits access to the lower levels of Myc protein in HFFs and thereby allows the transfected promoter to respond to E6 activation.
It is interesting that we found E6 protein associated with the endogenous hTERT promoter in both HFK and HFF cells, although there may be some quantitative differences (Fig. 6A). If Myc were the sole binding site on the hTERT promoter for E6, we should have found little E6 bound to the endogenous hTERT promoter. While forced expression of Myc does significantly enhance E6 binding, we speculate that E6 might interact with additional proteins (e.g., NFX1-91 or BRCA1) that have been reported to be found on the hTERT promoter and which repress gene transcription (12, 14, 62, 64). Activation of the hTERT promoter is clearly complex, and there is even a telomerase-defective E6 mutant that binds to the hTERT promoter without the activation of telomerase (51). Examining such mutants should provide insight into E6 transactivation.
In summary, the differential abilities of the HPV E6 and E7 genes to immortalize HFK and HFF cells correlates with Myc binding to the hTERT promoter, which provides the appropriate promoter environment for E6 responsiveness.
Acknowledgments
This work was supported by NIH grants R01CA106440 and R01CA53371 to R.S.
We thank Hang Yuan and Frank Suprynowicz for their advice and suggestions during this study.
Footnotes
Published ahead of print on 25 September 2008.
REFERENCES
- 1.Androphy, E. J., N. L. Hubbert, J. T. Schiller, and D. R. Lowy. 1987. Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J. 6989-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks, L., P. Spence, E. Androphy, N. Hubbert, G. Matlashewski, A. Murray, and L. Crawford. 1987. Identification of human papillomavirus type 18 E6 polypeptide in cells derived from human cervical carcinomas. J. Gen. Virol. 681351-1359. [DOI] [PubMed] [Google Scholar]
- 3.Bedard, K. M., M. P. Underbrink, H. L. Howie, and D. A. Galloway. 2008. The E6 oncoproteins from human betapapillomaviruses differentially activate telomerase through an E6AP-dependent mechanism and prolong the lifespan of primary keratinocytes. J. Virol. 823894-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benanti, J. A., M. L. Wang, H. E. Myers, K. L. Robinson, C. Grandori, and D. A. Galloway. 2007. Epigenetic down-regulation of ARF expression is a selection step in immortalization of human fibroblasts by c-Myc. Mol. Cancer Res. 51181-1189. [DOI] [PubMed] [Google Scholar]
- 5.Casillas, M. A., S. L. Brotherton, L. G. Andrews, J. M. Ruppert, and T. O. Tollefsbol. 2003. Induction of endogenous telomerase (hTERT) by c-Myc in WI-38 fibroblasts transformed with specific genetic elements. Gene 31657-65. [DOI] [PubMed] [Google Scholar]
- 6.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 111921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiPaolo, J. A., C. D. Woodworth, N. C. Popescu, V. Notario, and J. Doniger. 1989. Induction of human cervical squamous cell carcinoma by sequential transfection with human papillomavirus 16 DNA and viral Harvey ras. Oncogene 4395-399. [PubMed] [Google Scholar]
- 8.Dyson, N., P. Guida, K. Munger, and E. Harlow. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 666893-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243934-937. [DOI] [PubMed] [Google Scholar]
- 10.Fu, B., J. Quintero, and C. C. Baker. 2003. Keratinocyte growth conditions modulate telomerase expression, senescence, and immortalization by human papillomavirus type 16 E6 and E7 oncogenes. Cancer Res. 637815-7824. [PubMed] [Google Scholar]
- 11.Gallant, P. 2006. Myc/Max/Mad in invertebrates: the evolution of the Max network. Curr. Top. Microbiol. Immunol. 302235-253. [DOI] [PubMed] [Google Scholar]
- 12.Galloway, D. A., L. C. Gewin, H. Myers, W. Luo, C. Grandori, R. A. Katzenellenbogen, and J. K. McDougall. 2005. Regulation of telomerase by human papillomaviruses. Cold Spring Harbor Symp. Quant. Biol. 70209-215. [DOI] [PubMed] [Google Scholar]
- 13.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 757198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 182269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillison, M. L., W. M. Koch, R. B. Capone, M. Spafford, W. H. Westra, L. Wu, M. L. Zahurak, R. W. Daniel, M. Viglione, D. E. Symer, K. V. Shah, and D. Sidransky. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92709-720. [DOI] [PubMed] [Google Scholar]
- 16.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16653-699. [DOI] [PubMed] [Google Scholar]
- 17.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65337-365. [DOI] [PubMed] [Google Scholar]
- 18.Härle-Bachor, C., and P. Boukamp. 1996. Telomerase activity in the regenerative basal layer of the epidermis inhuman [sic] skin and in immortal and carcinoma-derived skin keratinocytes. Proc. Natl. Acad. Sci. USA 936476-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345458-460. [DOI] [PubMed] [Google Scholar]
- 20.Hawley-Nelson, P., K. H. Vousden, N. L. Hubbert, D. R. Lowy, and J. T. Schiller. 1989. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 83905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurlin, P. J., and S. Dezfouli. 2004. Functions of Myc:Max in the control of cell proliferation and tumorigenesis. Int. Rev. Cytol. 238183-226. [DOI] [PubMed] [Google Scholar]
- 22.James, M. A., J. H. Lee, and A. J. Klingelhutz. 2006. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 1191878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kampinga, H. H., M. A. Van Waarde-Verhagen, A. J. Van Assen-Bolt, B. Nieuwenhuis, H. P. Rodemann, K. R. Prowse, and M. H. Linskens. 2004. Reconstitution of active telomerase in primary human foreskin fibroblasts: effects on proliferative characteristics and response to ionizing radiation. Int. J. Rad. Biol. 80377-388. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, M. L., K. E. Keiger, C. J. Lee, and J. M. Huibregtse. 2005. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 793737-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 2662011-2015. [DOI] [PubMed] [Google Scholar]
- 26.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 39684-88. [DOI] [PubMed] [Google Scholar]
- 27.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 38079-82. [DOI] [PubMed] [Google Scholar]
- 28.Liu, X., G. L. Disbrow, H. Yuan, V. Tomaic, and R. Schlegel. 2007. Myc and human papillomavirus type 16 E7 genes cooperate to immortalize human keratinocytes. J. Virol. 8112689-12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X., J. Roberts, A. Dakic, Y. Zhang, and R. Schlegel. 2008. HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology 375611-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, X., H. Yuan, B. Fu, G. L. Disbrow, T. Apolinario, V. Tomaic, M. L. Kelley, C. C. Baker, J. Huibregtse, and R. Schlegel. 2005. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 28010807-10816. [DOI] [PubMed] [Google Scholar]
- 31.Lüscher, B. 2001. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 2771-14. [DOI] [PubMed] [Google Scholar]
- 32.McMurray, H. R., and D. J. McCance. 2003. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J. Virol. 779852-9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90785-795. [DOI] [PubMed] [Google Scholar]
- 34.Milyavsky, M., I. Shats, N. Erez, X. Tang, S. Senderovich, A. Meerson, Y. Tabach, N. Goldfinger, D. Ginsberg, C. C. Harris, and V. Rotter. 2003. Prolonged culture of telomerase-immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res. 637147-7157. [PubMed] [Google Scholar]
- 35.Münger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 7811451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Münger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89213-228. [DOI] [PubMed] [Google Scholar]
- 37.Münger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 634417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Münger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 84099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair, S. K., and S. K. Burley. 2006. Structural aspects of interactions within the Myc/Max/Mad network. Curr. Top. Microbiol. Immunol. 302123-143. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277955-959. [DOI] [PubMed] [Google Scholar]
- 41.Noble, J. R., Z. H. Zhong, A. A. Neumann, J. R. Melki, S. J. Clark, and R. R. Reddel. 2004. Alterations in the p16(INK4a) and p53 tumor suppressor genes of hTERT-immortalized human fibroblasts. Oncogene 233116-3121. [DOI] [PubMed] [Google Scholar]
- 42.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 755559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirisi, L., S. Yasumoto, M. Feller, J. Doniger, and J. A. DiPaolo. 1987. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J. Virol. 611061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirity, M., J. K. Blanck, and N. Schreiber-Agus. 2006. Lessons learned from Myc/Max/Mad knockout mice. Curr. Top. Microbiol. Immunol. 302205-234. [DOI] [PubMed] [Google Scholar]
- 45.Ramakrishnan, S., U. Eppenberger, H. Mueller, Y. Shinkai, and R. Narayanan. 1998. Expression profile of the putative catalytic subunit of the telomerase gene. Cancer Res. 58622-625. [PubMed] [Google Scholar]
- 46.Rottmann, S., and B. Luscher. 2006. The Mad side of the Max network: antagonizing the function of Myc and more. Curr. Top. Microbiol. Immunol. 30263-122. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75495-505. [DOI] [PubMed] [Google Scholar]
- 48.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 631129-1136. [DOI] [PubMed] [Google Scholar]
- 49.Schlegel, R., W. C. Phelps, Y. L. Zhang, and M. Barbosa. 1988. Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma. EMBO J. 73181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314111-114. [DOI] [PubMed] [Google Scholar]
- 51.Sekaric, P., J. J. Cherry, and E. J. Androphy. 2008. Binding of human papillomavirus type 16 E6 to E6AP is not required for activation of hTERT. J. Virol. 8271-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer. 33787-791. [DOI] [PubMed] [Google Scholar]
- 53.Smith, C. L., and G. L. Hager. 1997. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J. Biol. Chem. 27227493-27496. [DOI] [PubMed] [Google Scholar]
- 54.Stöppler, H., D. P. Hartmann, L. Sherman, and R. Schlegel. 1997. The human papillomavirus type 16 E6 and E7 oncoproteins dissociate cellular telomerase activity from the maintenance of telomere length. J. Biol. Chem. 27213332-13337. [DOI] [PubMed] [Google Scholar]
- 55.Takakura, M., S. Kyo, T. Kanaya, M. Tanaka, and M. Inoue. 1998. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 581558-1561. [PubMed] [Google Scholar]
- 56.Vaziri, H., F. Schachter, I. Uchida, L. Wei, X. Zhu, R. Effros, D. Cohen, and C. B. Harley. 1993. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am. J. Hum. Genet. 52661-667. [PMC free article] [PubMed] [Google Scholar]
- 57.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 754467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veldman, T., X. Liu, H. Yuan, and R. Schlegel. 2003. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 1008211-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, J., L. Y. Xie, S. Allan, D. Beach, and G. J. Hannon. 1998. Myc activates telomerase. Genes Dev. 121769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239197-201. [DOI] [PubMed] [Google Scholar]
- 61.Wu, K. J., C. Grandori, M. Amacker, N. Simon-Vermot, A. Polack, J. Lingner, and R. Dalla-Favera. 1999. Direct activation of TERT transcription by c-MYC. Nat. Genet. 21220-224. [DOI] [PubMed] [Google Scholar]
- 62.Xiong, J., S. Fan, Q. Meng, L. Schramm, C. Wang, B. Bouzahza, J. Zhou, B. Zafonte, I. D. Goldberg, B. R. Haddad, R. G. Pestell, and E. M. Rosen. 2003. BRCA1 inhibition of telomerase activity in cultured cells. Mol. Cell. Biol. 238668-8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasumoto, S., C. Kunimura, K. Kikuchi, H. Tahara, H. Ohji, H. Yamamoto, T. Ide, and T. Utakoji. 1996. Telomerase activity in normal human epithelial cells. Oncogene 13433-439. [PubMed] [Google Scholar]
- 64.Zhang, Y., S. Fan, Q. Meng, Y. Ma, P. Katiyar, R. Schlegel, and E. M. Rosen. 2005. BRCA1 interaction with human papillomavirus oncoproteins. J. Biol. Chem. 28033165-33177. [DOI] [PubMed] [Google Scholar]
- 65.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288F55-F78. [DOI] [PubMed] [Google Scholar]
- 66.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2342-350. [DOI] [PubMed] [Google Scholar]