Abstract
The importance of the N-terminal region of HIV gp120 conserved domain 1 (gp120-C1) to envelope function has been examined by alanine-scanning mutagenesis and subsequent characterization of the mutagenic effects on viral entry; envelope expression, processing, and incorporation; and gp120 association with gp41. With respect to the wild-type gp120, mutational effects on viral entry fall into two classes: functional, as defined by >20% entry with respect to wild type, and impaired, as defined by <20% entry with respect to wild type. Based on Western blot analyses of cell lysates and virions, the entry impairment of W35A, V38A, Y39A, Y40A, G41A, V42A, and I52A is due primarily to disruption of envelope processing. The entry impairment of P43A and W45A is apparently due to a combination of effects on processing and incorporation into virions. In contrast, the entry impairment of V44A and F53A is primarily due to disruption of the gp120-gp41 interaction, which results in dissociation of gp120 from the virion. We present a model for gp120-C1 interactions with gp120-C5 and the gp41 disulfide loop in unprocessed gp160 and processed gp120/gp41.
Human immunodeficiency virus (HIV)2 entry is mediated by the viral envelope proteins gp120 and gp41 (reviewed in Ref. 1). gp120 mediates attachment of the virus to the appropriate target cells through interactions with CD4 and chemokine co-receptors (2, 3). gp41, which is tethered to the viral membrane by a transmembrane domain, mediates fusion of the viral and target cell membranes (4). gp120 and gp41 are formed by cleavage of the precursor gp160 by cellular furin-like proteases (5, 6). Specifically, the amino acid sequence REKR located at the C terminus of gp120 forms the furin recognition site. Mutations of this site result in unprocessed gp160 and a loss of function (7, 8).
Based on amino acid sequence analyses, gp120 is composed of five conserved domains, termed C1–C5, and five variable domains, termed V1–V5 (9). There is substantial structural information for gp120 including the free state (10) and the CD4-bound state (11, 12). Unfortunately, the gp120 core structure observed in the crystal structures is missing large regions of C1 and C5. Specifically, residues 31–82 of gp120-C1 and residues 493–511 of gp120-C5 are missing with respect to the mature HIV gp120. However, the structure of HIV gp120-C5 (residues 489–511) has been determined as an isolated domain by NMR spectroscopy (13).
Mutagenesis studies have suggested that gp120-C1 and -C5 directly interact with gp41 in the processed form (14–16). Moreover, immunological studies have supported this model (17, 18). Recently, we have suggested that the gp120-C5 interacts directly with gp41 before furin processing (19–21). In the present study, we report the first systematic mutation study of the importance of HIV gp120-C1 residues to envelope function with the long term goal of identifying gp120 regions that are attractive sites for drug intervention. We present evidence that gp120-C1 plays an important role in the processing of the furin recognition site, which is located within gp120-C5. Moreover, we find two regions of gp120-C1 that are important to the association of gp120 with gp41.
MATERIALS AND METHODS
Mutagenesis and Viral Entry Assays—Mutants were prepared from plasmid pCONBgp160opt (22) using the Stratagene QuikChange II site-directed mutagenesis kit with subsequent verification by DNA sequencing. The functionality of gp120 mutants was determined in a luciferase-based entry assay (23). For this assay, plasmids pCONBgp160opt (bearing wild-type or mutant gp120) and pNL4-3.Luc.R-E- (24) were co-transfected by Lipofectamine 2000 (Invitrogen) into 293T cells, which were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin/streptomycin. Forty-eight hours post-transfection, the medium was harvested and filtered through a 0.45-μm filter to make the virus stock. For assay of viral entry, U87.CD4.CCR5 cells (3), which were maintained in Dulbecco's medium with 15% fetal bovine serum supplemented with 1 μg/ml puromycin, 300 μg/ml G418, 1% l-glutamine, and 1% penicillin/streptomycin, were seeded to 1 × 105 cells/well of a 12-well cell culture plate in a volume of 1 ml. The following day, 500 μl of the virus stock was added to each of the wells of the U87 cells after removal of the medium. The plates were incubated overnight at 37 °C in a CO2 incubator. After ∼16 h, the virus was aspirated and replaced with U87 medium, and the cells were allowed to rest for another 24 h. Luciferase activity was measured using the luciferase assay system from Promega and a Berthold FB12 luminometer running Sirius software. The experiments were run in triplicate from transfection to assay of luciferase activity, and thus the uncertainties represent all stages of the experiment. In all cases, the viral entry levels fell within the linear range of detection (i.e. the values of the wild-type and mutants never exceeded 3 × 106 relative light units) (14). Entry levels were normalized to relative p24 levels observed in the Western blot of the virus, as described below.
Western Blot Analysis—Cell lysates were collected from 293T producer cells using lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, and 0.1% SDS). The virus pellet was prepared by ultracentrifugation on a cushion of 20% sucrose at 55,000 rpm for 35 min using a Beckman SW55Ti rotor. The virus pellets were resuspended in lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, and 0.1% SDS). After SDS-PAGE, transfer, and blocking of the membranes with 5% milk/TBST for gp41 and SuperBlock (Pierce) for gp120, the blots were probed with either goat anti-HIV-1 gp120 polyclonal antibody (U. S. Biological) or with mouse anti-HIV-1 gp41 monoclonal antibody (Chessie 8; National Institutes of Health AIDS Research and Reference Reagent Program) (22). HIV-1 p24 was detected by probing the membrane with mouse anti-HIV-1-p24 antibody (U. S. Biological). The secondary antibody used was peroxidase-conjugated AffiniPure donkey anti-goat or goat anti-mouse IgG (H + L) from Jackson ImmunoResearch Laboratories, Inc. and developed using the ECL kit (Pierce). The relative amount of envelope or p24 on the Western blots was determined by a densitometric analysis. Briefly, digital scans of the blots were opened in Photoshop 7.0, the image was inverted, the appropriate bands were selected with the Lasso function, and the mean intensity and area were measured with the Histogram function. Subsequently, the relative percentage was determined by the following relationship,
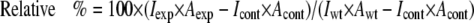 |
(Eq. 1) |
where I and A denote mean intensity and band area, respectively, and the subscripts exp, cont, and wt denote experimental, control (i.e. in the absence of envelope or pNL4-3.Luc.R-E- plasmid), and wild-type, respectively. The mean intensity and band area values used in this analysis are shown in Tables S1 and S2.
RESULTS
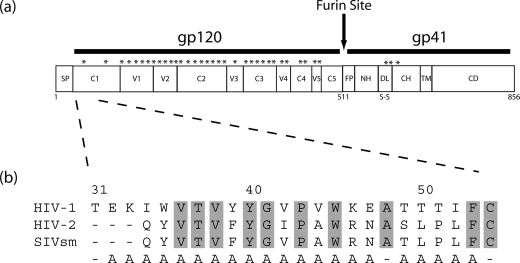
Design of the HIV gp120-C1 Mutants—The location of gp120-C1 within the context of gp160 is shown in Fig. 1a. The N-terminal region of gp120-C1 is relatively conserved between HIV-1, HIV-2, and SIV with ∼50% sequence identity (9), with the most conserved region, composed of hydrophobic residues, spanning residues 36–45. In the present study, 21 alanine substitutions of HIV-1 gp120-C1 were generated by site-directed mutagenesis (Fig. 1b). We have chosen to use the CONB envelope sequence, a consensus sequence of subtype B that is optimized for expression in mammalian cells and uses CCR5 as the co-receptor (22).
FIGURE 1.
a, organization of HIV envelope proteins (9). Putative glycosylation sites are denoted by asterisks. SP, signal peptide; C1–C5, conserved domains 1–5; V1–V5, variable domains 1–5; FP, fusion peptide; NH, N helix; DL, disulfide loop; CH, C helix; TM, transmembrane domain; CD, cytoplasmic domain. b, amino acid sequence alignment of HIV-1, HIV-2, and SIVsm gp120-C1 (9). Conserved residues of gp120-C1 are highlighted by gray shading. Residues that are substituted by alanine in the present study are denoted below. The numbering corresponds to that of HIV-1 HXB2.
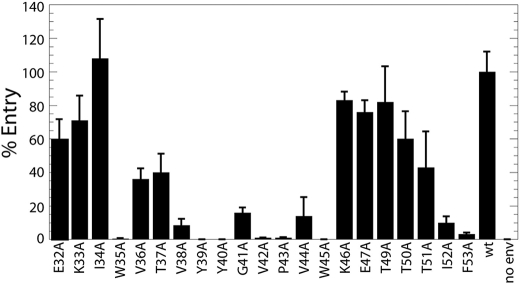
Viral Entry of the HIV gp120-C1 Mutants—The effects of the gp120 mutations on viral entry were first tested by a luciferase-based assay, in which viral entry is proportional to the observed luciferase activity (14, 23). Viral entry levels of wild-type and mutants are shown in Fig. 2. The mutational effects on viral entry can be divided into two classes: 1) functional, as defined by >20% entry with respect to wild type (E32A, K33A, I34A, V36A, T37A, K46A, E47A, T49A, T50A, and T51A); 2) impaired, as defined by <20% entry with respect to wild type (W35A, V38A, Y39A, Y40A, G41A, V42A, P43A, V44A, W45A, I52A, and F53A). With respect to the functional mutants, Val36 and Thr37 are highly conserved among HIV-1, HIV-2, and SIV; however, substitution of these residues with alanine is a relatively conservative change. With respect to the impaired mutations, Val38, Tyr40, Gly41, Pro43, Trp45, and Phe53 are highly conserved among HIV-1, HIV-2, and SIV. Interestingly, many of the largest effects to entry occur in the hydrophobic region encompassing Val38–Trp45.
FIGURE 2.
Mutational effects on viral entry with respect to the HIV-1 wild type (wt), based on the luciferase reporter assay for entry into U87.CD4.CCR5 cells. The error bars represent the S.D. of three separate experiments from the transfection step.
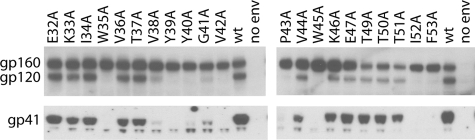
Envelope Expression and Processing—A Western blot analysis of wild-type and mutant envelope present in cell lysates was carried out to probe for mutational effects on expression and processing, as shown in Fig. 3. First note that the wild type shows the characteristic bands of gp160, gp120, and gp41, indicating that gp160 is expressed and processed by furin-like proteases to form mature gp120 and gp41. In the case of the mutants, the relatively high levels of gp160 present in cell lysates suggest that envelope expression has not been significantly affected by any of the substitutions. On the other hand, many of the mutations have reduced gp160 processing, as evidenced by reduced levels of gp120 and gp41. For example, W35A, V38A, Y39A, Y40A, G41A, V42A, P43A, W45A, I52A, and F53A exhibit undetectable or greatly reduced levels of gp120 and gp41 in the cell lysates, presumably due to a lack of processing by furin-like proteases. Not surprisingly, many of the mutants that display reduced gp160 processing also are impaired for viral entry (i.e. entry levels are <20% with respect to the wild type). V44A is a notable exception for which gp160 processing is not significantly affected but it nonetheless is impaired for viral entry.
FIGURE 3.
Western blot analysis of envelope expression and processing for HIV-1 wild type (wt) and the gp120-C1 mutants in lysates from 293T cells. The positions of gp41, gp120, and gp160 are shown.
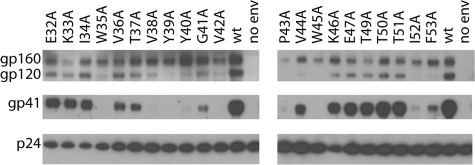
Virion Incorporation and gp120 Association with gp41—A Western blot analysis of wild-type and mutant envelope present in purified virions was carried out to probe for mutational effects on envelope incorporation into virions and gp120 association with gp41 as well as to confirm the expression and processing results observed in cell lysates. Note that virion incorporation requires surface expression of envelope, which has been shown to be dependent upon proper folding and oligomerization (26–28). The Western blot analysis of wild-type and mutant virions is shown in Fig. 4. The relative amounts of p24 were assayed as a loading control. The wild-type virions show the presence of gp160, gp120, and gp41. The presence of gp160 in virions has been previously attributed to saturation of furin-like proteases by overproduction of envelope (22, 29). Not surprisingly, the functional mutants (E32A, K33A, I34A, V36A, T37A, K46A, E47A, T49A, T50A, and T51A) exhibit the wild-type pattern, suggesting that envelope is incorporated into the virion and that gp120 remains noncovalently associated with gp41. The impaired mutants P43A and W45A exhibit lower levels of virion incorporation of envelope as evidenced by the presence of p24 and the absence of envelope, and thus these mutants may be less efficiently transported to the cell surface due to disruption of folding and/or oligomerization. Moreover, the absence of gp120 and gp41 in the P43A and W45A mutants implies that envelope processing has also been disrupted. The impaired mutants W35A, V38A, Y39A, Y40A, G41A, V42A, and I52A exhibit gp160, suggesting virion incorporation, but the absence or greatly reduced level of gp120 and gp41 suggests a processing defect, which is in agreement with the result for the Western blot of the cell lysates (Fig. 3). Impaired mutants V44A and F53A display bands for gp160, suggesting virion incorporation, and gp41, suggesting processing. In the case of F53A, the processing observed in virions is in disagreement with the lack of processing observed in cell lysates (compare Figs. 3 and 4); the underlying reasons for this discrepancy are not apparent at this time. Interestingly, the absence of gp120 in the impaired mutants V44A and F53A suggests that the gp120-gp41 interaction has been disrupted to result in “shedding.”
FIGURE 4.
Western blot analysis of virion incorporation and gp120-gp41 association for HIV-1 wild type (wt) and the gp120-C1 mutants. The positions of p24, gp41, gp120, and gp160 are shown.
DISCUSSION
Summary of the gp120-C1 Mutational Effects—The effects of the gp120-C1 alanine substitutions on viral entry; envelope expression, processing, and virion incorporation; and gp120-gp41 association are summarized in Table 1. Based on the data presented herein, a phenotype can be given for each of the mutants. For example, mutants E32A, K33A, I34A, V36A, T37A, K46A, E47A, T49A, T50A, and T51A are functional for viral entry (defined as >20% entry with respect to the wild type), and they exhibit the wild type pattern of gp160, gp120, and gp41 in cell extracts and virions. Consequently, this group is assigned a wild-type phenotype (Table 1). Impaired mutants P43A and W45A show reduced levels of virion incorporation and processing, and thus they exhibit a mixed phenotype. Impaired mutants W35A, V38A, Y39A, Y40A, G41A, V42A, and I52A exhibit a processing defect, as evidenced by decreased levels of gp120 and gp41 in cell lysates and virus (cf. Figs. 3 and 4 and Table 1). Note that the Y40A substitution is found in some HIV-1 subtype O strains, which suggests that other mutations compensate to allow processing in these strains. Impaired mutants V44A and F53A exhibit a defect in gp120-gp41 association. In the case of V44A, the alanine substitution occurs in the HIV-2 and SIV envelope (Fig. 1), which suggests that other mutations compensate to stabilize gp120-gp41 association.
TABLE 1.
Summary of viral entry and the level of envelope detected in cell lysates and virus of the wild-type and alanine mutants of HIV gp120-C1 Entry levels are reported as the percentage of entry with respect to the wild-type virus. The reported levels are normalized to p24 levels observed in the Western blot of the virus (see “Materials and Methods”). The reported errors correspond to the S.D. of three separate experiments. Envelope levels are based on a densitometric analysis of Western blots. –, <20%; +, 20–60%; ++, 60–140%; +++, >140% (cf. supplemental Tables S1 and S2).
| Mutant | Entry (percentage of wild type) | Cell gp160 | Cell gp120 | Cell gp41 | Virus gp160 | Virus gp120 | Virus gp41 | Phenotype |
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| Wild type | 100 ± 12 | ++ | ++ | ++ | ++ | ++ | ++ | Wild type |
| E32A | 60 ± 12 | +++ | +++ | ++ | ++ | + | + | Wild type |
| K33A | 71 ± 15 | +++ | ++ | + | + | + | + | Wild type |
| I34A | 108 ± 24 | +++ | +++ | ++ | + | + | + | Wild type |
| W35A | 0.4 ± 0.1 | +++ | – | – | + | – | – | Processing |
| V36A | 36 ± 6 | +++ | ++ | + | + | + | + | Wild type |
| T37A | 40 ± 11 | +++ | ++ | + | ++ | + | + | Wild type |
| V38A | 8.4 ± 4.0 | +++ | + | – | ++ | + | – | Processing |
| Y39A | 0.0 ± 0.1 | +++ | – | – | ++ | – | – | Processing |
| Y40A | 0.0 ± 0.1 | +++ | – | – | ++ | + | – | Processing |
| G41A | 16 ± 3 | ++ | + | – | ++ | + | + | Processing |
| V42A | 1.0 ± 0.2 | ++ | – | – | + | – | – | Processing |
| P43A | 1.0 ± 0.4 | +++ | – | – | + | – | – | Processing/Incorporation |
| V44A | 14 ± 11 | +++ | + | + | ++ | – | + | Association/Processing |
| W45A | 0.0 ± 0.1 | +++ | – | – | + | – | – | Processing/Incorporation |
| K46A | 83 ± 5 | +++ | + | ++ | ++ | + | + | Wild type |
| E47A | 76 ± 7 | +++ | + | ++ | + | + | + | Wild type |
| T49A | 82 ± 21 | ++ | + | ++ | + | + | ++ | Wild type |
| T50A | 53 ± 16 | ++ | + | ++ | ++ | + | ++ | Wild type |
| T51A | 60 ± 21 | ++ | + | + | ++ | + | ++ | Wild type |
| I52A | 10 ± 4 | ++ | – | – | ++ | – | – | Processing |
| F53A | 3.2 ± 0.9 | ++ | – | – | ++ | – | + | Association/Processing |
Previous Studies of gp120-C1—A number of previous studies have characterized the effects of mutations in HIV gp120-C1 on viral function, and thus it is of interest to compare the conclusions of these studies in light of the present work. For example, Ivey-Hoyle et al. (14) observed that deletion of the 31 aminoterminal residues of gp120 (residues 31–57 of gp120-C1) (Fig. 1) did not disrupt envelope processing or CD4 binding but did result in dissociation of gp120 from gp41, which suggested that the gp120-C1 domain directly interacts with gp41. The V44A and F53A mutants of the present study, which are found within this region, also disrupt gp120 association with gp41 and thus support this notion. In another study, Helseth et al. (15) found that single-site mutants V36L and Y40D were processed and bound CD4 but resulted in dissociation of gp120 from gp41. In the present study, the analogous mutant V36A exhibited a wild-type phenotype, with the difference presumably due to the nature of the substituting group (e.g. the leucine substitution is more bulky than valine and may result in steric hindrance). Moreover, the Y40A mutant of the present study disrupted envelope processing, with the difference probably due to the smaller and more hydrophobic nature of alanine with respect to aspartate. In addition, Helseth et al. (15) found that mutant W45S reduced processing and gp120 association with gp41. In agreement, the analogous W45A mutant of the present study also reduced processing; however, the effects on gp120-gp41 association could not be assayed due to the complete absence of processing in this mutant. Finally, we note that the present study represents the first single site mutations to Glu32, Lys33, Ile34, Trp35, Thr37, Val38, Tyr39, Gly41, Val42, Pro43, Val44, Lys46, Glu47, Thr49, Thr50, Thr51, Ile52, and Phe53. Of the novel mutations, mutants W35A, V38A, Y49A, V42A, and I52A exhibit significantly impaired function due to processing defects, mutant P43A exhibits impaired function due to defects in processing and virion incorporation, and mutants V44A and F53A exhibit impaired function due to disruption of the gp120-gp41 association. Taken together, the gp120-C1 region plays important roles in envelope processing and stabilization of the gp120-gp41 interaction.
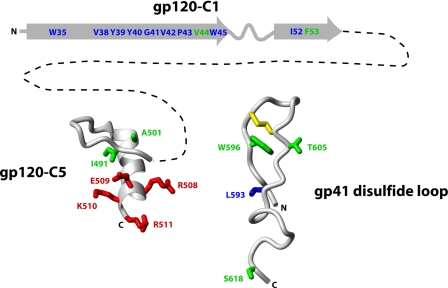
Interactions of gp120-C1 in gp120/gp41—As mentioned in the Introduction, large regions of the gp120-C1 and gp120-C5 domains are missing from the available structures of the gp120 core (10–12), and thus mutagenesis and immunological studies play important roles in the characterization of these domains, which are highly conserved between HIV-1, HIV-2, and SIV (9). However, it is important to note that single site mutational effects can be due to direct interactions or interactions propagated to more distant sites. Nonetheless, we would like to consider intermolecular interactions between gp120-C1 and gp120-C5 with the gp41 disulfide loop in processed gp160 (Fig. 5). The gp120-C1 domain is shown with the secondary structure predicted by Hansen et al. (30). The gp120-C5 structure is taken from Guilhaudis et al. (13), and the gp41 disulfide loop structure is taken from Refs. 25 and 31. Note that the gp120-C5 structure has the caveat that it was determined in the presence of 40% trifluoroethanol, a co-solvent that stabilizes helical structure, and in the absence of other gp120 and gp41 domains (13). The gp41 disulfide loop has the caveat that it is based on the postfusion form of gp41 (i.e. the six-helix bundle); however, we are not aware of any evidence for a large structural change of the disulfide loop during HIV entry. In Fig. 5, residues implicated by mutagenesis studies in forming intermolecular interactions in processed gp160 are shown in green. The interaction between Val44 and Phe53 of gp120-C1 and gp41 is based on the present work, in which single site mutations resulted in disruption of the gp120-gp41 interaction. The intermolecular interactions of Ile491 of gp120-C5 and Trp596 and Ser618 of the gp41 disulfide loop are based on previous mutagenesis studies, suggested by dissociation of gp120 from gp41 (19, 21). The interaction between A501 of gp120-C5 and Thr605 of the gp41 disulfide loop is based on the SOS mutant of Binley et al. (16), in which the double mutant A501C/T605C was shown to form a nonnative disulfide bond, and thus a direct interaction between these residues was implied. As noted above, Val36, Tyr40, and Trp45 have also been previously implicated in forming intermolecular interactions with gp41, based on gp120 dissociation from gp41 (15). Importantly, immunological studies have also suggested interactions between the gp120-C1, gp120-C5, and the gp41 disulfide loop (17, 18), thereby supporting the schematic model of processed gp160. Finally, note that the topology of the gp120 core structures imply that gp120-C1 and -C5 are in close proximity (10–12).
FIGURE 5.
Schematic model depicting long range interactions between gp120-C1, gp120-C5, and the gp41 disulfide loop, deduced from site-directed mutagenesis studies. Residues implicated in forming long range intermolecular interactions in processed gp160 are shown in green; residues implicated in forming long range intramolecular interactions with the furin recognition site in unprocessed gp160 are shown in blue. The furin recognition site (REKR of gp120-C5) is shown in red. The dotted line between gp120-C1 and -C5 represents the 457 intervening residues. For reference, the disulfide bond of the gp41 disulfide loop is shown in yellow. The N and C termini of gp120 and the gp41 disulfide loop are denoted. The gp120-C1 domain is shown with the secondary structure predicted by Hansen et al. (30). The gp120-C5 structure is taken from Guilhaudis et al. (13), and the gp41 disulfide loop structure is based on Refs. 25 and 31.
Interactions of gp120-C1 in gp160—It is next of interest to examine potential long range intramolecular interactions between gp120-C1, gp120-C5, and the gp41 disulfide loop in unprocessed gp160. In Fig. 5, residues implicated in forming long range interactions with the furin recognition site in unprocessed gp160 are shown in blue. Specifically, the interaction between Trp35, Val38, Tyr39, Tyr40, Gly41, Val42, Pro43, Trp45, and Ile52 of gp120-C1 and the furin recognition site of gp120-C5 is based on the data presented herein, in which alanine substitutions resulted in severely decreased envelope processing. The interaction between Leu593 of the gp41 disulfide loop and the furin recognition site is based on a previous mutagenesis study (19). The notion that the gp41 disulfide bond interacts with gp120-C5 in gp160 is also supported by the mutagenesis study of Sen et al. (20), who showed that the disulfide bond within the loop was important to gp160 processing. Interestingly, the present study suggests that long range interactions between gp120-C1 and gp120-C5, which encompasses the furin recognition site, occur in unprocessed gp160. Indeed, the impaired entry of many of the gp120-C1 mutants was due to a lack of processing, underscoring the importance of gp120-C1 residues to proper presentation of the gp120-C5 furin recognition site, a site that is ∼450 residues away (cf. Fig. 1). Ivey-Hoyle et al. (14) have shown that deletion of the N-terminal 31 residues of gp120 (residues 31–57 of gp120-C1) (Fig. 1) does not significantly reduce envelope processing, which may appear to be in disagreement with the results presented herein. It is tempting to speculate that deletion of gp120-C1, in contrast to single site mutations, may remove residues with the potential to reduce access to the furin recognition site, thereby allowing envelope processing.
In summary, the extreme sensitivity of gp120-C1 to alanine substitutions suggests that this region is an attractive and novel target for future drug discovery efforts. For example, compounds that bind to gp120-C1 may be expected to disrupt gp120 function and hence HIV entry, by disrupting either gp160 processing or gp120 association with gp41.
Supplementary Material
Acknowledgments
Reagents pNL4-3.Luc.R-E-, pCONBgp160opt, U87.CD4.CCR5, and HIV-1 gp41 Hybridoma (Chessie 8) were obtained through the National Institutes of Health AIDS Research and Reference Program. We thank Caitlin Ondracek for aid in the preparation of the gp120-C1 mutants.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 AI47674 (to M. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: HIV, human immunodeficiency virus; SIV, simian immunodeficiency virus.
References
- 1.Gallo, S., Finnegan, C., Viard, M., Raviv, Y., Dimitrov, A., Rawat, S., Puri, A., Durrell, S., and Blumenthal, R. (2003) Biochim. Biophys. Acta 1614 36-50 [DOI] [PubMed] [Google Scholar]
- 2.Bazan, H., Alkhatib, G., Broder, C., and Berger, E. (1998) J. Virol. 72 4485-4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björndal, A., Deng, H., Jansson, M., Fiore, J. R., Colognesi, C., Karlsson, A., Albert, J., Scarlatti, G., Littman, D. R., and Fenyo, E. M. (1997) J. Virol. 71 7478-7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed, E., and Martin, M. (1995) J. Biol. Chem. 270 23833-23836 [DOI] [PubMed] [Google Scholar]
- 5.McCune, J. M., Rabin, L. B., Feinberg, M. B., Lieberman, M., Kosek, J. C., Reyes, G. R., and Weissman, I. L. (1988) Cell 53 55-67 [DOI] [PubMed] [Google Scholar]
- 6.Moulard, M., Hallenberger, S., Garten, W., and Klenk, H. (1999) Virus Res. 60 55-65 [DOI] [PubMed] [Google Scholar]
- 7.Freed, E. O., Myers, D. J., and Risser, R. (1989) J. Virol. 63 4670-4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch, V., and Pawlita, M. (1990) J. Virol. 64 2337-2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas, N., Munro, G., and Daniels, R. (1997) J. Mol. Biol. 273 122-149 [DOI] [PubMed] [Google Scholar]
- 10.Chen, B., Vogan, E., Gong, H., Skehel, J., Wiley, D., and Harrison, S. (2005) Nature 433 834-841 [DOI] [PubMed] [Google Scholar]
- 11.Kwong, P., Wyatt, R., Robinson, T., Sweet, R., Sodroski, J., and Hendrickson, W. (1998) Nature 393 648-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, C., Tang, M., Zhang, M., Majeed, S., Montabana, E., Stanfield, R., Dimitrov, D., Korber, B., Sodroski, J., and Wilson, I. (2005) Science 310 1025-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilhaudis, L., Jacobs, A., and Caffrey, M. (2002) Eur. J. Biochem. 269 4860-4867 [DOI] [PubMed] [Google Scholar]
- 14.Ivey-Hoyle, M., Clark, R., and Rosenberg, M. (1991) J. Virol. 65 2682-2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helseth, E., Olshevsky, U., Furman, C., and Sodroski, J. (1991) J. Virol. 65 2119-2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binley, J., Sanders, R., Clas, B., Schuelke, N., Master, A., Guo, Y., Kajumo, F., Anselma, J., Maddon, P., Olson, W., and Moore, J. (2000) J. Virol. 74 627-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore, J., Willey, R., Lewis, G., Robinson, J., and Sodroski, J. (1994) J. Virol. 68 6836-6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt, R., Desjardin, E., Olshevsky, U., Nixon, C., Binley, J., Olshevsky, V., and Sodroski, J. (1997) J. Virol. 71 9722-9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, A., Sen, J., Rong, L., and Caffrey, M. (2005) J. Biol. Chem. 280 27284-27288 [DOI] [PubMed] [Google Scholar]
- 20.Sen, J., Jacobs, A., Jiang, H., Rong, L., and Caffrey, M. (2007) Protein Sci. 16 1236-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen, J., Jacobs, A., and Caffrey, M. (2008) Biochemistry 47 7788-7795 [DOI] [PubMed] [Google Scholar]
- 22.Kothe, D., Decker, J., Li, Y., Weng, Z., Bibollet-Ruche, F., Zammit, K., Salazar, M., Chen, Y., Salazar-Gonzalez, J., Moldoveanu, Z., Mestecky, J., Gao, F., Haynes, B., Shaw, G., Muldoon, M., Korber, B., and Hahn, B. (2007) Virology 360 218-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page, K., Landau, N., and Littman, D. (1990) J. Virol. 64 5270-5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connor R., Chen, B., Choe, S., and Landau, N. (1995) Virology 206 935-944 [DOI] [PubMed] [Google Scholar]
- 25.Caffrey, M. (2001) Biochim. Biophys. Acta 1536 116-122 [DOI] [PubMed] [Google Scholar]
- 26.Bird, C., Gleeson, P., and McCluskey, J. (1990) J. Biol. Chem. 265 19151-19157 [PubMed] [Google Scholar]
- 27.Otteken, A., Earl, P., and Moss, B. (1996) J. Virol. 70 3407-3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eligard, L., and Helenius, A. (2003) Nat. Rev. Mol. Cell. Biol. 4 181-191 [DOI] [PubMed] [Google Scholar]
- 29.Binley, J., Sanders, R., Master, A., Cayanan, C., Wiley, C., Schiffner, L., Travis, B., Kuhmann, S., Burton, D., Hu, S., Olson, W., and Moore, J. (2002) J. Virol. 76 2606-2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen, J., Lund, O., Nielsen, J., Brunak, S., and Hansen, J. (1996) Proteins 25 1-11 [DOI] [PubMed] [Google Scholar]
- 31.Caffrey, M., Cai, M., Kaufman, J., Stahl, S., Wingfield, P., Gronenborn, A., and Clore, G. (1998) EMBO J. 17 4572-4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.